Lumpy Skin Disease Outbreaks in Africa, Europe, and Asia (2005–2022): Multiple Change Point Analysis and Time Series Forecast
Abstract
:1. Introduction
2. Materials and Methods
2.1. LSD Outbreak Data
2.2. Change Point Analysis
2.3. Forecasting of LSD Outbreaks
3. Results
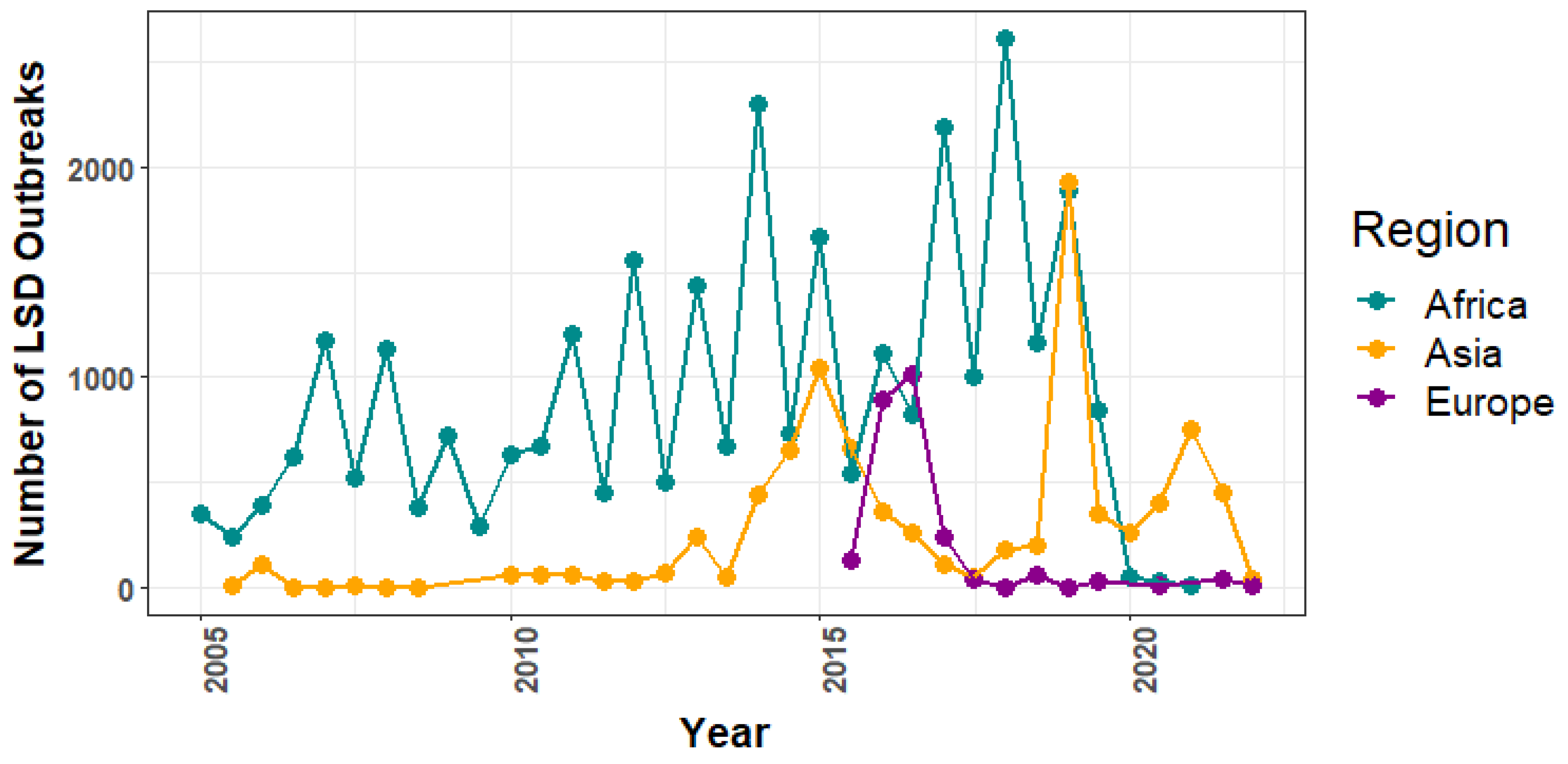
3.1. Lumpy Skin Disease Outbreak Reports
3.2. Change Points in the Time Series Data of Lumpy Skin Disease Outbreak Reports
3.3. Forecasts of LSD Outbreaks
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tuppurainen, E.; Venter, E.H.; Shisler, J.; Gari, G.; Mekonnen, G.; Juleff, N.; Lyons, N.; De Clercq, K.; Upton, C.; Bowden, T. Capripoxvirus diseases: Current status and opportunities for control. Transbound. Emerg. Dis. 2017, 64, 729–745. [Google Scholar] [CrossRef] [PubMed]
- Tuppurainen, E.; Alexandrov, T.; Beltrán-Alcrudo, D. Lumpy Skin Disease—A Manual for Veterinarians; FAO Animal Production and Health Manual; FAO: Rome, Italy, 2017. [Google Scholar]
- Sudhakar, S.B.; Mishra, N.; Kalaiyarasu, S.; Jhade, S.K.; Hemadri, D.; Sood, R.; Bal, G.C.; Nayak, M.K.; Pradhan, S.K.; Singh, V.P. Lumpy skin disease (LSD) outbreaks in cattle in Odisha state, India in August 2019: Epidemiological features and molecular studies. Transbound. Emerg. Dis. 2020, 67, 2408–2422. [Google Scholar] [CrossRef] [PubMed]
- Tuppurainen, E.S.; Stoltsz, W.H.; Troskie, M.; Wallace, D.B.; Oura, C.; Mellor, P.S.; Coetzer, J.A.; Venter, E.H. A potential role for ixodid (hard) tick vectors in the transmission of lumpy skin disease virus in cattle. Transbound. Emerg. Dis. 2011, 58, 93–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lubinga, J.; Tuppurainen, E.; Mahlare, R.; Coetzer, J.; Stoltsz, W.; Venter, E. Evidence of Transstadial and Mechanical Transmission of Lumpy Skin Disease Virus by A mblyomma hebraeum Ticks. Transbound. Emerg. Dis. 2015, 62, 174–182. [Google Scholar] [CrossRef]
- Lubinga, J.; Tuppurainen, E.; Stoltsz, W.; Ebersohn, K.; Coetzer, J.; Venter, E. Detection of lumpy skin disease virus in saliva of ticks fed on lumpy skin disease virus-infected cattle. Exp. Appl. Acarol. 2013, 61, 129–138. [Google Scholar] [CrossRef]
- Issimov, A.; Kutumbetov, L.; Orynbayev, M.B.; Khairullin, B.; Myrzakhmetova, B.; Sultankulova, K.; White, P.J. Mechanical Transmission of Lumpy Skin Disease Virus by Stomoxys spp. (Stomoxys calsitrans, Stomoxys sitiens, Stomoxys indica), Diptera: Muscidae. Animals 2020, 10, 477. [Google Scholar] [CrossRef] [Green Version]
- Abutarbush, S.; Ababneh, M.; Al Zoubi, I.; Al Sheyab, O.; Al Zoubi, M.; Alekish, M.; Al Gharabat, R. Lumpy Skin Disease in Jordan: Disease Emergence, Clinical Signs, Complications and Preliminary-associated Economic Losses. Transbound. Emerg. Dis. 2015, 62, 549–554. [Google Scholar] [CrossRef]
- Babiuk, S.; Bowden, T.; Boyle, D.; Wallace, D.B.; Kitching, R. Capripoxviruses: An emerging worldwide threat to sheep, goats and cattle. Transbound. Emerg. Dis. 2008, 55, 263–272. [Google Scholar] [CrossRef] [Green Version]
- Sprygin, A.; Artyuchova, E.; Babin, Y.; Prutnikov, P.; Kostrova, E.; Byadovskaya, O.; Kononov, A. Epidemiological characterization of lumpy skin disease outbreaks in Russia in 2016. Transbound. Emerg. Dis. 2018, 65, 1514–1521. [Google Scholar] [CrossRef]
- Tuppurainen, E.; Oura, C. Lumpy skin disease: An emerging threat to Europe, the Middle East and Asia. Transbound. Emerg. Dis. 2012, 59, 40–48. [Google Scholar] [CrossRef]
- Mercier, A.; Arsevska, E.; Bournez, L.; Bronner, A.; Calavas, D.; Cauchard, J.; Falala, S.; Caufour, P.; Tisseuil, C.; Lefrançois, T. Spread rate of lumpy skin disease in the Balkans, 2015–2016. Transbound. Emerg. Dis. 2018, 65, 240–243. [Google Scholar] [CrossRef]
- Şevik, M.; Doğan, M. Epidemiological and molecular studies on lumpy skin disease outbreaks in Turkey during 2014–2015. Transbound. Emerg. Dis. 2017, 64, 1268–1279. [Google Scholar] [CrossRef]
- Beard, P.M. Lumpy skin disease: A direct threat to Europe. Vet. Rec. 2016, 178, 557–558. [Google Scholar] [CrossRef] [Green Version]
- Panel, E.A. Statement: Urgent advice on lumpy skin disease. EFSA J. 2016, 14, 4573. [Google Scholar]
- Authority, E.F.S. Lumpy skin disease: I. Data collection and analysis. EFSA J. 2017, 15, e04773. [Google Scholar]
- Ripani, A.; Pacholek, X. Lumpy Skin Disease: Emerging disease in the Middle East-Threat to EuroMed countries. In Proceedings of the 10th Meeting of the REMESA Joint Permanent Committee, Heraklion, Greece, 17 March 2015; pp. 16–17. [Google Scholar]
- Tasioudi, K.; Antoniou, S.; Iliadou, P.; Sachpatzidis, A.; Plevraki, E.; Agianniotaki, E.; Fouki, C.; Mangana-Vougiouka, O.; Chondrokouki, E.; Dile, C. Emergence of lumpy skin disease in Greece, 2015. Transbound. Emerg. Dis. 2016, 63, 260–265. [Google Scholar] [CrossRef]
- Wainwright, S.; El Idrissi, A.; Mattioli, R.; Tibbo, M.; Njeumi, F.; Raizman, E. Emergence of lumpy skin disease in the Eastern Mediterranean Basin countries. FAO Empres Watch 2013, 29, 1–6. [Google Scholar]
- Khalil, M.I.; Sarker, M.F.R.; Hasib, F.Y.; Chowdhury, S. Outbreak investigation of lumpy skin disease in dairy farms at Barishal, Bangladesh. Turk. J. Agric. Food Sci. Technol. 2021, 9, 205–209. [Google Scholar] [CrossRef]
- Lu, G.; Xie, J.; Luo, J.; Shao, R.; Jia, K.; Li, S. Lumpy skin disease outbreaks in China, since 3 August 2019. Transbound. Emerg. Dis. 2021, 68, 216–219. [Google Scholar] [CrossRef]
- Maw, M.T.; Khin, M.M.; Hadrill, D.; Meki, I.K.; Settypalli, T.B.K.; Kyin, M.M.; Myint, W.W.; Thein, W.Z.; Aye, O.; Palamara, E. First Report of Lumpy Skin Disease in Myanmar and Molecular Analysis of the Field Virus Isolates. Microorganisms 2022, 10, 897. [Google Scholar] [CrossRef]
- Tran, H.T.T.; Truong, A.D.; Dang, A.K.; Ly, D.V.; Nguyen, C.T.; Chu, N.T.; Hoang, T.V.; Nguyen, H.T.; Nguyen, V.T.; Dang, H.V. Lumpy skin disease outbreaks in vietnam, 2020. Transbound. Emerg. Dis. 2021, 68, 977–980. [Google Scholar] [CrossRef]
- Koirala, P.; Meki, I.K.; Maharjan, M.; Settypalli, B.K.; Manandhar, S.; Yadav, S.K.; Cattoli, G.; Lamien, C.E. Molecular Characterization of the 2020 Outbreak of Lumpy Skin Disease in Nepal. Microorganisms 2022, 10, 539. [Google Scholar] [CrossRef]
- Arjkumpa, O.; Suwannaboon, M.; Boonrod, M.; Punyawan, I.; Liangchaisiri, S.; Laobannue, P.; Lapchareonwong, C.; Sansri, C.; Kuatako, N.; Panyasomboonying, P. The first lumpy skin disease outbreak in Thailand (2021): Epidemiological features and spatio-temporal analysis. Front. Vet. Sci. 2021, 8, 799065. [Google Scholar] [CrossRef]
- Gargoum, S.A.; Gargoum, A.S. Limiting mobility during COVID-19, when and to what level? An international comparative study using change point analysis. J. Transp. Health 2021, 20, 101019. [Google Scholar] [CrossRef]
- Küchenhoff, H.; Günther, F.; Höhle, M.; Bender, A. Analysis of the early COVID-19 epidemic curve in Germany by regression models with change points. Epidemiol. Infect. 2021, 149, e68. [Google Scholar] [CrossRef]
- Nuño, M.; García, Y.; Rajasekar, G.; Pinheiro, D.; Schmidt, A.J. COVID-19 hospitalizations in five California hospitals: A retrospective cohort study. BMC Infect. Dis. 2021, 21, 938. [Google Scholar] [CrossRef]
- Pradhan, A.; Anasuya, A.; Pradhan, M.M.; Ak, K.; Kar, P.; Sahoo, K.C.; Panigrahi, P.; Dutta, A. Trends in Malaria in Odisha, India—An analysis of the 2003–2013 time-series data from the national vector borne disease control program. PLoS ONE 2016, 11, e0149126. [Google Scholar] [CrossRef] [Green Version]
- Tuppurainen, E.; Oura, C. Lumpy Skin Disease (LSD) an Emerging Threat to Europe, the Middle East and Asia; Institute for Animal Health, Pirbright: Surrey, UK, 2011. [Google Scholar]
- Das, M.; Chowdhury, M.S.R.; Akter, S.; Mondal, A.K.; Jamal, M. An updated review on lumpy skin disease: Perspective of Southeast Asian countries. J. Adv. Biotechnol. Exp. Ther. 2021, 4, 322–333. [Google Scholar] [CrossRef]
- Khan, Y.R.; Ali, A.; Hussain, K.; Ijaz, M.; Rabbani, A.H.; Khan, R.L.; Abbas, S.N.; Aziz, M.U.; Ghaffar, A.; Sajid, H.A. A review: Surveillance of lumpy skin disease (LSD) a growing problem in Asia. Microb. Pathog. 2021, 158, 105050. [Google Scholar] [CrossRef] [PubMed]
- Gupta, T.; Patial, V.; Bali, D.; Angaria, S.; Sharma, M.; Chahota, R. A review: Lumpy skin disease and its emergence in India. Vet. Res. Commun. 2020, 44, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Kayesh, M.E.H.; Hussan, M.T.; Hashem, M.A.; Eliyas, M.; Anower, A.M. Lumpy skin disease virus infection: An emerging threat to cattle health in Bangladesh. Hosts Viruses 2020, 7, 97. [Google Scholar] [CrossRef]
- Papastefanopoulos, V.; Linardatos, P.; Kotsiantis, S. COVID-19: A comparison of time series methods to forecast percentage of active cases per population. Appl. Sci. 2020, 10, 3880. [Google Scholar] [CrossRef]
- Perone, G. Comparison of ARIMA, ETS, NNAR, TBATS and hybrid models to forecast the second wave of COVID-19 hospitalizations in Italy. Eur. J. Health Econ. 2022, 23, 917–940. [Google Scholar] [CrossRef]
- Punyapornwithaya, V.; Mishra, P.; Sansamur, C.; Pfeiffer, D.; Arjkumpa, O.; Prakotcheo, R.; Damrongwatanapokin, T.; Jampachaisri, K. Time-Series Analysis for the Number of Foot and Mouth Disease Outbreak Episodes in Cattle Farms in Thailand Using Data from 2010–2020. Viruses 2022, 14, 1367. [Google Scholar] [CrossRef]
- Kane, M.J.; Price, N.; Scotch, M.; Rabinowitz, P. Comparison of ARIMA and Random Forest time series models for prediction of avian influenza H5N1 outbreaks. BMC Bioinform. 2014, 15, 276. [Google Scholar] [CrossRef]
- Killick, R.; Eckley, I. Changepoint: An R package for changepoint analysis. J. Stat. Softw. 2014, 58, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Khandelwal, I.; Adhikari, R.; Verma, G. Time series forecasting using hybrid ARIMA and ANN models based on DWT decomposition. Procedia Comput. Sci. 2015, 48, 173–179. [Google Scholar] [CrossRef] [Green Version]
- Sun, J. Forecasting COVID-19 pandemic in Alberta, Canada using modified ARIMA models. Comput. Methods Programs Biomed. Update 2021, 1, 100029. [Google Scholar] [CrossRef]
- Hyndman, R.J.; Athanasopoulos, G. Forecasting: Principles and Practice; OTexts: Melbourne, Australia, 2018. [Google Scholar]
- Büyükşahin, Ü.Ç.; Ertekin, Ş. Improving forecasting accuracy of time series data using a new ARIMA-ANN hybrid method and empirical mode decomposition. Neurocomputing 2019, 361, 151–163. [Google Scholar] [CrossRef] [Green Version]
- Alabdulrazzaq, H.; Alenezi, M.N.; Rawajfih, Y.; Alghannam, B.A.; Al-Hassan, A.A.; Al-Anzi, F.S. On the accuracy of ARIMA based prediction of COVID-19 spread. Results Phys. 2021, 27, 104509. [Google Scholar] [CrossRef]
- Punyapornwithaya, V.; Jampachaisri, K.; Klaharn, K.; Sansamur, C. Forecasting of Milk Production in Northern Thailand Using Seasonal Autoregressive Integrated Moving Average, Error Trend Seasonality, and Hybrid Models. Front. Vet. Sci. 2021, 8, 775114. [Google Scholar] [CrossRef]
- Kruse, H.; Kirkemo, A.-M.; Handeland, K. Wildlife as source of zoonotic infections. Emerg. Infect. Dis. 2004, 10, 2067–2072. [Google Scholar] [CrossRef]
- Swiswa, S.; Masocha, M.; Pfukenyi, D.M.; Dhliwayo, S.; Chikerema, S.M. Long-term changes in the spatial distribution of lumpy skin disease hotspots in Zimbabwe. Trop. Anim. Health Prod. 2017, 49, 195–199. [Google Scholar] [CrossRef]
- Gari, G.; Waret-Szkuta, A.; Grosbois, V.; Jacquiet, P.; Roger, F. Risk factors associated with observed clinical lumpy skin disease in Ethiopia. Epidemiol. Infect. 2010, 138, 1657–1666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayelet, G.; Haftu, R.; Jemberie, S.; Belay, A.; Gelaye, E.; Sibhat, B.; Skjerve, E.; Asmare, K. Lumpy skin disease in cattle in central Ethiopia: Outbreak investigation and isolation and molecular detection of the virus. Rev. Sci. Tech. 2014, 33, 877–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molla, W.; Frankena, K.; Gari, G.; de Jong, M.C. Field study on the use of vaccination to control the occurrence of lumpy skin disease in Ethiopian cattle. Prev. Vet. Med. 2017, 147, 34–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allam, A.M.; Elbayoumy, M.K.; Abdel-Rahman, E.H.; Hegazi, A.G.; Farag, T.K. Molecular characterization of the 2018 outbreak of lumpy skin disease in cattle in Upper Egypt. Vet. World 2020, 13, 1262–1268. [Google Scholar] [CrossRef] [PubMed]
- Selim, A.; Manaa, E.; Khater, H. Seroprevalence and risk factors for lumpy skin disease in cattle in Northern Egypt. Trop. Anim. Health Prod. 2021, 53, 350. [Google Scholar] [CrossRef]
- Tuppurainen, E.; Antoniou, S.; Tsiamadis, E.; Topkaridou, M.; Labus, T.; Debeljak, Z.; Plavšić, B.; Miteva, A.; Alexandrov, T.; Pite, L. Field observations and experiences gained from the implementation of control measures against lumpy skin disease in South-East Europe between 2015 and 2017. Prev. Vet. Med. 2020, 181, 104600. [Google Scholar] [CrossRef]
- Ander, M.; Troell, K.; Chirico, J. Barcoding of biting midges in the genus Culicoides: A tool for species determination. Med. Vet. Entomol. 2013, 27, 323–331. [Google Scholar] [CrossRef]
- Authority, E.F.S.; Calistri, P.; De Clercq, K.; Gubbins, S.; Klement, E.; Stegeman, A.; Cortiñas Abrahantes, J.; Marojevic, D.; Antoniou, S.E.; Broglia, A. Lumpy skin disease epidemiological report IV: Data collection and analysis. EFSA J. 2020, 18, e06010. [Google Scholar]
- Sprygin, A.; Pestova, Y.; Bjadovskaya, O.; Prutnikov, P.; Zinyakov, N.; Kononova, S.; Ruchnova, O.; Lozovoy, D.; Chvala, I.; Kononov, A. Evidence of recombination of vaccine strains of lumpy skin disease virus with field strains, causing disease. PLoS ONE 2020, 15, e0232584. [Google Scholar] [CrossRef]
- Kononov, A.; Byadovskaya, O.; Kononova, S.; Yashin, R.; Zinyakov, N.; Mischenko, V.; Perevozchikova, N.; Sprygin, A. Detection of vaccine-like strains of lumpy skin disease virus in outbreaks in Russia in 2017. Arch. Virol. 2019, 164, 1575–1585. [Google Scholar] [CrossRef]
- Authority, E.F.S.; Calistri, P.; DeClercq, K.; Gubbins, S.; Klement, E.; Stegeman, A.; Cortiñas Abrahantes, J.; Antoniou, S.E.; Broglia, A.; Gogin, A. Lumpy skin disease: III. Data collection and analysis. EFSA J. 2019, 17, e05638. [Google Scholar]
- Al-Salihi, K.A.; Hassan, I.Q. Lumpy Skin Disease in Iraq: Study of the Disease Emergence. Transbound. Emerg. Dis. 2015, 62, 457–462. [Google Scholar] [CrossRef]
- Azeem, S.; Sharma, B.; Shabir, S.; Akbar, H.; Venter, E. Lumpy skin disease is expanding its geographic range: A challenge for Asian livestock management and food security. Vet. J. 2022, 279, 105785. [Google Scholar] [CrossRef]
- Punyapornwithaya, V.; Seesupa, S.; Phuykhamsingha, S.; Arjkumpa, O.; Sansamur, C.; Jarassaeng, C. Spatio-temporal patterns of lumpy skin disease outbreaks in dairy farms in northeastern Thailand. Front. Vet. Sci. 2022, 9, 957306. [Google Scholar] [CrossRef]
- Singhla, T.; Boonsri, K.; Kreausukon, K.; Modethed, W.; Pringproa, K.; Sthitmatee, N.; Punyapornwithaya, V.; Vinitchaikul, P. Molecular Characterization and Phylogenetic Analysis of Lumpy Skin Disease Virus Collected from Outbreaks in Northern Thailand in 2021. Vet. Sci. 2022, 9, 194. [Google Scholar] [CrossRef]
- Chibssa, T.R.; Sombo, M.; Lichoti, J.K.; Adam, T.I.B.; Liu, Y.; Elraouf, Y.A.; Grabherr, R.; Settypalli, T.B.K.; Berguido, F.J.; Loitsch, A. Molecular analysis of East African lumpy skin disease viruses reveals a mixed isolate with features of both vaccine and field isolates. Microorganisms 2021, 9, 1142. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anwar, A.; Na-Lampang, K.; Preyavichyapugdee, N.; Punyapornwithaya, V. Lumpy Skin Disease Outbreaks in Africa, Europe, and Asia (2005–2022): Multiple Change Point Analysis and Time Series Forecast. Viruses 2022, 14, 2203. https://doi.org/10.3390/v14102203
Anwar A, Na-Lampang K, Preyavichyapugdee N, Punyapornwithaya V. Lumpy Skin Disease Outbreaks in Africa, Europe, and Asia (2005–2022): Multiple Change Point Analysis and Time Series Forecast. Viruses. 2022; 14(10):2203. https://doi.org/10.3390/v14102203
Chicago/Turabian StyleAnwar, Ayesha, Kannika Na-Lampang, Narin Preyavichyapugdee, and Veerasak Punyapornwithaya. 2022. "Lumpy Skin Disease Outbreaks in Africa, Europe, and Asia (2005–2022): Multiple Change Point Analysis and Time Series Forecast" Viruses 14, no. 10: 2203. https://doi.org/10.3390/v14102203





