Host Plants Shape the Codon Usage Pattern of Turnip Mosaic Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus Isolates
2.2. Recombination and Phylogenetic Analysis
2.3. Nucleotide Composition Analysis
2.4. Relative Synonymous Codon Usage (RSCU) Analysis
2.5. Principal Component Analysis (PCA)
2.6. Effective Number of Codons Analysis (ENC)
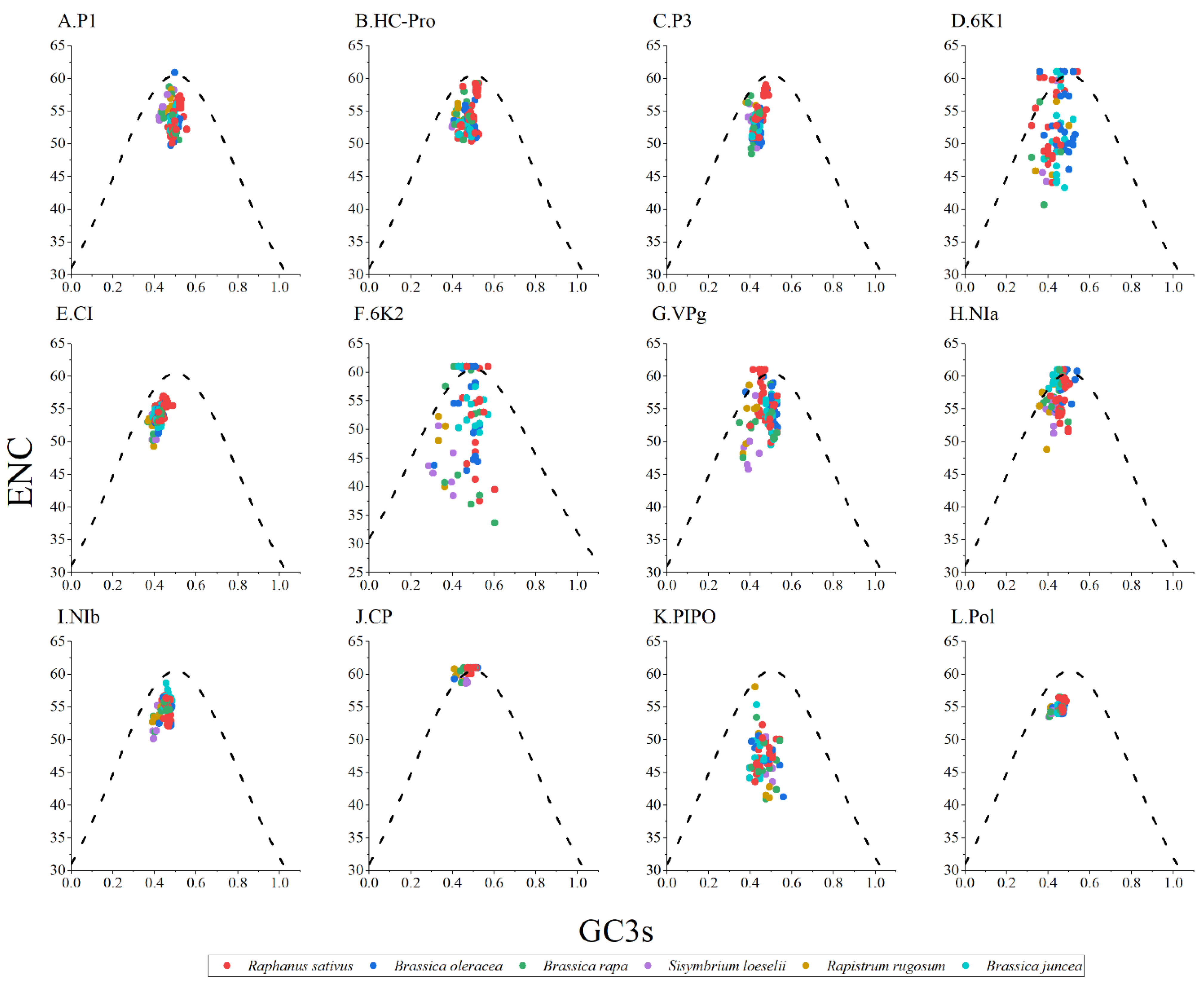
2.7. ENC-Plot Analysis
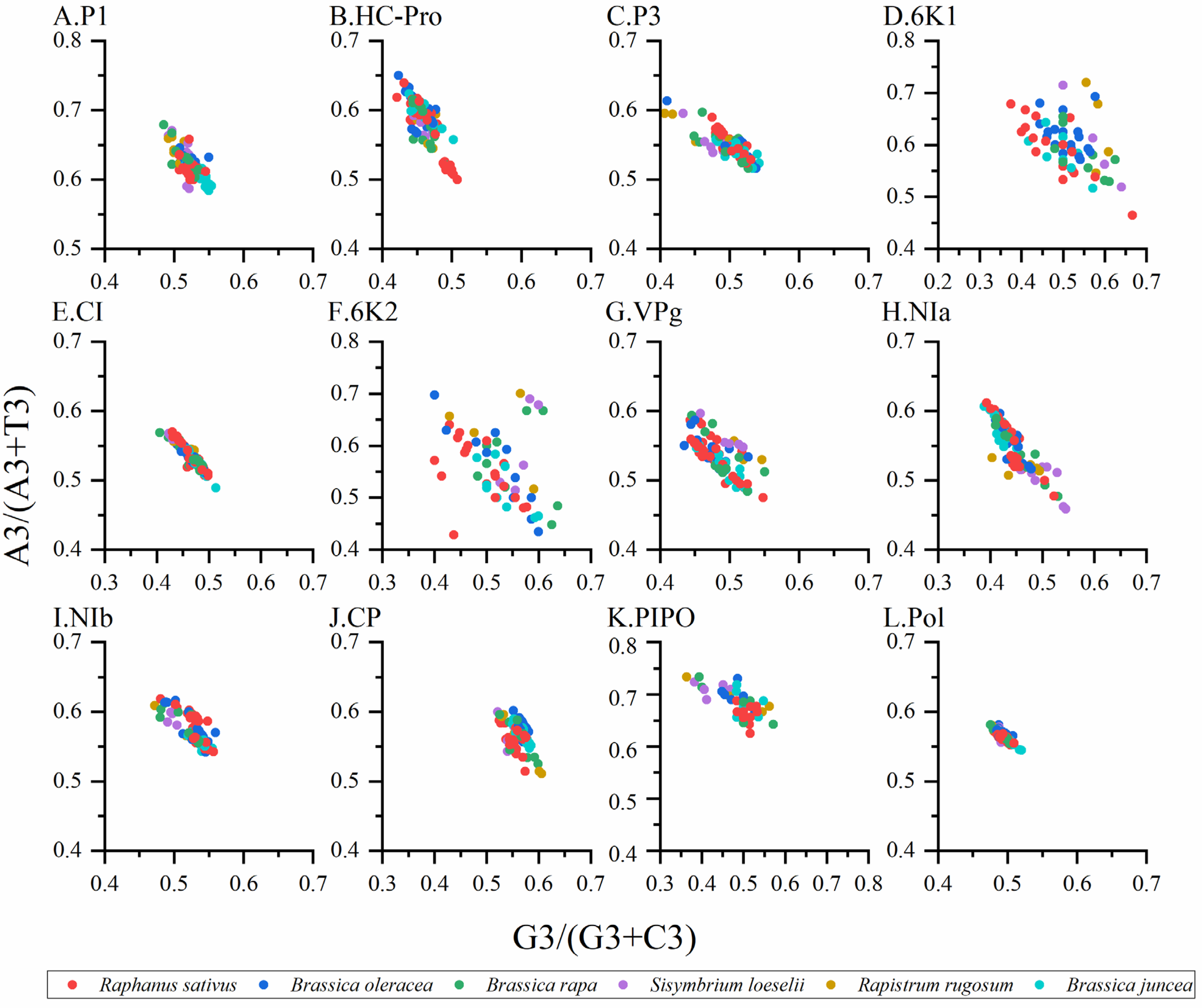
2.8. Parity Rule 2 Analysis (PR2)
2.9. Neutrality Analysis
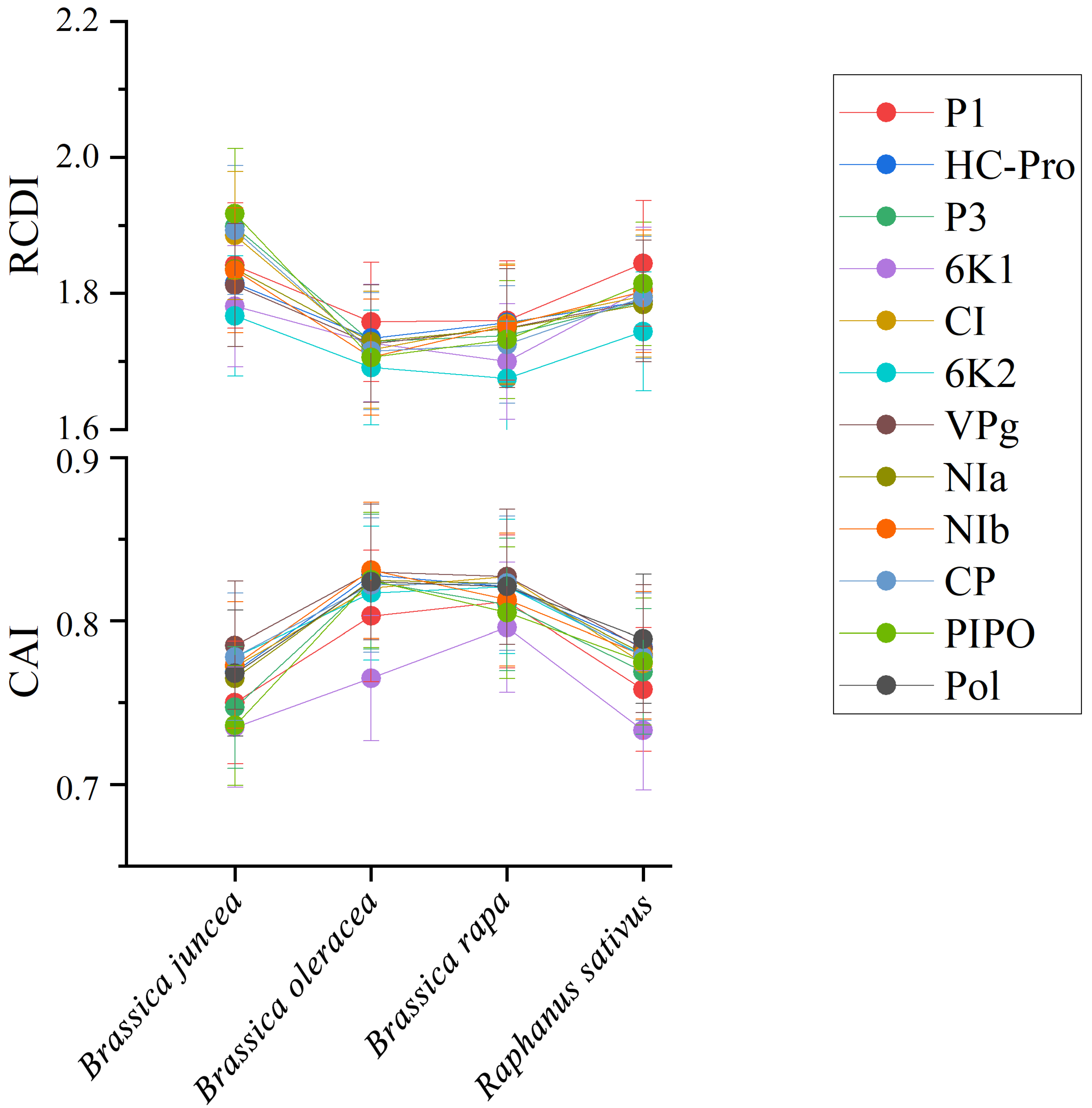
2.10. Codon Adaptation Index (CAI) Analysis
2.11. Relative Codon Deoptimization Index (RCDI) Analysis
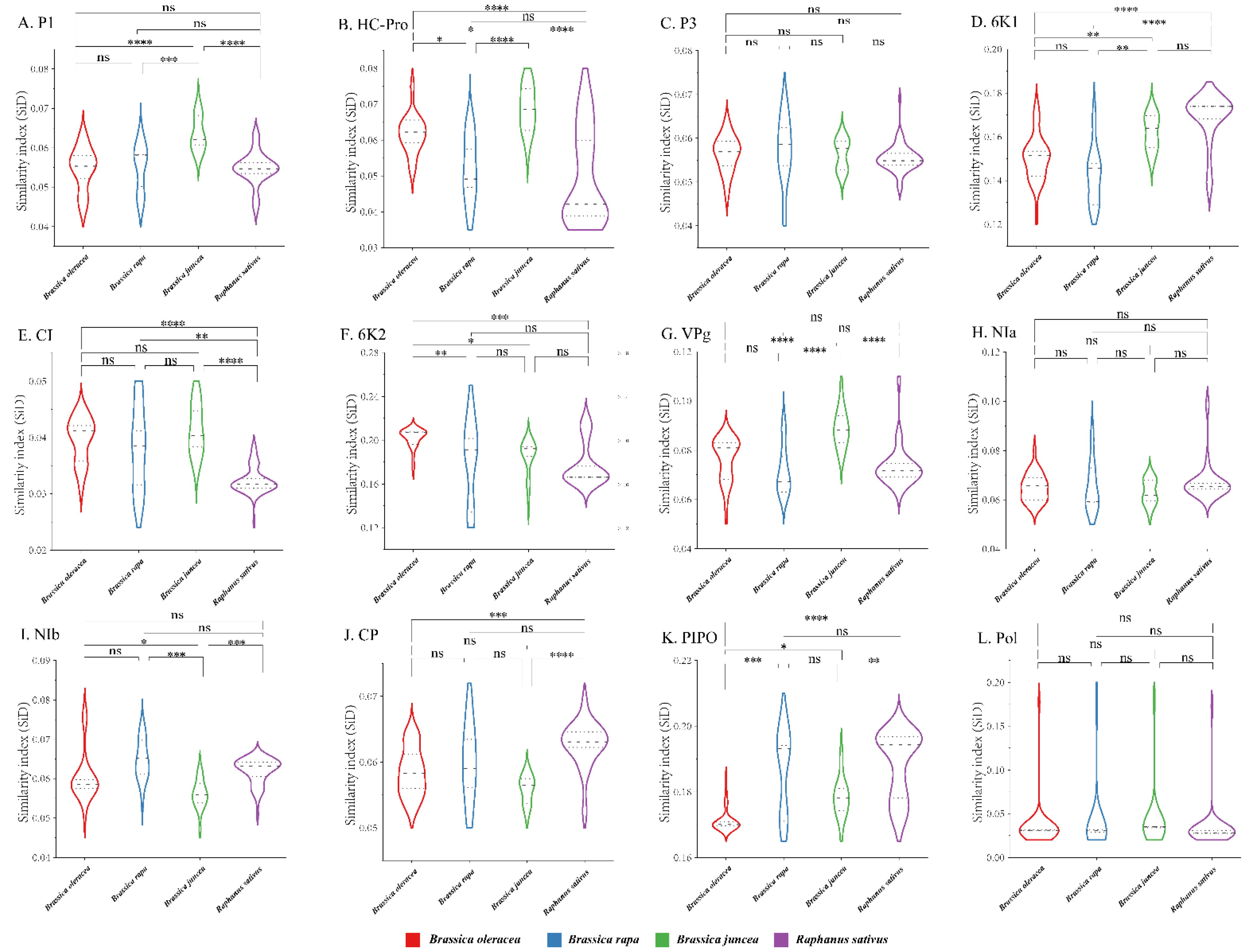
2.12. Similarity Index (SiD) Analysis
3. Results
3.1. Recombination and Phylogenetic Analysis
3.2. Nucleotide Bias Analysis
3.3. Relative Synonymous Codon Usage Analysis of TuMV and Its Hosts
3.4. Trends in Codon Usage Variations
3.5. Codon Usage Bias of TuMV
3.6. ENC-Plot Analysis
3.7. Neutrality Plot
3.8. Parity Analysis
3.9. Codon Usage Adaptation in TuMV
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wylie, S.J.; Adams, M.; Chalam, C.; Kreuze, J.; López-Moya, J.J.; Ohshima, K.; Praveen, S.; Rabenstein, F.; Stenger, D.; Wang, A.; et al. ICTV Virus Taxonomy Profile: Potyviridae. J. Gen. Virol. 2017, 98, 352–354. [Google Scholar] [CrossRef] [PubMed]
- Walsh, J.A.; Jenner, C.E. Turnip Mosaic Virus and the Quest for Durable Resistance. J. Mol. Plant. Pathol. 2002, 3, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.Y.W.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An Overlapping Essential Gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, M.; Yasunaga, T.; Miyata, T. Secondary Structure of MS2 Phage RNA and Bias in Code Word Usage. Nucleic. Acids. Res. 1979, 7, 2073–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharp, P.M.; Tuohy, T.M.F.; Mosurski, K.R. Codon Usage in Yeast: Cluster Analysis Clearly Differentiates Highly and Lowly Expressed Genes. Nucleic. Acids. Res. 1986, 14, 5125–5143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sueoka, N. Directional Mutation Pressure and Neutral Molecular Evolution. Proc. Natl. Acad. Sci. USA 1988, 85, 2653–2657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharp, P.M.; Cowe, E. Synonymous Codon Usage in Saccharomyces Cerevisiae. Yeast 1991, 7, 657–678. [Google Scholar] [CrossRef] [PubMed]
- Comeron, J.M.; Aguadé, M. An Evaluation of Measures of Synonymous Codon Usage Bias. J.Mol. Evol. 1998, 47, 268–274. [Google Scholar] [CrossRef]
- Hershberg, R.; Petrov, D.A. Selection on Codon Bias. Annu. Rev. Genet. 2008, 42, 287–299. [Google Scholar] [CrossRef] [Green Version]
- Kyte, J.; Doolittle, R.F. A Simple Method for Displaying the Hydropathic Character of a Protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef]
- Sueoka, N. Translation-Coupled Violation of Parity Rule 2 in Human Genes Is Not the Cause of Heterogeneity of the DNA G+C Content of Third Codon Position. Gene. 1999, 238, 53–58. [Google Scholar] [CrossRef]
- Duret, L.; Mouchiroud, D. Expression Pattern and, Surprisingly, Gene Length Shape Codon Usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc. Natl. Acad. Sci. USA. 1999, 96, 4482–4487. [Google Scholar] [CrossRef] [Green Version]
- Fuglsang, A. Accounting for Background Nucleotide Composition When Measuring Codon Usage Bias: Brilliant Idea, Difficult in Practice. Mol. Biol. Evol. 2006, 23, 1345–1347. [Google Scholar] [CrossRef]
- Coleman, J.R.; Papamichail, D.; Skiena, S.; Futcher, B.; Wimmer, E.; Mueller, S. Virus Attenuation by Genome-Scale Changes in Codon Pair Bias. Science 2008, 320, 1784–1787. [Google Scholar] [CrossRef] [Green Version]
- Ohshima, K.; Yamaguchi, Y.; Hirota, R.; Hamamoto, T.; Tomimura, K.; Tan, Z.; Sano, T.; Azuhata, F.; Walsh, J.A.; Fletcher, J.; et al. Molecular Evolution of Turnip Mosaic Virus: Evidence of Host Adaptation, Genetic Recombination and Geographical Spread. J. Gen. Virol. 2002, 83, 1511–1521. [Google Scholar] [CrossRef]
- Tomimura, K.; Gibbs, A.J.; Jenner, C.E.; Walsh, J.A.; Ohshima, K. The Phylogeny of Turnip Mosaic Virus; Comparisons of 38 Genomic Sequences Reveal a Eurasian Origin and a Recent “emergence” in East Asia. Mol. Ecol. 2003, 12, 2099–2111. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Tomitaka, Y.; Ho, S.Y.W.; Duchêne, S.; Vetten, H.J.; Lesemann, D.; Walsh, J.A.; Gibbs, A.J.; Ohshima, K. Turnip Mosaic Potyvirus Probably First Spread to Eurasian Brassica Crops from Wild Orchids about 1000 Years Ago. PLoS ONE 2013, 8, e55336. [Google Scholar] [CrossRef]
- Nguyen, H.D.; Tran, H.T.N.; Ohshima, K. Genetic Variation of the Turnip Mosaic Virus Population of Vietnam: A Case Study of Founder, Regional and Local Influences. Virus. Res. 2013, 171, 138–149. [Google Scholar] [CrossRef]
- Gibbs, A.J.; Nguyen, H.D.; Ohshima, K. The “emergence” of Turnip Mosaic Virus Was Probably a “Gene-for-Quasi-Gene” Event. Curr. Opin. Virol. 2015, 10, 20–26. [Google Scholar] [CrossRef]
- Yasaka, R.; Ohba, K.; Schwinghamer, M.W.; Fletcher, J.; Ochoa-Corona, F.M.; Thomas, J.E.; Ho, S.Y.W.; Gibbs, A.J.; Ohshima, K. Phylodynamic Evidence of the Migration of Turnip Mosaic Potyvirus from Europe to Australia and New Zealand. J. Gen. Virol. 2015, 96, 701–713. [Google Scholar] [CrossRef]
- Yasaka, R.; Fukagawa, H.; Ikematsu, M.; Soda, H.; Korkmaz, S.; Golnaraghi, A.; Katis, N.; Ho, S.Y.W.; Gibbs, A.J.; Ohshima, K. The Timescale of Emergence and Spread of Turnip Mosaic Potyvirus. Sci. Rep. 2017, 7, 4240. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawakubo, S.; Gao, F.; Li, S.; Tan, Z.; Huang, Y.-K.; Adkar-Purushothama, C.R.; Gurikar, C.; Maneechoat, P.; Chiemsombat, P.; Aye, S.S.; et al. Genomic Analysis of the Brassica Pathogen Turnip Mosaic Potyvirus Reveals Its Spread along the Former Trade Routes of the Silk Road. Proc. Natl. Acad. Sci. USA 2021, 118, e2021221118. [Google Scholar] [CrossRef] [PubMed]
- Kawakubo, S.; Tomitaka, Y.; Tomimura, K.; Koga, R.; Matsuoka, H.; Uematsu, S.; Yamashita, K.; Ho, S.Y.W.; Ohshima, K. The Recombinogenic History of Turnip Mosaic Potyvirus Reveals Its Introduction to Japan in the 19th Century. Virus Evol. 2022, 8, veac060. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and Analysis of Recombination Patterns in Virus Genomes. Virus. Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, D.; Rybicki, E. RDP: Detection of Recombination amongst Aligned Sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawyer, S.A. GENECONV: A Computer Package for the Statistical Detection of Gene Conversion; Department of Mathematics, Washington University in Louis: Washington, WA, USA, 1999. [Google Scholar]
- Salminen, M.O.; Carr, J.K.; Burke, D.S.; Mccutchan, F.E. Identification of Breakpoints in Intergenotypic Recombinants of HIV Type 1 by Bootscanning. AIDS Res. Hum. Retroviruses. 1995, 11, 1423–1425. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.M. Analyzing the Mosaic Structure of Genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. Evaluation of Methods for Detecting Recombination from DNA Sequences: Computer Simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef] [Green Version]
- Boni, M.F.; Posada, D.; Feldman, M.W. An Exact Nonparametric Method for Inferring Mosaic Structure in Sequence Triplets. Genetics 2007, 176, 1035–1047. [Google Scholar] [CrossRef]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-Scanning: A Monte Carlo Procedure for Assessing Signals in Rebombinant Sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Page, R.D.M. Treeview: An Application to Display Phylogenetic Trees on Personal Computers. Bioinformatics 1996, 12, 357–358. [Google Scholar] [CrossRef] [Green Version]
- Sharp, P.M.; Li, W.H. An Evolutionary Perspective on Synonymous Codon Usage in Unicellular Organisms. J. Mol. Evol. 1986, 24, 28–38. [Google Scholar] [CrossRef]
- Wright, F. The “effective Number of Codons” Used in a Gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Gerton, J.L.; DeRisi, J.; Shroff, R.; Lichten, M.; Brown, P.O.; Petes, T.D. Global Mapping of Meiotic Recombination Hotspots and Coldspots in the Yeast Saccharomyces Cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 11383–11390. [Google Scholar] [CrossRef] [Green Version]
- Steel, M. The Phylogenetic Handbook: A Practical Approach to Phylogenetic Analysis and Hypothesis Testing, 2nd ed.; Lemey, P., Salemi, M., Vandamme, A.M., Eds.; Cambridge University Press: Cambridge, UK, 2010; Volume 66, pp. 324–325. [Google Scholar]
- Fajinmi, A.A. Interactive Effect of Blackeye Cowpea Mosaic Virus and Cucumber Mosaic Virus on Vigna Unguiculata. Hortic. Plant. J. 2019, 5, 88–92. [Google Scholar] [CrossRef]
- Yasaka, R.; Nguyen, H.D.; Ho, S.Y.W.; Duchêne, S.; Korkmaz, S.; Katis, N.; Takahashi, H.; Gibbs, A.J.; Ohshima, K. The Temporal Evolution and Global Spread of Cauliflower Mosaic Virus, a Plant Pararetrovirus. PLoS ONE 2014, 9, e85641. [Google Scholar] [CrossRef]
- He, Z.; Dong, T.; Wang, T.; Chen, W.; Liu, X.; Li, L. Genetic Variation of the Novel Badnaviruses Infecting Nelumbo Nucifera Based on the RT/RNase H Coding Region Sequences. Hortic. Plant. J. 2020, 6, 335–342. [Google Scholar] [CrossRef]
- Butt, A.M.; Nasrullah, I.; Qamar, R.; Tong, Y. Evolution of Codon Usage in Zika Virus Genomes Is Host and Vector Specific. Emerg. Microbes. Infect. 2016, 5, 107. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Wang, H.; Wang, S.; Xing, G.; Zhang, C.; Zhang, W.; Liu, J.; Zhang, J.; Su, S.; Zhou, J. Insights into the Genetic and Host Adaptability of Emerging Porcine Circovirus 3. Virulence 2018, 9, 1301–1313. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Zhao, J.; Xing, G.; Li, G.; Wang, R.; Wang, Z.; Zhang, C.; Franzo, G.; Su, S.; Zhou, J. Genetic Analysis and Evolutionary Changes of Porcine Circovirus 2. Mol. Phylogenet. Evol. 2019, 139, 106520. [Google Scholar] [CrossRef]
- Yan, Z.; Wang, R.; Zhang, L.; Shen, B.; Wang, N.; Xu, Q.; He, W.; He, W.; Li, G.; Su, S. Evolutionary Changes of the Novel Influenza D Virus Hemagglutinin-Esterase Fusion Gene Revealed by the Codon Usage Pattern. Virulence 2019, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Zhang, L.; He, W.; Zhang, X.; Wen, B.; Wang, C.; Xu, Q.; Li, G.; Zhou, J.; Veit, M.; et al. Genetic Evolution and Molecular Selection of the HE Gene of Influenza C Virus. Viruses 2019, 11, 167. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.Z.; Liu, Q.P.; Fan, L.J.; Cui, X.F.; Zhou, X.P. Analysis of Synonymous Codon Usage and Evolution of Begomoviruses. Zhejiang. Univ. Sci. B. 2008, 9, 667–674. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Qin, L.; Xu, X.; Ding, S. Evolution and Host Adaptability of Plant RNA Viruses: Research Insights on Compositional Biases. Comput. Struct. Biotechnol. J. 2022, 20, 2600–2610. [Google Scholar] [CrossRef]
- Biswas, K.K.; Palchoudhury, S.; Chakraborty, P.; Bhattacharyya, U.K.; Ghosh, D.K.; Debnath, P.; Ramadugu, C.; Keremane, M.L.; Khetarpal, R.K.; Lee, R.F. Codon Usage Bias Analysis of Citrus Tristeza Virus: Higher Codon Adaptation to Citrus Reticulata Host. Viruses 2019, 11, 331. [Google Scholar] [CrossRef] [Green Version]
- He, M.; Guan, S.Y.; He, C.Q. Evolution of Rice Stripe Virus. Mol. Phylogenet. Evol. 2017, 109, 343–350. [Google Scholar] [CrossRef]
- Chakraborty, P.; Das, S.; Saha, B.; Sarkar, P.; Karmakar, A.; Saha, A.; Saha, D.; Saha, A. Phylogeny and Synonymous Codon Usage Pattern of Papaya Ringspot Virus Coat Protein Gene in the Sub-Himalayan Region of North-East India. Can. J. Microbiol. 2015, 61, 555–564. [Google Scholar] [CrossRef]
- He, Z.; Gan, H.; Liang, X. Analysis of Synonymous Codon Usage Bias in Potato Virus M and Its Adaption to Hosts. Viruses 2019, 11, 752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Dong, Z.; Gan, H. Genetic Changes and Host Adaptability in Sugarcane Mosaic Virus Based on Complete Genome Sequences. Mol. Phylogenet. Evol. 2020, 149, 106848. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Dong, Z.; Qin, L.; Gan, H. Phylodynamics and Codon Usage Pattern Analysis of Broad Bean Wilt Virus 2. Viruses 2021, 13, 198. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Dong, Z.; Gan, H. Comprehensive Codon Usage Analysis of Rice Black-Streaked Dwarf Virus Based on P8 and P10 Protein Coding Sequences. Infect. Genet. Evol. 2020, 86, 104601. [Google Scholar] [CrossRef]
- He, Z.; Ding, S.; Guo, J.; Qin, L.; Xu, X. Synonymous Codon Usage Analysis of Three Narcissus Potyviruses. Viruses 2022, 14, 846. [Google Scholar] [CrossRef]
- Puigbò, P.; Aragonès, L.; Garcia-Vallvé, S. RCDI/ERCDI: A Web-Server to Estimate Codon Usage Deoptimization. BMC. Res. Notes 2010, 3, 87. [Google Scholar] [CrossRef]



| Codon | aa | TuMV | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | HC | P3 | 6K1 | CI | 6K2 | Vpg | Nla | Nlb | CP | PIPO | Pol | ||
| TTT | F | 0.98 | 0.66 | 1.02 | 1.41 | 0.95 | 0.37 | 0.79 | 0.90 | 0.81 | 0.91 | 1.75 | 0.87 |
| TTC | F | 1.02 | 1.34 | 0.98 | 0.59 | 1.05 | 1.63 | 1.21 | 1.10 | 1.19 | 1.09 | 0.24 | 1.13 |
| TTA | L | 0.65 | 0.72 | 1.35 | 0.81 | 0.62 | 0.49 | 0.49 | 1.04 | 0.50 | 1.32 | 2.35 | 0.80 |
| TTG | L | 1.76 | 1.31 | 1.44 | 0.78 | 0.90 | 1.16 | 1.08 | 1.31 | 1.41 | 1.02 | 2.40 | 1.25 |
| CTT | L | 0.66 | 0.82 | 0.68 | 1.00 | 1.73 | 1.69 | 1.13 | 0.89 | 0.89 | 0.93 | 0.07 | 1.03 |
| CTC | L | 1.29 | 0.97 | 0.68 | 2.53 | 1.31 | 0.75 | 1.68 | 1.08 | 1.12 | 1.15 | 0.01 | 1.14 |
| CTA | L | 0.79 | 1.23 | 1.11 | 0.70 | 0.98 | 1.07 | 0.57 | 1.26 | 0.87 | 0.83 | 0.44 | 0.98 |
| CTG | L | 0.86 | 0.95 | 0.75 | 0.17 | 0.46 | 0.85 | 1.05 | 0.42 | 1.19 | 0.75 | 0.74 | 0.80 |
| ATT | I | 1.22 | 0.65 | 0.87 | 0.81 | 0.81 | 1.45 | 0.85 | 1.11 | 0.74 | 1.20 | 0.26 | 0.90 |
| ATC | I | 1.13 | 1.12 | 1.36 | 0.62 | 1.06 | 1.11 | 0.89 | 0.76 | 1.15 | 0.84 | 1.73 | 1.08 |
| ATA | I | 0.65 | 1.23 | 0.77 | 1.57 | 1.12 | 0.44 | 1.26 | 1.13 | 1.10 | 0.96 | 1.01 | 1.02 |
| GTT | V | 1.05 | 1.11 | 0.90 | 0.94 | 1.14 | 1.31 | 0.67 | 0.68 | 1.03 | 0.90 | 0.09 | 1.02 |
| GTC | V | 0.94 | 1.18 | 1.08 | 1.57 | 0.86 | 0.22 | 0.47 | 1.10 | 1.02 | 0.66 | 2.81 | 0.95 |
| GTA | V | 0.79 | 0.70 | 0.97 | 0.39 | 0.90 | 0.82 | 0.52 | 0.67 | 0.98 | 1.20 | 0.32 | 0.84 |
| GTG | V | 1.23 | 1.01 | 1.05 | 1.11 | 1.11 | 1.65 | 2.34 | 1.56 | 0.97 | 1.25 | 0.78 | 1.19 |
| TCT | S | 0.42 | 0.56 | 0.67 | 1.56 | 0.78 | 0.88 | 1.61 | 0.49 | 0.21 | 1.18 | 0.47 | 0.62 |
| TCC | S | 0.69 | 0.51 | 0.39 | 1.55 | 0.49 | 0.83 | 0.46 | 0.71 | 0.32 | 1.16 | 0.37 | 0.55 |
| TCA | S | 1.38 | 1.73 | 1.06 | 0.09 | 1.53 | 1.24 | 1.07 | 1.18 | 1.78 | 0.60 | 1.64 | 1.38 |
| TCG | S | 0.38 | 0.71 | 0.50 | 0.00 | 0.53 | 0.23 | 0.06 | 0.47 | 1.27 | 0.09 | 1.00 | 0.57 |
| AGT | S | 1.61 | 1.10 | 1.88 | 1.15 | 1.20 | 0.34 | 0.61 | 1.34 | 1.11 | 1.61 | 1.02 | 1.35 |
| AGC | S | 1.53 | 1.40 | 1.50 | 1.65 | 1.47 | 2.48 | 2.19 | 1.81 | 1.32 | 1.36 | 1.50 | 1.52 |
| CCT | P | 0.67 | 0.44 | 0.62 | 2.30 | 0.62 | 1.63 | 0.81 | 0.42 | 0.87 | 0.16 | 0.00 | 0.62 |
| CCC | P | 0.60 | 0.32 | 0.33 | 1.54 | 0.34 | 2.28 | 0.42 | 0.24 | 0.38 | 0.65 | 0.39 | 0.43 |
| CCA | P | 2.15 | 2.70 | 2.64 | 0.15 | 2.45 | 0.09 | 2.26 | 2.54 | 2.41 | 1.79 | 2.61 | 2.36 |
| CCG | P | 0.58 | 0.53 | 0.41 | 0.00 | 0.59 | 0.00 | 0.51 | 0.81 | 0.34 | 1.40 | 0.02 | 0.59 |
| ACT | T | 0.66 | 0.84 | 1.03 | 1.33 | 0.87 | 0.42 | 0.54 | 0.89 | 0.89 | 0.65 | 0.00 | 0.81 |
| ACC | T | 1.33 | 0.57 | 0.66 | 0.25 | 0.57 | 1.38 | 0.74 | 0.81 | 0.72 | 0.76 | 0.40 | 0.76 |
| ACA | T | 1.54 | 1.65 | 1.49 | 2.13 | 2.01 | 0.76 | 2.16 | 1.21 | 1.80 | 1.11 | 2.22 | 1.67 |
| ACG | T | 0.47 | 0.93 | 0.82 | 0.30 | 0.55 | 1.45 | 0.56 | 1.08 | 0.59 | 1.48 | 0.83 | 0.76 |
| GCT | A | 0.76 | 0.78 | 0.83 | 0.29 | 0.89 | 0.00 | 1.50 | 0.77 | 0.89 | 0.83 | 1.21 | 0.85 |
| GCC | A | 0.88 | 0.45 | 0.81 | 0.82 | 0.61 | 0.07 | 1.15 | 0.69 | 0.30 | 0.52 | 0.19 | 0.64 |
| GCA | A | 1.81 | 2.15 | 1.65 | 2.20 | 1.81 | 0.03 | 0.92 | 2.09 | 2.13 | 2.04 | 2.35 | 1.88 |
| GCG | A | 0.55 | 0.62 | 0.71 | 0.69 | 0.69 | 0.03 | 0.43 | 0.45 | 0.69 | 0.61 | 0.24 | 0.63 |
| TAT | Y | 0.73 | 0.75 | 0.63 | 0.63 | 1.07 | 0.00 | 0.75 | 0.90 | 1.13 | 0.61 | 0.58 | 0.88 |
| TAC | Y | 1.27 | 1.25 | 1.37 | 1.37 | 0.93 | 0.00 | 1.25 | 1.10 | 0.87 | 1.39 | 1.42 | 1.12 |
| CAT | H | 0.68 | 0.94 | 1.17 | 1.31 | 0.79 | 0.44 | 0.67 | 0.00 | 0.60 | 1.24 | 0.00 | 0.87 |
| CAC | H | 1.32 | 1.06 | 0.83 | 0.69 | 1.21 | 1.56 | 1.33 | 0.97 | 1.40 | 0.76 | 0.00 | 1.13 |
| CAA | Q | 0.95 | 1.02 | 1.24 | 1.53 | 0.85 | 1.02 | 0.88 | 1.33 | 1.03 | 0.80 | 1.33 | 1.02 |
| CAG | Q | 1.05 | 0.98 | 0.76 | 0.47 | 1.15 | 0.97 | 1.12 | 0.00 | 0.97 | 1.20 | 0.67 | 0.98 |
| AAT | N | 0.53 | 0.79 | 1.11 | 0.20 | 0.91 | 0.82 | 0.90 | 0.69 | 0.76 | 0.83 | 0.03 | 0.83 |
| AAC | N | 1.47 | 1.21 | 0.89 | 1.76 | 1.09 | 1.18 | 1.10 | 1.31 | 1.24 | 1.17 | 1.69 | 1.17 |
| AAA | K | 0.73 | 0.97 | 0.97 | 0.60 | 1.02 | 1.09 | 0.83 | 1.11 | 0.91 | 0.89 | 1.31 | 0.92 |
| AAG | K | 1.27 | 1.03 | 1.03 | 1.40 | 0.98 | 0.91 | 1.17 | 0.89 | 1.09 | 1.11 | 0.69 | 1.08 |
| GAT | D | 0.55 | 1.10 | 1.23 | 1.14 | 1.02 | 0.83 | 1.08 | 1.02 | 1.19 | 0.92 | 1.21 | 1.05 |
| GAC | D | 1.45 | 0.90 | 0.77 | 0.86 | 0.98 | 1.17 | 0.92 | 0.98 | 0.81 | 1.08 | 0.79 | 0.95 |
| GAA | E | 0.94 | 0.92 | 1.08 | 1.23 | 0.94 | 1.80 | 1.20 | 0.70 | 1.18 | 0.90 | 1.15 | 1.01 |
| GAG | E | 1.06 | 1.08 | 0.92 | 0.77 | 1.06 | 0.20 | 0.80 | 1.30 | 0.82 | 1.10 | 0.85 | 0.99 |
| TGT | C | 0.33 | 1.08 | 1.36 | 1.64 | 1.24 | 1.77 | 1.79 | 1.16 | 1.01 | 0.00 | 1.96 | 1.12 |
| TGC | C | 1.66 | 0.92 | 0.64 | 0.36 | 0.76 | 0.23 | 0.21 | 0.84 | 0.99 | 2.00 | 0.03 | 0.88 |
| CGT | R | 0.59 | 0.53 | 0.41 | 1.08 | 0.53 | 1.07 | 0.85 | 0.56 | 0.36 | 1.19 | 0.34 | 0.63 |
| CGC | R | 0.86 | 1.33 | 1.10 | 0.70 | 0.37 | 0.49 | 0.50 | 0.21 | 1.03 | 0.75 | 1.48 | 0.77 |
| CGA | R | 0.75 | 0.71 | 0.53 | 1.52 | 1.17 | 2.39 | 1.12 | 1.56 | 1.90 | 1.15 | 0.70 | 1.09 |
| CGG | R | 0.21 | 0.32 | 0.10 | 0.62 | 0.44 | 1.21 | 0.13 | 1.14 | 0.72 | 0.43 | 0.12 | 0.41 |
| AGA | R | 2.07 | 1.81 | 2.30 | 1.43 | 2.44 | 0.48 | 1.66 | 1.85 | 1.23 | 1.43 | 3.24 | 1.90 |
| AGG | R | 1.52 | 1.29 | 1.56 | 0.65 | 1.05 | 0.36 | 1.74 | 0.69 | 0.77 | 1.06 | 0.13 | 1.20 |
| GGT | G | 0.66 | 0.95 | 1.11 | 0.11 | 1.07 | 0.74 | 1.03 | 1.25 | 0.95 | 1.31 | 0.48 | 1.01 |
| GGC | G | 0.63 | 1.15 | 1.33 | 0.02 | 0.67 | 0.83 | 0.80 | 0.74 | 0.72 | 1.23 | 3.52 | 0.86 |
| GGA | G | 1.89 | 1.40 | 1.23 | 3.41 | 1.53 | 2.34 | 1.72 | 1.24 | 1.67 | 1.18 | 0.01 | 1.54 |
| GGG | G | 0.82 | 0.50 | 0.32 | 0.46 | 0.73 | 0.08 | 0.45 | 0.77 | 0.66 | 0.27 | 1.75 | 0.59 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qin, L.; Ding, S.; Wang, Z.; Jiang, R.; He, Z. Host Plants Shape the Codon Usage Pattern of Turnip Mosaic Virus. Viruses 2022, 14, 2267. https://doi.org/10.3390/v14102267
Qin L, Ding S, Wang Z, Jiang R, He Z. Host Plants Shape the Codon Usage Pattern of Turnip Mosaic Virus. Viruses. 2022; 14(10):2267. https://doi.org/10.3390/v14102267
Chicago/Turabian StyleQin, Lang, Shiwen Ding, Zhilei Wang, Runzhou Jiang, and Zhen He. 2022. "Host Plants Shape the Codon Usage Pattern of Turnip Mosaic Virus" Viruses 14, no. 10: 2267. https://doi.org/10.3390/v14102267
APA StyleQin, L., Ding, S., Wang, Z., Jiang, R., & He, Z. (2022). Host Plants Shape the Codon Usage Pattern of Turnip Mosaic Virus. Viruses, 14(10), 2267. https://doi.org/10.3390/v14102267







