The Network of Interactions between the Porcine Epidemic Diarrhea Virus Nucleocapsid and Host Cellular Proteins
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Antibodies and Reagents
2.3. Plasmid Construction and Cell Transfection
2.4. SDS-PAGE and Western Blotting
2.5. Co-Immunoprecipitation (Co-IP) Assays
2.6. Liquid Chromatography Mass Spectrometry (LC-MS)
2.7. Construction and Analysis of the Protein-Protein Interactions Network
2.8. GO and KEGG Pathway Analyses
2.9. Confocal Microscopy
3. Results
3.1. Identification of PEDV N Protein-Host Protein Interactions by Liquid Chromatography-Mass Spectrometry
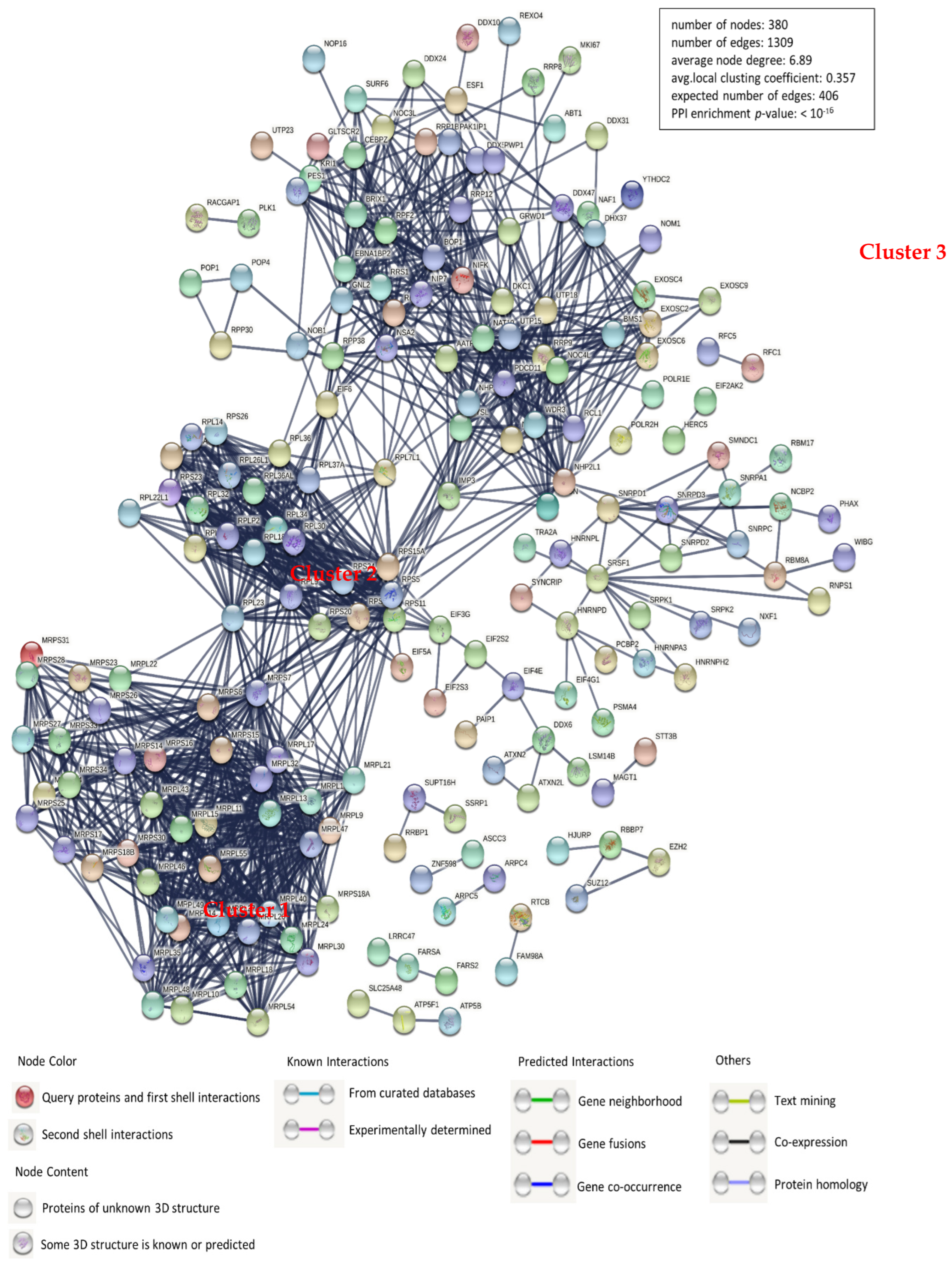
3.2. Construction of a Protein-Protein Interaction Network
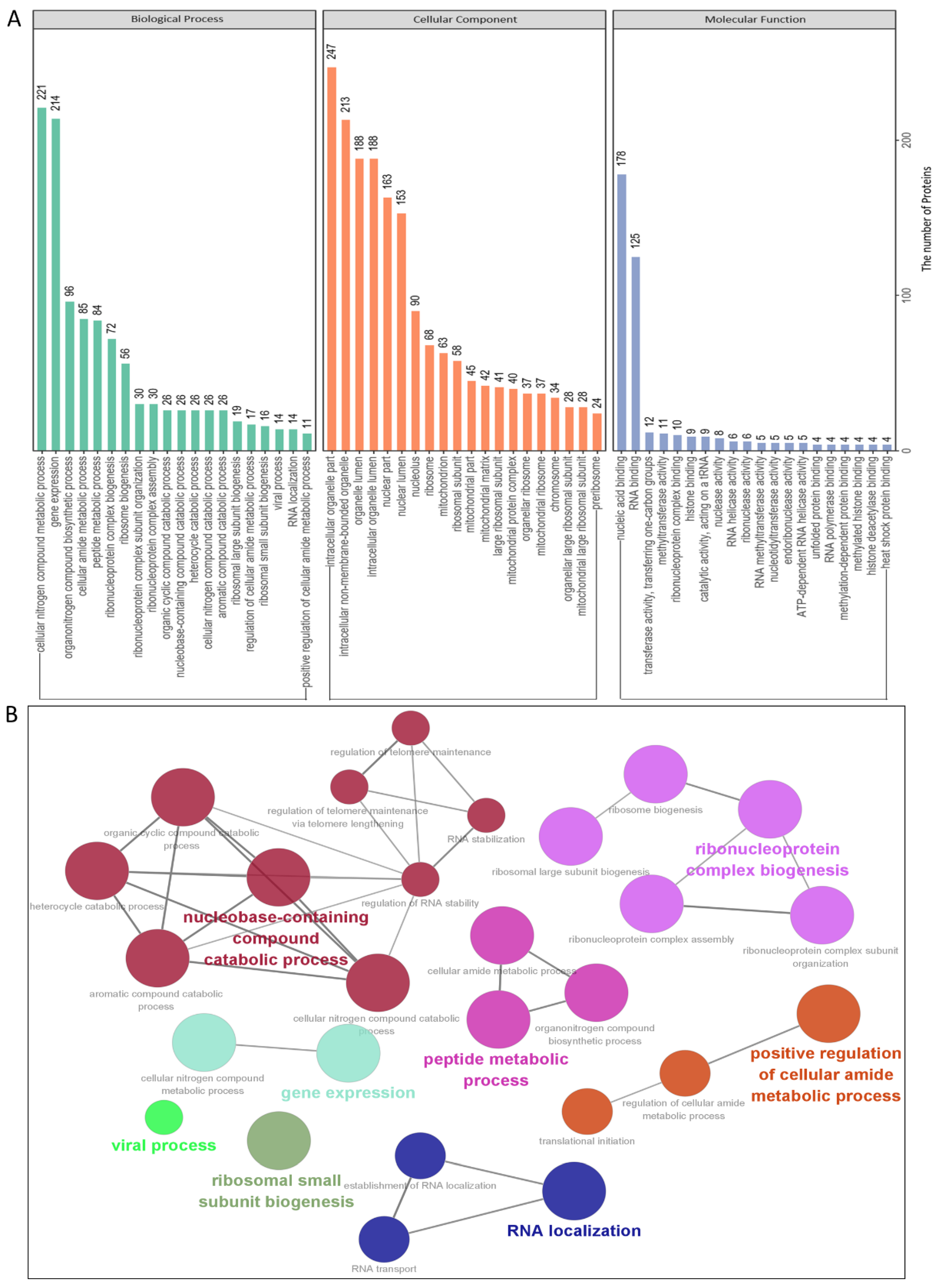
3.3. Gene Ontology Annotation and Analysis
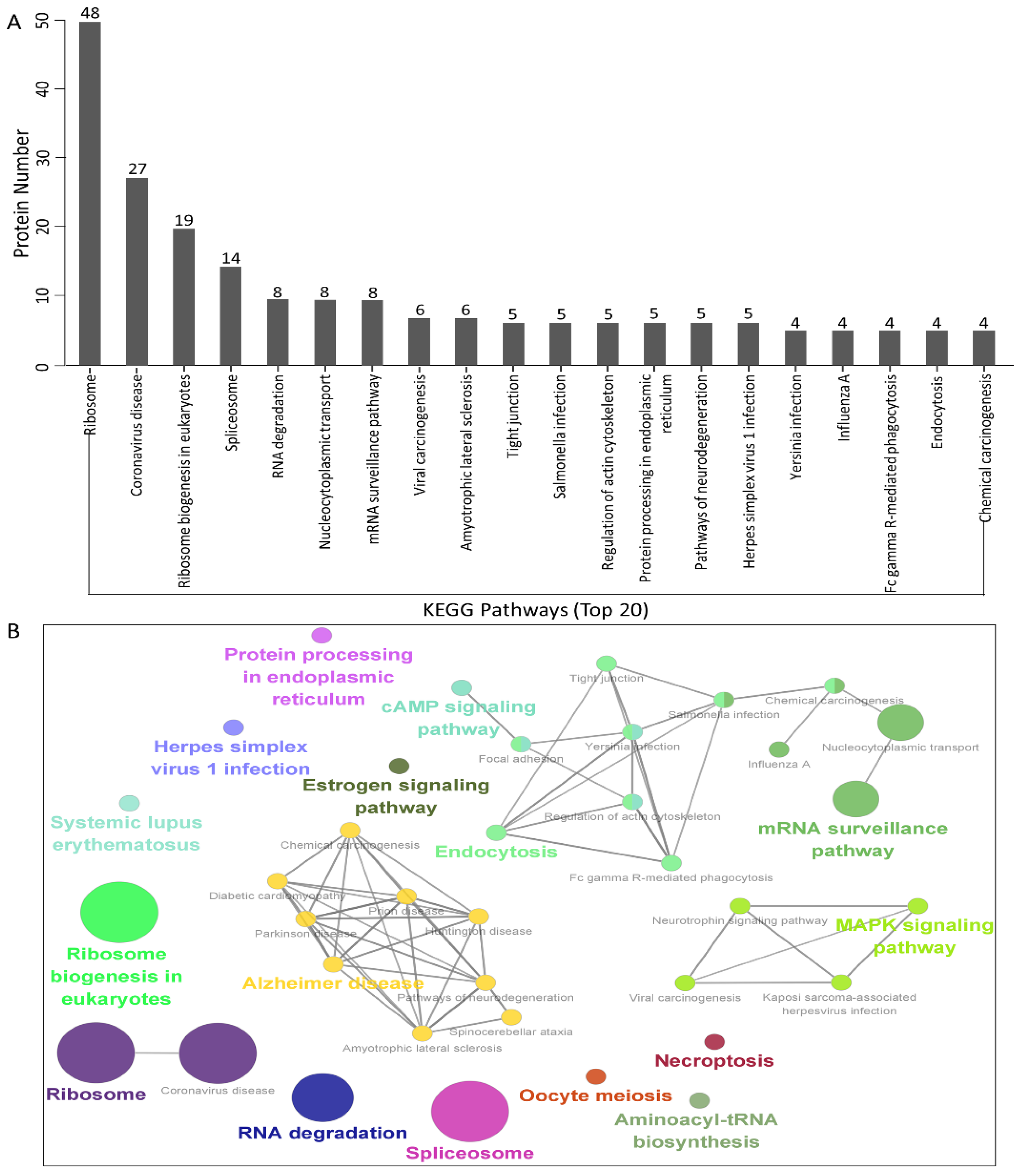
3.4. KEGG Pathway Enrichment Analysis
3.5. Validation of the Interactions between the Host Proteins and the PEDV Nucleocapsid Protein
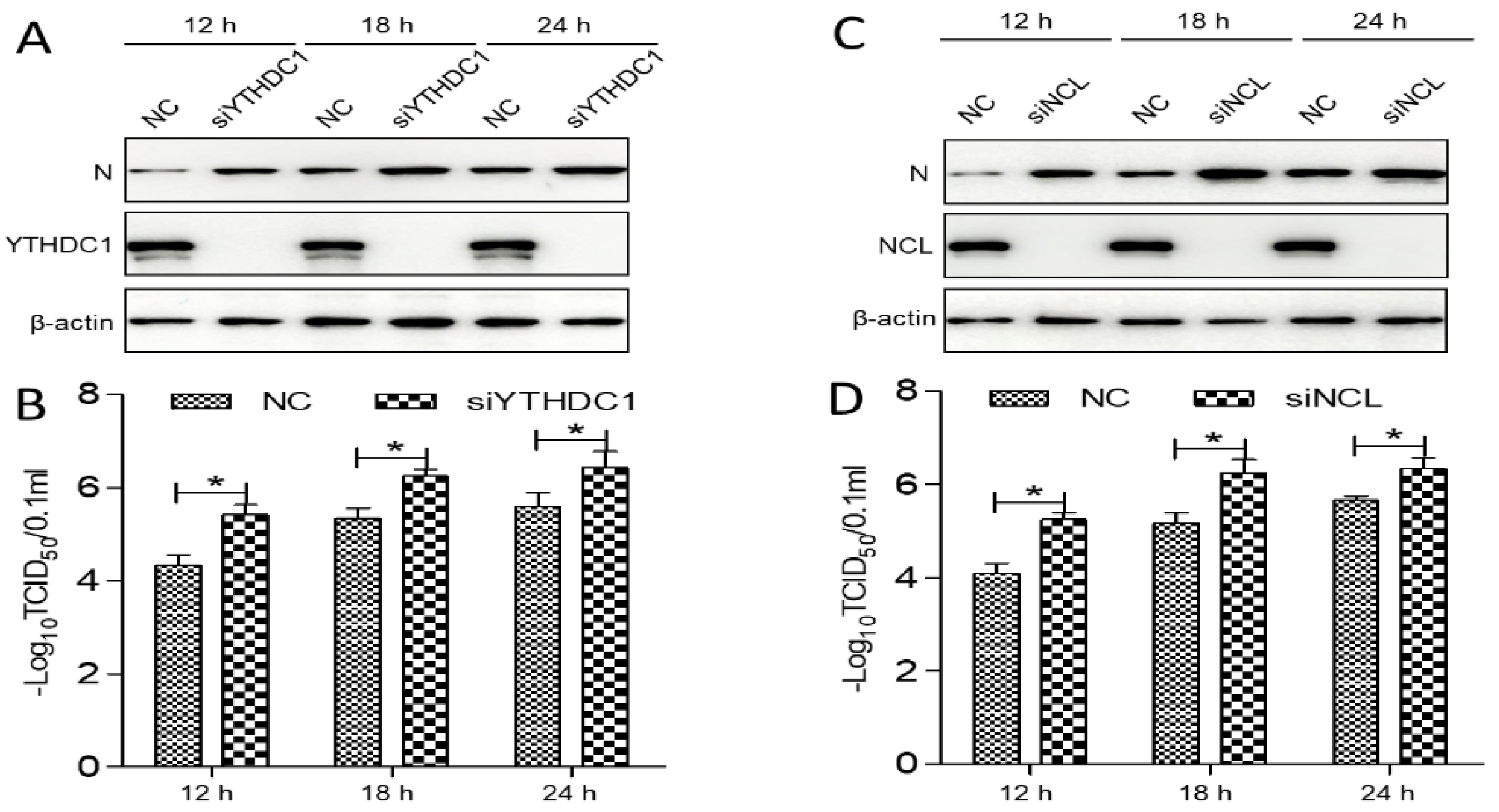
3.6. PEDV Infection Resulted in the Redistribution of YTHDC1 and NCL
3.7. The YTHDC1 and NCL Expression Inhibited PEDV Replication
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brian, D.A.; Baric, R.S. Coronavirus genome structure and replication. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2005; Volume 287, pp. 1–30. [Google Scholar] [CrossRef] [Green Version]
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Li, H.; Liu, Y.; Pan, Y.; Deng, F.; Song, Y.; Tang, X.; He, Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012, 18, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Wang, X.; Wei, S.; Chen, J.; Feng, L. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: A mini-review. J. Vet. Med. Sci. 2016, 78, 355–363. [Google Scholar] [CrossRef] [Green Version]
- Wang, E.; Guo, D.; Li, C.; Wei, S.; Wang, Z.; Liu, Q.; Zhang, B.; Kong, F.; Feng, L.; Sun, D. Molecular Characterization of the ORF3 and S1 Genes of Porcine Epidemic Diarrhea Virus Non S-INDEL Strains in Seven Regions of China, 2015. PLoS ONE 2016, 11, e0160561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef]
- Su, M.; Li, C.; Qi, S.; Yang, D.; Jiang, N.; Yin, B.; Guo, D.; Kong, F.; Yuan, D.; Feng, L.; et al. A molecular epidemiological investigation of PEDV in China: Characterization of co-infection and genetic diversity of S1-based genes. Transbound. Emerg. Dis. 2020, 67, 1129–1140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duarte, M.; Gelfi, J.; Lambert, P.; Rasschaert, D.; Laude, H. Genome organization of porcine epidemic diarrhoea virus. Adv. Exp. Med. Biol. 1993, 342, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Kocherhans, R.; Bridgen, A.; Ackermann, M.; Tobler, K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes 2001, 23, 137–144. [Google Scholar] [CrossRef] [Green Version]
- McBride, R.; van Zyl, M.; Fielding, B.C. The coronavirus nucleocapsid is a multifunctional protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, M.E.; Fehr, A.R.; Athmer, J.; Perlman, S. The coronavirus nucleocapsid protein is ADP-ribosylated. Virology 2018, 517, 62–68. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, H.; Zhang, Q.; Huang, Y.; Dong, J.; Liang, Y.; Liu, H.J.; Tong, D. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet. Microbiol. 2013, 164, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Fang, L.; Jing, H.; Zeng, S.; Wang, D.; Liu, L.; Zhang, H.; Luo, R.; Chen, H.; Xiao, S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014, 88, 8936–8945. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, D.; Shi, H.; Sun, D.; Chen, J.; Zhang, X.; Wang, X.; Zhang, J.; Ji, Z.; Liu, J.; Cao, L.; et al. Nucleocapsid Interacts with NPM1 and Protects it from Proteolytic Cleavage, Enhancing Cell Survival, and is Involved in PEDV Growth. Sci. Rep. 2017, 7, 39700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emmott, E.; Munday, D.; Bickerton, E.; Britton, P.; Rodgers, M.A.; Whitehouse, A.; Zhou, E.M.; Hiscox, J.A. The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J. Virol. 2013, 87, 9486–9500. [Google Scholar] [CrossRef] [Green Version]
- Crua Asensio, N.; Munoz Giner, E.; de Groot, N.S.; Torrent Burgas, M. Centrality in the host-pathogen interactome is associated with pathogen fitness during infection. Nat. Commun. 2017, 8, 14092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fermin, G.; Tennant, P. HostÀVirus Interactions: Battles Between Viruses and Their Hosts. In Viruses: Molecular Biology, Host Interactions, and Applications to Biotechnology; Academic Press: Cambridge, MA, USA, 2018; p. 245. [Google Scholar]
- Su, M.; Shi, D.; Xing, X.; Qi, S.; Yang, D.; Zhang, J.; Han, Y.; Zhu, Q.; Sun, H.; Wang, X.; et al. Coronavirus Porcine Epidemic Diarrhea Virus Nucleocapsid Protein Interacts with p53 To Induce Cell Cycle Arrest in S-Phase and Promotes Viral Replication. J. Virol. 2021, 95, e0018721. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Kong, N.; Jiao, Y.; Dong, S.; Sun, D.; Chen, X.; Zheng, H.; Tong, W.; Yu, H.; Yu, L.; et al. EGR1 Suppresses Porcine Epidemic Diarrhea Virus Replication by Regulating IRAV To Degrade Viral Nucleocapsid Protein. J. Virol. 2021, 95, e0064521. [Google Scholar] [CrossRef]
- Xu, J.; Mao, J.; Han, X.; Shi, F.; Gao, Q.; Wang, T.; Zhang, Z.; Shan, Y.; Fang, W.; Li, X. Porcine Epidemic Diarrhea Virus Inhibits HDAC1 Expression To Facilitate Its Replication via Binding of Its Nucleocapsid Protein to Host Transcription Factor Sp1. J. Virol. 2021, 95, e0085321. [Google Scholar] [CrossRef] [PubMed]
- Kong, N.; Shan, T.; Wang, H.; Jiao, Y.; Zuo, Y.; Li, L.; Tong, W.; Yu, L.; Jiang, Y.; Zhou, Y.; et al. BST2 suppresses porcine epidemic diarrhea virus replication by targeting and degrading virus nucleocapsid protein with selective autophagy. Autophagy 2020, 16, 1737–1752. [Google Scholar] [CrossRef]
- Huan, C.C.; Wang, H.X.; Sheng, X.X.; Wang, R.; Wang, X.; Liao, Y.; Liu, Q.F.; Tong, G.Z.; Ding, C.; Fan, H.J.; et al. Porcine epidemic diarrhea virus nucleoprotein contributes to HMGB1 transcription and release by interacting with C/EBP-beta. Oncotarget 2016, 7, 75064–75080. [Google Scholar] [CrossRef] [Green Version]
- Jiao, Y.; Kong, N.; Wang, H.; Sun, D.; Dong, S.; Chen, X.; Zheng, H.; Tong, W.; Yu, H.; Yu, L.; et al. PABPC4 Broadly Inhibits Coronavirus Replication by Degrading Nucleocapsid Protein through Selective Autophagy. Microbiol. Spectr. 2021, 9, e0090821. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zeng, W.; Ye, S.; Lv, J.; Nie, A.; Zhang, B.; Sun, Y.; Han, H.; He, Q. Cellular hnRNP A1 Interacts with Nucleocapsid Protein of Porcine Epidemic Diarrhea Virus and Impairs Viral Replication. Viruses 2018, 10, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Chi, H.; Fu, Y.; Cao, A.; Shi, J.; Zhu, M.; Zhang, L.; Hua, D.; Huang, J. The antiviral protein viperin interacts with the viral N protein to inhibit proliferation of porcine epidemic diarrhea virus. Arch. Virol. 2020, 165, 2279–2289. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, X.; Kong, N.; Jiao, Y.; Sun, D.; Dong, S.; Qin, W.; Zhai, H.; Yu, L.; Zheng, H.; et al. TRIM21 inhibits porcine epidemic diarrhea virus proliferation by proteasomal degradation of the nucleocapsid protein. Arch. Virol. 2021, 166, 1903–1911. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Li, H.; Yu, T.; Li, J.; Dong, W.; Ojha, N.K.; Jin, Y.; Gu, J.; Zhou, J. Protein Interactions Network of Porcine Circovirus Type 2 Capsid With Host Proteins. Front. Microbiol. 2020, 11, 1129. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.W.; Dai, Y.D.; Lin, C.; Zhang, Y.; Feng, Z.X.; Dong, W.R.; Jin, Y.L.; Yan, Y.; Zhou, J.Y.; Gu, J.Y. Nucleolar protein NPM1 is essential for circovirus replication by binding to viral capsid. Virulence 2020, 11, 1379–1393. [Google Scholar] [CrossRef]
- Zhou, J.; Li, J.; Li, H.; Zhang, Y.; Dong, W.; Jin, Y.; Yan, Y.; Gu, J.; Zhou, J. The serine-48 residue of nucleolar phosphoprotein nucleophosmin-1 plays critical role in subcellular localization and interaction with porcine circovirus type 3 capsid protein. Vet. Res. 2021, 52, 4. [Google Scholar] [CrossRef]
- Zhou, J.; Qiu, Y.; Zhu, N.; Zhou, L.; Dai, B.; Feng, X.; Hou, L.; Liu, J. The Nucleolar Localization Signal of Porcine Circovirus Type 4 Capsid Protein Is Essential for Interaction With Serine-48 Residue of Nucleolar Phosphoprotein Nucleophosmin-1. Front. Microbiol. 2021, 12, 751382. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, Y.; Qiu, Y.; Wang, Y.; Yang, X.; Liu, C.; Shi, Y.; Feng, X.; Hou, L.; Liu, J. Contribution of DEAD-Box RNA Helicase 21 to the Nucleolar Localization of Porcine Circovirus Type 4 Capsid Protein. Front. Microbiol. 2022, 13, 802740. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.R.; Leibowitz, J.L. Coronavirus pathogenesis. Adv. Virus Res. 2011, 81, 85–164. [Google Scholar] [CrossRef]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Stevenson, G.W.; Hoang, H.; Schwartz, K.J.; Burrough, E.R.; Sun, D.; Madson, D.; Cooper, V.L.; Pillatzki, A.; Gauger, P.; Schmitt, B.J.; et al. Emergence of Porcine epidemic diarrhea virus in the United States: Clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Investig. 2013, 25, 649–654. [Google Scholar] [CrossRef] [Green Version]
- Song, D.; Park, B. Porcine epidemic diarrhoea virus: A comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 2012, 44, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Fan, C.; Liu, S.; Zhou, J.; Jin, Y.; Zheng, X.; Wang, Q.; Liu, J.; Yang, H.; Gu, J.; et al. Cellular proteomic analysis of porcine circovirus type 2 and classical swine fever virus coinfection in porcine kidney-15 cells using isobaric tags for relative and absolute quantitation-coupled LC-MS/MS. Electrophoresis 2017, 38, 1276–1291. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.W.; Seo, J.P.; Jung, G. Heat shock protein 70 and heat shock protein 90 synergistically increase hepatitis B viral capsid assembly. Biochem. Biophys. Res. Commun. 2018, 503, 2892–2898. [Google Scholar] [CrossRef] [PubMed]
- Geller, R.; Taguwa, S.; Frydman, J. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim. Biophys. Acta 2012, 1823, 698–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Moniruzzaman, M.; Shuffle, E.; Lourie, R.; Hasnain, S.Z. Immune regulation of the unfolded protein response at the mucosal barrier in viral infection. Clin. Transl. Immunol. 2018, 7, e1014. [Google Scholar] [CrossRef]
- Will, C.L.; Luhrmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.K.; Chen, C.J.; Wu, C.C.; Chen, S.W.; Shih, S.R.; Kuo, R.L. Cellular hnRNP A2/B1 interacts with the NP of influenza A virus and impacts viral replication. PLoS ONE 2017, 12, e0188214. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, J.; Du, Y. hnRNP A2/B1 interacts with influenza A viral protein NS1 and inhibits virus replication potentially through suppressing NS1 RNA/protein levels and NS1 mRNA nuclear export. Virology 2014, 449, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Gordon, H.; Ajamian, L.; Valiente-Echeverria, F.; Levesque, K.; Rigby, W.F.; Mouland, A.J. Depletion of hnRNP A2/B1 overrides the nuclear retention of the HIV-1 genomic RNA. RNA Biol. 2013, 10, 1714–1725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, X.; Wang, L.; Zou, W.; Chen, X.; Roizman, B.; Zhou, G.G. hnRNPA2B1 Associated with Recruitment of RNA into Exosomes Plays a Key Role in Herpes Simplex Virus 1 Release from Infected Cells. J. Virol. 2020, 94, e00367-20. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wen, M.; Cao, X. Nuclear hnRNPA2B1 initiates and amplifies the innate immune response to DNA viruses. Science 2019, 365, eaav0758. [Google Scholar] [CrossRef]
- Casaca, A.; Fardilha, M.; da Cruz e Silva, E.; Cunha, C. The heterogeneous ribonuclear protein C interacts with the hepatitis delta virus small antigen. Virol. J. 2011, 8, 358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dechtawewat, T.; Songprakhon, P.; Limjindaporn, T.; Puttikhunt, C.; Kasinrerk, W.; Saitornuang, S.; Yenchitsomanus, P.T.; Noisakran, S. Role of human heterogeneous nuclear ribonucleoprotein C1/C2 in dengue virus replication. Virol. J. 2015, 12, 14. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Singh, N.; Sengupta, N.; Fatima, M.; Seth, P.; Mahadevan, A.; Shankar, S.K.; Bhattacharyya, A.; Basu, A. Japanese encephalitis virus induces human neural stem/progenitor cell death by elevating GRP78, PHB and hnRNPC through ER stress. Cell Death Dis. 2017, 8, e2556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, L.; Liu, S.; Li, Y.; Yang, G.; Luo, Y.; Li, S.; Du, H.; Zhao, Y.; Wang, D.; Chen, J.; et al. The Nuclear Matrix Protein SAFA Surveils Viral RNA and Facilitates Immunity by Activating Antiviral Enhancers and Super-enhancers. Cell Host Microbe. 2019, 26, 369–384.e8. [Google Scholar] [CrossRef]



| Gene Product | Sense Primer (5′ to 3′) | Antisense Primer (5′ to 3′) |
|---|---|---|
| PEDV N | ATGGCTTCTGTCAGTTTTCAGGAT | TTAATTTCCTGTGTCGAAGATCT |
| TRIM21 | ATGGCTTCAGCAGCACGCTTGACAA | TCAATAGTCAGTGGATCCTTGTGATC |
| DDX24 | ATGAAGTTGAAGGACACAAAATCAAG | TTAATTTGCACTTGTACTTGGCTGTG |
| G3BP1 | ATGGTGATGGAGAAGCCTAGTCCCC | TCACTGCCGTGGCGCAAGCC |
| HSPA8 | ATGTCCAAGGGACCTGCAGTTGGTAT | TTAATCAACCTCTTCAATAGTGGGCC |
| HSP90AB1 | ATGCCTGAGGAAGTGCACCATGGAGA | CTAATCGACTTCTTCCATGCGAGACG |
| YTHDC1 | ATGGCGGCCGACAGTCGGGAGGA | TTATCTTCTATATCGACCTCTCTCC |
| NCL | ATGGTGAAGCTCGCGAAGGCAGGTA | CTATTCAAACTTCGTCTTCTTTCCTT |
| YBX1 | ATGAGCAGCGAGGCCGAGACCCA | TTACTCAGCCCCGCCCTGCTCAGC |
| Vimentin | ATGACCACCAGGTCCGTGTCCTCGT | TTATTCAAGGTCATCGTGATGCTGAG |
| hnRNPA2/B1 | ATGGAGAAAACTTTAGAAACTGTTCC | TCAGTATCGGCTCCTCCCACCATAA |
| KPNA1 | ATGACCACCCCAGGAAAAGAGAACTT | TCAAAGCTGGAAACCTTCCATAGGAGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Qiu, Y.; Zhao, J.; Wang, Y.; Zhu, N.; Wang, D.; Cui, Y.; Guo, J.; Sun, T.; Ji, Y.; et al. The Network of Interactions between the Porcine Epidemic Diarrhea Virus Nucleocapsid and Host Cellular Proteins. Viruses 2022, 14, 2269. https://doi.org/10.3390/v14102269
Zhou J, Qiu Y, Zhao J, Wang Y, Zhu N, Wang D, Cui Y, Guo J, Sun T, Ji Y, et al. The Network of Interactions between the Porcine Epidemic Diarrhea Virus Nucleocapsid and Host Cellular Proteins. Viruses. 2022; 14(10):2269. https://doi.org/10.3390/v14102269
Chicago/Turabian StyleZhou, Jianwei, Yonghui Qiu, Jie Zhao, Yongxia Wang, Ning Zhu, Dedong Wang, Yongqiu Cui, Jinshuo Guo, Tong Sun, Ying Ji, and et al. 2022. "The Network of Interactions between the Porcine Epidemic Diarrhea Virus Nucleocapsid and Host Cellular Proteins" Viruses 14, no. 10: 2269. https://doi.org/10.3390/v14102269






