Antibody Response to Rotavirus C Pre-Farrow Natural Planned Exposure to Gilts and Their Piglets
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design, NPE Material and Sampling
2.2. Generation of Rotavirus C VP7 and VP4* Expression Constructs
2.3. Recombinant Protein Expression, Purification and Validation
2.4. Development of Recombinant Protein ELISAs to Quantitate RVC Antibodies
2.5. Screening Serum and Milk of Gilts and Piglet Serum for RVC Antibodies Using Genotype-Specific ELISA
2.6. Next Generation Sequencing and Real-Time PCR to Detect RVC in NPE Material and Piglet Feces
2.7. Statistical Analysis
3. Results
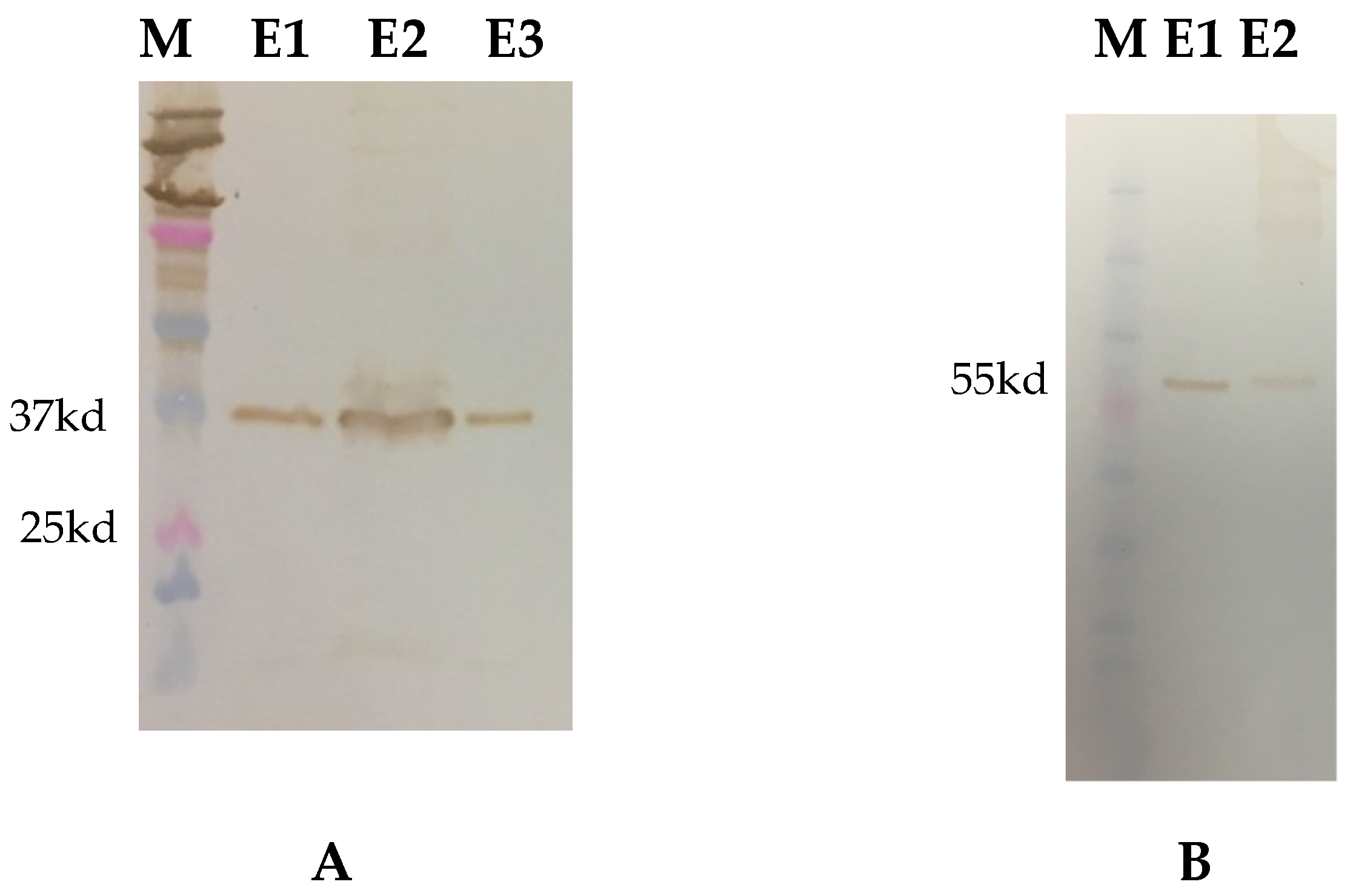
3.1. Recombinant Protein Expression
3.2. Antibody Response to RVC NPE
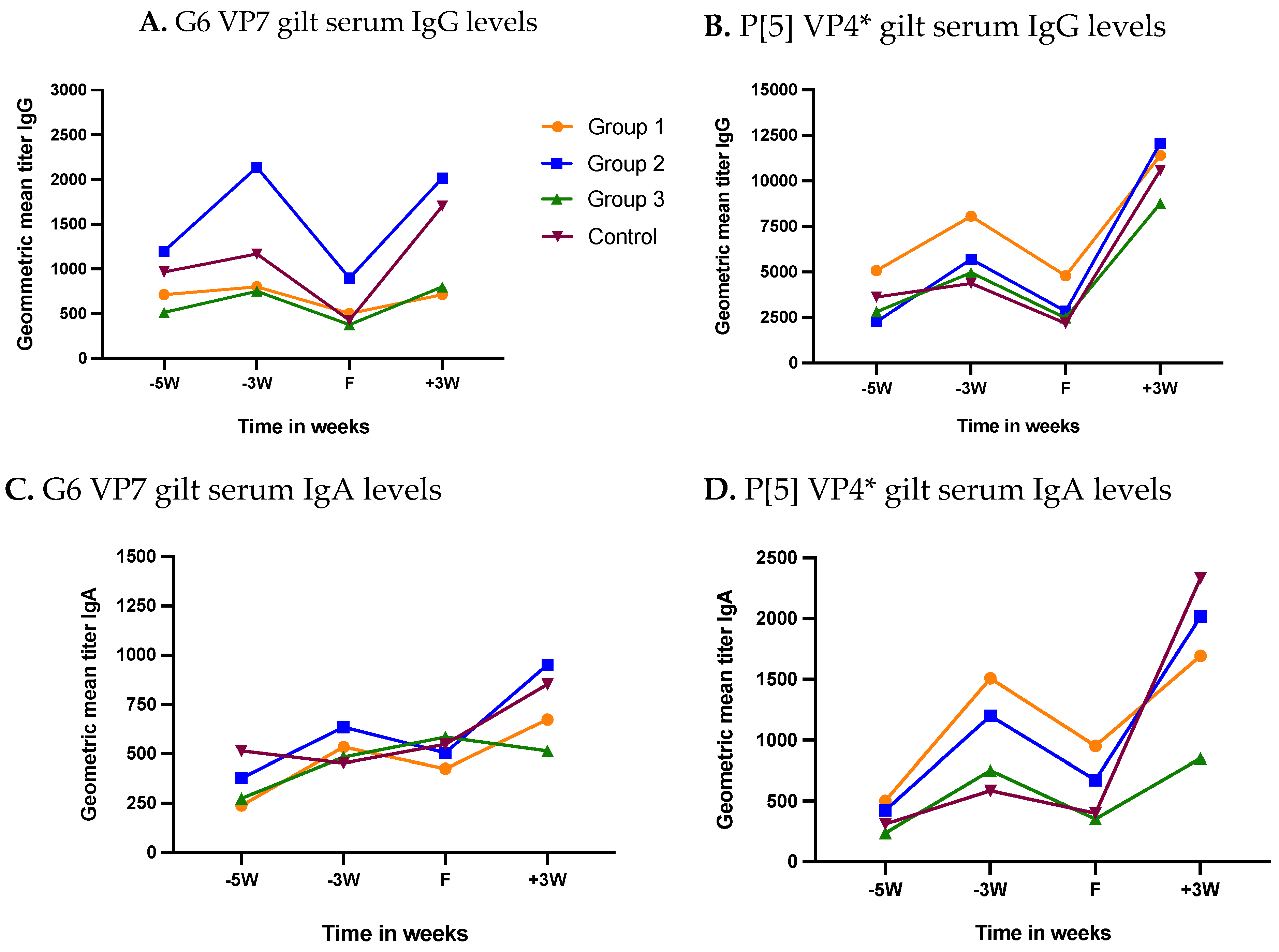
3.2.1. Gilt Serum
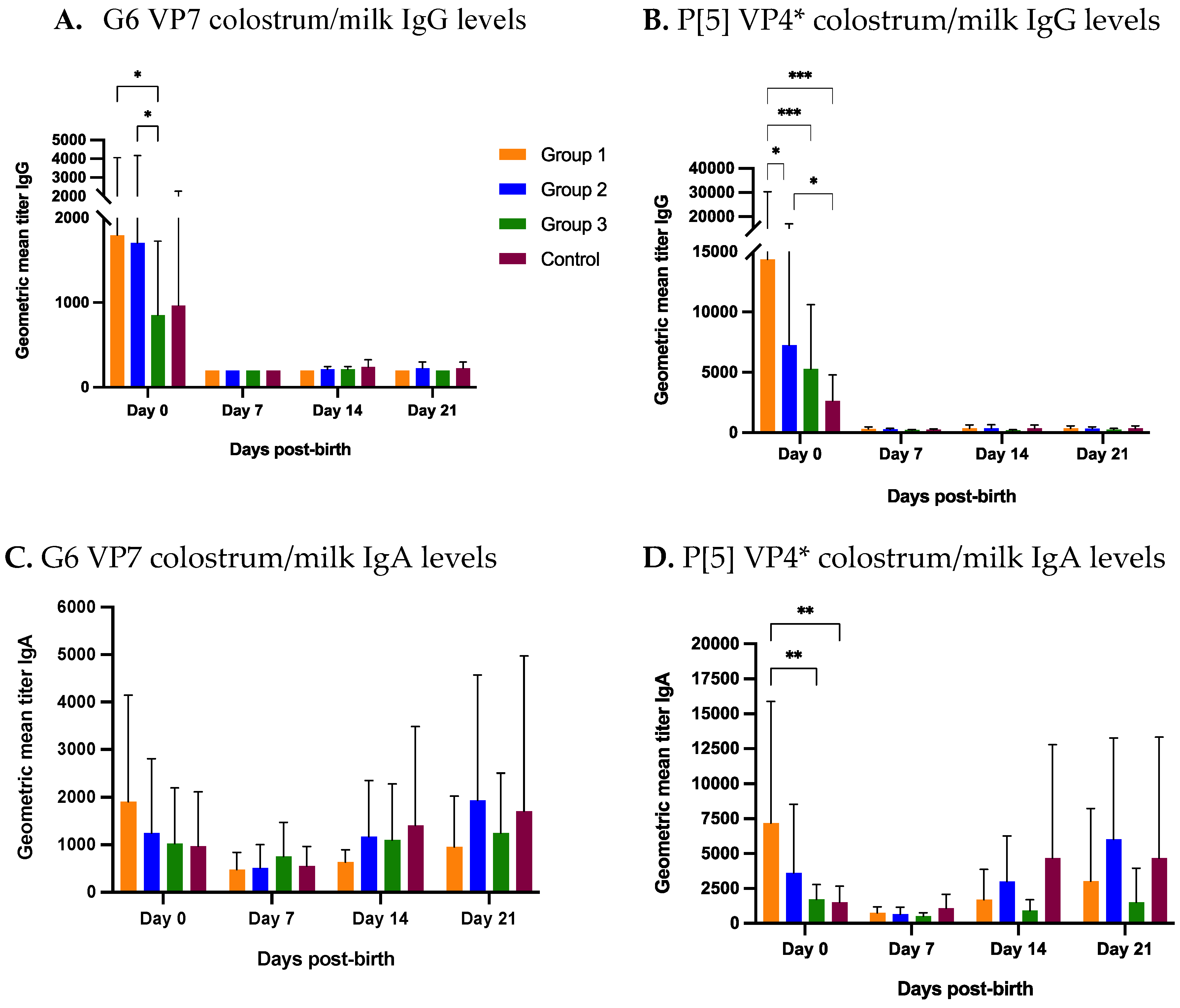
3.2.2. Colostrum and Milk
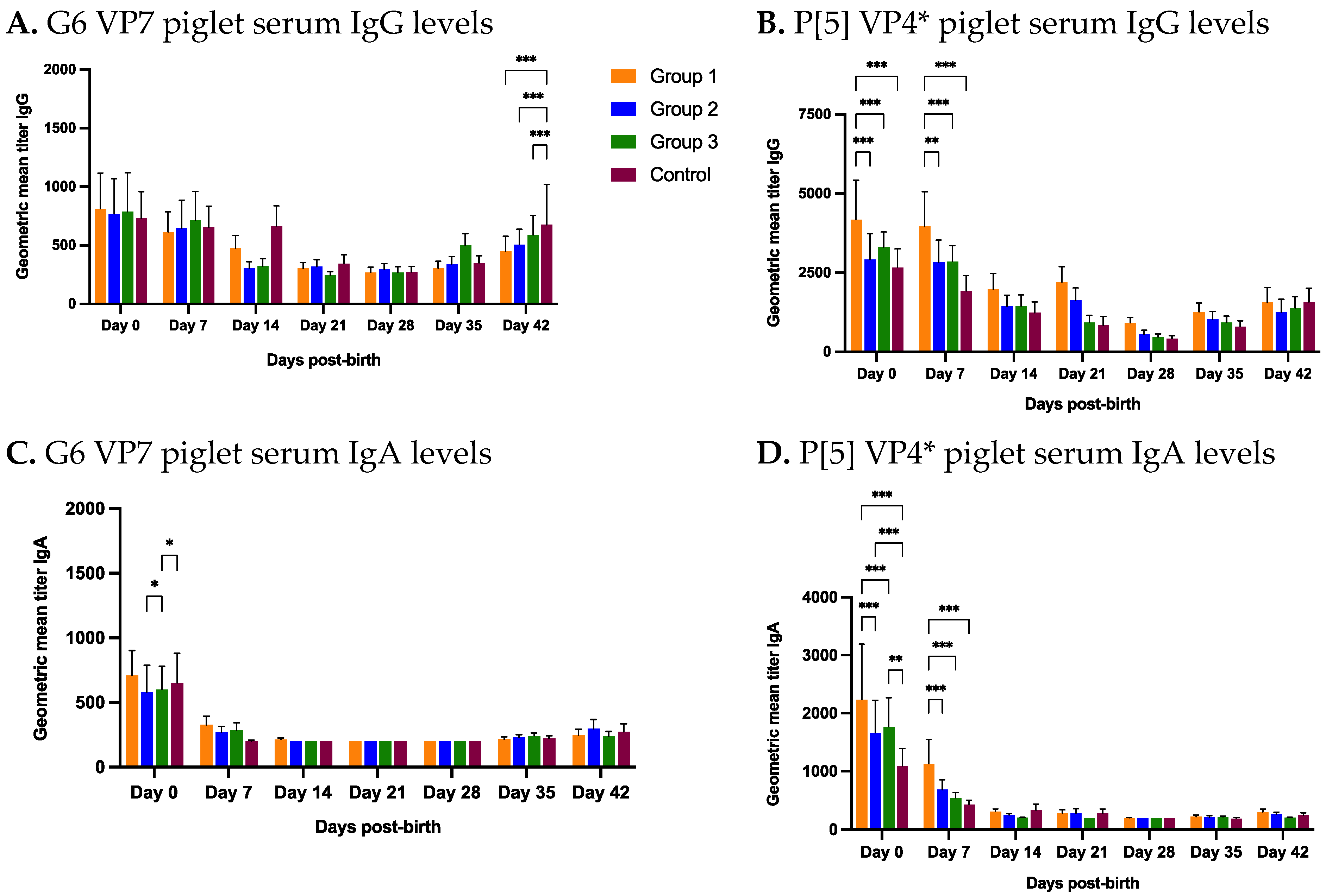
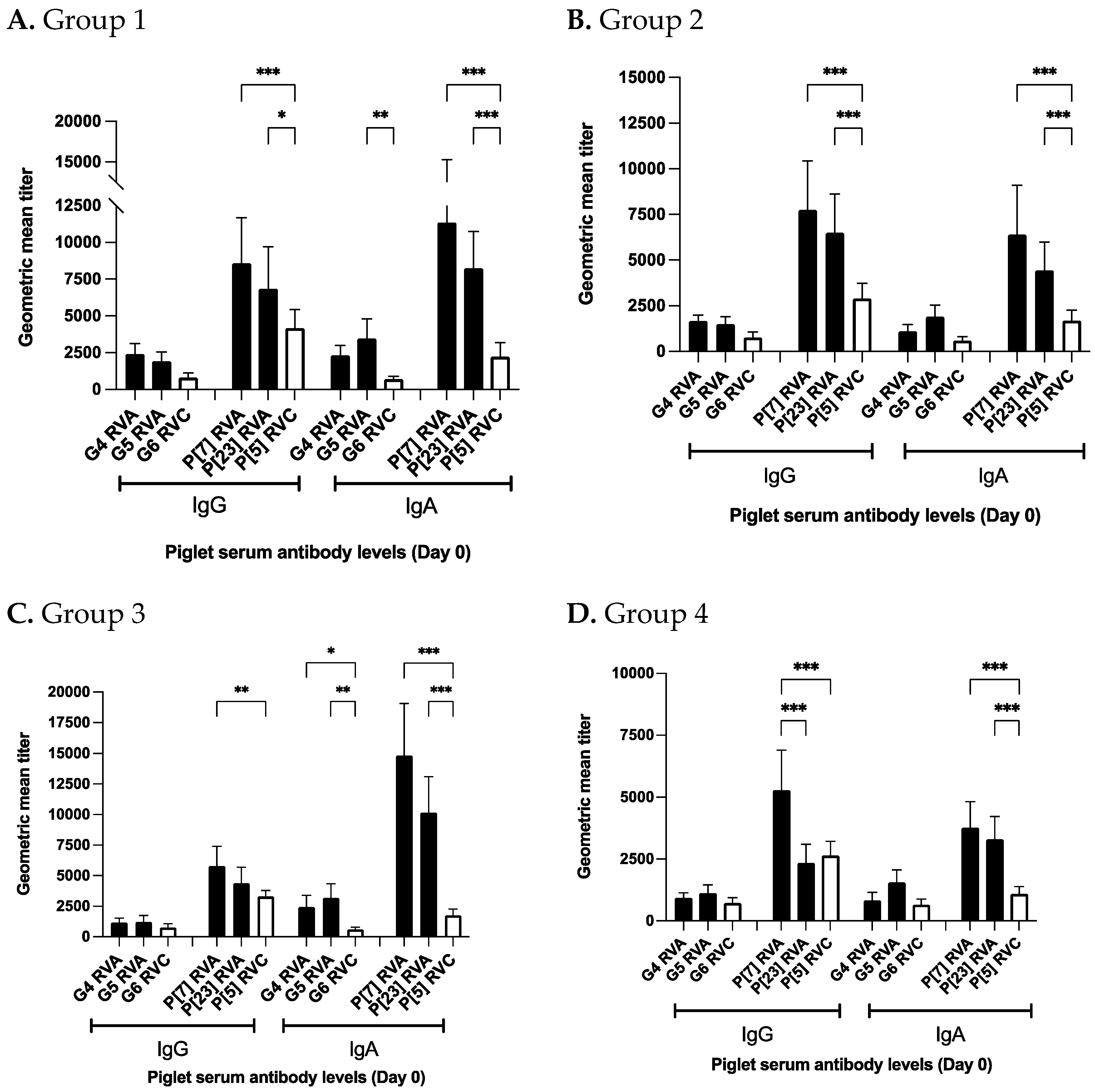
3.2.3. Piglet Serum
3.3. Levels of Antibodies against RVC Were Lower Than RVA in Colostrum
3.4. Piglet Serum at Birth Has Lower Antibodies against RVC Than RVA
3.5. RVC Fecal Shedding in Piglets and Antibody Response
3.6. Sequence Analysis of RVC from NPE and Piglet Feces
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Desselberger, U. Differences of Rotavirus Vaccine Effectiveness by Country: Likely Causes and Contributing Factors. Pathogens 2017, 6, 65. [Google Scholar] [CrossRef] [Green Version]
- Lestari, F.B.; Vongpunsawad, S.; Wanlapakorn, N.; Poovorawan, Y. Rotavirus infection in children in Southeast Asia 2008–2018: Disease burden, genotype distribution, seasonality, and vaccination. J. Biomed. Sci. 2020, 27, 66. [Google Scholar] [CrossRef] [PubMed]
- Shaw, R.D.; Vo, P.T.; Offit, P.A.; Coulsont, B.S.; Greenberg, H.B. Antigenic mapping of the surface proteins of rhesus rotavirus. Virology 1986, 155, 434–451. [Google Scholar] [CrossRef]
- Clarke, E.; Desselberger, U. Correlates of protection against human rotavirus disease and the factors influencing protection in low-income settings. Mucosal Immunol. 2015, 8, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramani, S.; Hu, L.; Prasad, B.V.; Estes, M.K. Diversity in Rotavirus–Host Glycan Interactions: A “Sweet” Spectrum. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 263–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angel, J.; Franco, M.A.; Greenberg, H. Rotavirus vaccines: Recent developments and future considerations. Nat. Rev. Genet. 2007, 5, 529–539. [Google Scholar] [CrossRef]
- Ludert, J.E.; Ruiz, M.C.; Hidalgo, C.; Liprandi, F. Antibodies to Rotavirus Outer Capsid Glycoprotein VP7 Neutralize Infectivity by Inhibiting Virion Decapsidation. J. Virol. 2002, 76, 6643–6651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matthijnssens, J.; Ciarlet, M.; Rahman, M.; Attoui, H.; Bányai, K.; Estes, M.K.; Gentsch, J.R.; Iturriza-Gómara, M.; Kirkwood, C.D.; Martella, V.; et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 2008, 153, 1621–1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bányai, K.; Kemenesi, G.; Budinski, I.; Földes, F.; Zana, B.; Marton, S.; Varga-Kugler, R.; Oldal, M.; Kurucz, K.; Jakab, F. Candidate new rotavirus species in Schreiber’s bats, Serbia. Infect. Genet. Evol. 2016, 48, 19–26. [Google Scholar] [CrossRef]
- Alekseev, K.P.; Penin, A.A.; Mukhin, A.N.; Khametova, K.M.; Grebennikova, T.V.; Yuzhakov, A.G.; Moskvina, A.S.; Musienko, M.I.; Raev, S.A.; Mishin, A.M.; et al. Genome Characterization of a Pathogenic Porcine Rotavirus B Strain Identified in Buryat Republic, Russia in 2015. Pathogens 2018, 7, 46. [Google Scholar] [CrossRef]
- Chasey, D.; Bridger, J.C.; McCrae, M.A. A new type of atypical rotavirus in pigs. Arch. Virol. 1986, 89, 235–243. [Google Scholar] [CrossRef]
- Homwong, N.; Diaz, A.; Rossow, S.; Ciarlet, M.; Marthaler, D. Three-Level Mixed-Effects Logistic Regression Analysis Reveals Complex Epidemiology of Swine Rotaviruses in Diagnostic Samples from North America. PLoS ONE 2016, 11, e0154734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marthaler, D.; Rossow, K.; Culhane, M.; Goyal, S.; Collins, J.; Matthijnssens, J.; Nelson, M.; Ciarlet, M. Widespread Rotavirus H in Commercially Raised Pigs, United States. Emerg. Infect. Dis. 2014, 20, 1203–1206. [Google Scholar] [CrossRef] [Green Version]
- Kattoor, J.J.; Saurabh, S.; Malik, Y.S.; Sircar, S.; Dhama, K.; Ghosh, S.; Bányai, K.; Kobayashi, N.; Singh, R.K. Unexpected detection of porcine rotavirus C strains carrying human origin VP6 gene. Vet. Q. 2017, 37, 252–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marthaler, D.; Homwong, N.; Rossow, K.; Culhane, M.; Goyal, S.; Collins, J.; Matthijnssens, J.; Ciarlet, M. Rapid detection and high occurrence of porcine rotavirus A, B, and C by RT-qPCR in diagnostic samples. J. Virol. Methods 2014, 209, 30–34. [Google Scholar] [CrossRef]
- Medici, K.; Barry, A.; Alfieri, A.; Alfieri, A. Porcine rotavirus groups A, B, and C identified by polymerase chain reaction in a fecal sample collection with inconclusive results by polyacrylamide gel electrophoresis. J. Swine Health Prod. 2018, 19, 146–150. [Google Scholar]
- Kumazaki, M.; Usuku, S. Epidemiological and genetic analysis of human group C rotaviruses isolated from outbreaks of acute gastroenteritis in Yokohama, Japan, between 2006 and 2012. Arch. Virol. 2013, 159, 761–771. [Google Scholar] [CrossRef]
- Tiku, V.R.; Jiang, B.; Kumar, P.; Aneja, S.; Bagga, A.; Bhan, M.K.; Ray, P. First study conducted in Northern India that identifies group C rotavirus as the etiological agent of severe diarrhea in children in Delhi. Virol. J. 2017, 14, 100. [Google Scholar] [CrossRef] [Green Version]
- Soma, J.; Tsunemitsu, H.; Miyamoto, T.; Suzuki, G.; Sasaki, T.; Suzuki, T. Whole-genome analysis of two bovine rotavirus C strains: Shintoku and Toyama. J. Gen. Virol. 2013, 94, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wise, A.G.; Smedley, R.C.; Kiupel, M.; Maes, R.K. Detection of Group C Rotavirus in Juvenile Ferrets (Mustela putorius furo) with Diarrhea by Reverse Transcription Polymerase Chain Reaction: Sequencing and Analysis of the Complete Coding Region of the VP6 Gene. Vet. Pathol. 2009, 46, 985–991. [Google Scholar] [CrossRef] [PubMed]
- Otto, P.H.; Rosenhain, S.; Elschner, M.C.; Hotzel, H.; Machnowska, P.; Trojnar, E.; Hoffmann, K.; Johne, R. Detection of rotavirus species A, B and C in domestic mammalian animals with diarrhoea and genotyping of bovine species A rotavirus strains. Vet. Microbiol. 2015, 179, 168–176. [Google Scholar] [CrossRef]
- Marton, S.; Mihalov-Kovács, E.; Dóró, R.; Csata, T.; Fehér, E.; Oldal, M.; Jakab, F.; Matthijnssens, J.; Martella, V.; Banyai, K. Canine rotavirus C strain detected in Hungary shows marked genotype diversity. J. Gen. Virol. 2015, 96, 3059–3071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saif, L.J.; Bohl, E.H.; Theil, K.W.; Cross, R.F.; House, J.A. Rotavirus-Like, Calicivirus-Like, and 23-nm Virus-Like Particles Associated with Diarrhea in Young Pigs. J. Clin. Microbiol. 1980, 12, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Marthaler, D.; Rossow, K.; Culhane, M.; Collins, J.; Goyal, S.; Ciarlet, M.; Matthijnssens, J. Identification, phylogenetic analysis and classification of porcine group C rotavirus VP7 sequences from the United States and Canada. Virology 2013, 446, 189–198. [Google Scholar] [CrossRef] [Green Version]
- Theuns, S.; Conceição-Neto, N.; Zeller, M.; Heylen, E.; Roukaerts, I.D.; Desmarets, L.M.; Van Ranst, M.; Nauwynck, H.J.; Matthijnssens, J. Characterization of a genetically heterogeneous porcine rotavirus C, and other viruses present in the fecal virome of a non-diarrheic Belgian piglet. Infect. Genet. Evol. 2016, 43, 135–145. [Google Scholar] [CrossRef]
- Chepngeno, J.; Diaz, A.; Paim, F.C.; Saif, L.J.; Vlasova, A.N. Rotavirus C: Prevalence in suckling piglets and development of virus-like particles to assess the influence of maternal immunity on the disease development. Vet. Res. 2019, 50, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langel, S.N.; Wang, Q.; Vlasova, A.N.; Saif, L.J. Host Factors Affecting Generation of Immunity Against Porcine Epidemic Diarrhea Virus in Pregnant and Lactating Swine and Passive Protection of Neonates. Pathogens 2020, 9, 130. [Google Scholar] [CrossRef] [Green Version]
- Gellberg, H.B.; Patterson, J.S.; Woode, G.N. A longitudinal study of rotavirus antibody titers in swine in a closed specific pathogen-free herd. Vet. Microbiol. 1991, 28, 231–242. [Google Scholar] [CrossRef]
- Shepherd, F. Enhancing Control of Porcine Rotavirus through the Identification of Candidate B Cell Epitopes. Ph.D. Thesis, University of Minnesota, Minneapolis, MN, USA, 2020. Retrieved from the University of Minnesota Digital Conservancy. Available online: https://hdl.handle.net/11299/217156 (accessed on 20 September 2022).
- Ward, L.A.; Rich, E.D.; Besser, T.E. Role of Maternally Derived Circulating Antibodies in Protection of Neonatal Swine against Porcine Group A Rotavirus. J. Infect. Dis. 1996, 174, 276–282. [Google Scholar] [CrossRef]
- Mantis, N.J.; Rol, N.; Corthésy, B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011, 4, 603–611. [Google Scholar] [CrossRef] [Green Version]
- Pittman, J. Field Experiences with Interventions for Rotavirus Control. AASV. 2016. Available online: https://www.aasv.org/library/swineinfo/ (accessed on 20 September 2022).
- Shepherd, F.K.; Dvorak, C.M.T.; Murtaugh, M.P.; Marthaler, D.G. Leveraging a Validated in silico Approach to Elucidate Genotype-Specific VP7 Epitopes and Antigenic Relationships of Porcine Rotavirus A. Front. Genet. 2020, 11, 828. [Google Scholar] [CrossRef]
- Kumar, D.; Anderson, A.; Pittman, J.; Springer, N.; Mallen, K.; Roman-Sosa, G.; Sangewar, N.; Casey-Moore, M.; Bowen, M.D.; Marthaler, D.; et al. Role of pre-farrow natural planned exposure to gilts in shaping the passive antibody response to rotavirus A in piglets. 2022; manuscript in preparation. [Google Scholar]
- Maan, S.; Rao, S.; Maan, N.S.; Anthony, S.J.; Attoui, H.; Samuel, A.R.; Mertens, P.P.C. Rapid cDNA synthesis and sequencing techniques for the genetic study of bluetongue and other dsRNA viruses. J. Virol. Methods 2007, 143, 132–139. [Google Scholar] [CrossRef]
- Klobasa, F.; Habe, F.; Werhahn, E.; Butler, J. Changes in the concentrations of serum IgG, IgA and IgM of sows throughout the reproductive cycle. Vet. Immunol. Immunopathol. 1985, 10, 341–353. [Google Scholar] [CrossRef]
- Bourne, F.J.; Curtis, J. The transfer of immunoglobins IgG, IgA and IgM from serum to colostrum and milk in the sow. Immunology 1973, 24, 157–162. [Google Scholar]
- Matías, J.; Berzosa, M.; Pastor, Y.; Irache, J.M.; Gamazo, C. Maternal Vaccination. Immunization of Sows during Pregnancy against ETEC Infections. Vaccines 2017, 5, 48. [Google Scholar] [CrossRef] [Green Version]
- Moffett, A.; Loke, C. Immunology of placentation in eutherian mammals. Nat. Rev. Immunol. 2006, 6, 584–594. [Google Scholar] [CrossRef]
- Fu, Z.; Hampson, D.; Wilks, C. Transfer of maternal antibody against group A rotavirus from sows to piglets and serological responses following natural infection. Res. Vet. Sci. 1990, 48, 365–373. [Google Scholar] [CrossRef]
- Nair, N.; Feng, N.; Blum, L.K.; Sanyal, M.; Ding, S.; Jiang, B.; Sen, A.; Morton, J.M.; He, X.S.; Robinson, W.H.; et al. VP4- and VP7-specific antibodies mediate heterotypic immunity to rotavirus in humans. Sci. Transl. Med. 2017, 9, eaam5434. [Google Scholar] [CrossRef]


| −5W | −4W | −3W | 0W (Farrowing) | 1W | 2W | 3W (Weaning) | 4W | 5W | 6W | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Group 1 | Gilts (n = 12) | X * | * | X * | X+ | + | + | X+ | |||
| 5 piglets/gilt | X | X | X | X | X | X | X | ||||
| Group 2 | Gilts (n = 12) | X * | X * | X+ | + | + | X+ | ||||
| 5 piglets/gilt | X | X | X | X | X | X | X | ||||
| Group 3 | Gilts (n = 11) | X * | X | X+ | + | + | X+ | ||||
| 5 piglets/gilt | X | X | X | X | X | X | X | ||||
| Group 4 | Gilts (n = 11) | X | X | X+ | + | + | X+ | ||||
| 5 piglets/gilt | X | X | X | X | X | X | X |
| Litter ID | Group | Week | RVC Ct | Genome Constellation |
|---|---|---|---|---|
| 40960 | 2 | 1 | 17.66 | G6-P[5]-I5-R1-C1-M1-A1-N6-Tx-Ex-H1 |
| 2 | 18.39 | G6-P[5]-I5-R1-C1-M1-A1- N6-Tx-Ex-H1 | ||
| 41045 | 2 | 2 | 22.57 | G6-P[5]-I5-R1-C1-M1-A1- N6-Tx-Ex-H1 |
| 3 | 21.7 | G6-P[5]-I5-R1-C1-M1-A1- N6-Tx-Ex-H1 | ||
| 41009 | 3 | 2 | 17.48 | G6-P[5]-I5-R1-C1-M1-A1- N6-Tx-Ex-H1 |
| 3 | 19.68 | G6-P[5]-I5-R1-C1-M1-A1- N6-Tx-Ex-H1 | ||
| 41285 | 3 | 2 | 19.64 | G6-P[5]-I5-R1-C1-M1-A1- N6-Tx-Ex-H1 |
| 3 | 22.49 | G6-P[5]-I5-R1-C1-M1-A1- N6-Tx-Ex-H1 | ||
| 41014 | 4 | 1 | 17.94 | G6-P[5]-I5-R1-C1-M1-A1- N6-Tx-Ex-H1 |
| 41174 | 4 | 1 | 17 | G6-P[5]-I5-R1-C1-M1-A1- N6-Tx-Ex-H1 |
| 41025 | 4 | 2 | 21.13 | G6-P[5]-I5-R1-C1-M1-A1- N6-Tx-Ex-H1 |
| Amino Acid Positions in VP7 | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 84 | 88 | 89 | 90 | 91 | 92 | 150 | 151 | 152 | 153 | 154 | 155 | 156 | 194 | 195 | 197 | 226 | 245 | 246 | 247 | 248 | 249 | 250 | 251 | 252 | |
| G6-Parent Sequence | A | S | P | G | P | G | E | P | K | N | S | E | A | E | D | D | D | S | S | S | L | N | Q | L | Q |
| RVC/PIG/USA/S8/2017/G6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| RVC/PIG/USA/S18/2017/G6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| RVC/PIG/USA/S20/2017/G6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | -- | -- | -- | I | . | . | . | . |
| RVC/PIG/USA/S31/2017/G6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | -- | -- | -- | I | . | . | . | . |
| RVC/PIG/USA/S21/2017/G6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| RVC/PIG/USA/S32/2017/G6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | -- | -- | -- | I | . | . | . | . |
| RVC/PIG/USA/S22/2017/G6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| RVC/PIG/USA/S33/2017/G6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| RVC/PIG/USA/S12/2017/G6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | -- | -- | -- | I | . | . | . | . |
| RVC/PIG/USA/S13/2017/G6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
| RVC/PIG/USA/S24/2017/G6 | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . | . |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, D.; Anderson, A.V.; Pittman, J.; Springer, N.L.; Marthaler, D.G.; Mwangi, W. Antibody Response to Rotavirus C Pre-Farrow Natural Planned Exposure to Gilts and Their Piglets. Viruses 2022, 14, 2250. https://doi.org/10.3390/v14102250
Kumar D, Anderson AV, Pittman J, Springer NL, Marthaler DG, Mwangi W. Antibody Response to Rotavirus C Pre-Farrow Natural Planned Exposure to Gilts and Their Piglets. Viruses. 2022; 14(10):2250. https://doi.org/10.3390/v14102250
Chicago/Turabian StyleKumar, Deepak, Amanda V. Anderson, Jeremy Pittman, Nora L. Springer, Douglas G. Marthaler, and Waithaka Mwangi. 2022. "Antibody Response to Rotavirus C Pre-Farrow Natural Planned Exposure to Gilts and Their Piglets" Viruses 14, no. 10: 2250. https://doi.org/10.3390/v14102250





