Comprehensive Analysis of Ubiquitome Changes in Nicotiana benthamiana after Rice Stripe Virus Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials, Agroinfiltration, and Virus Inoculation
2.2. Plasmid Construction
2.3. Western Blotting and Antibodies
2.4. Protein Extraction and Trypsin Digestion
2.5. Affinity Enrichment of Ubiquitinated Peptides
2.6. Proteome and Ubiquitome Analysis by LC–MS/MS
2.7. Database Search
2.8. Bioinformatics Analysis
2.9. Statistical Analysis
2.10. Real-Time Quantitative PCR
2.11. Immunoprecipitation Assay
3. Results
3.1. General Features of the Quantitative Proteome and Ubiquitome Datasets
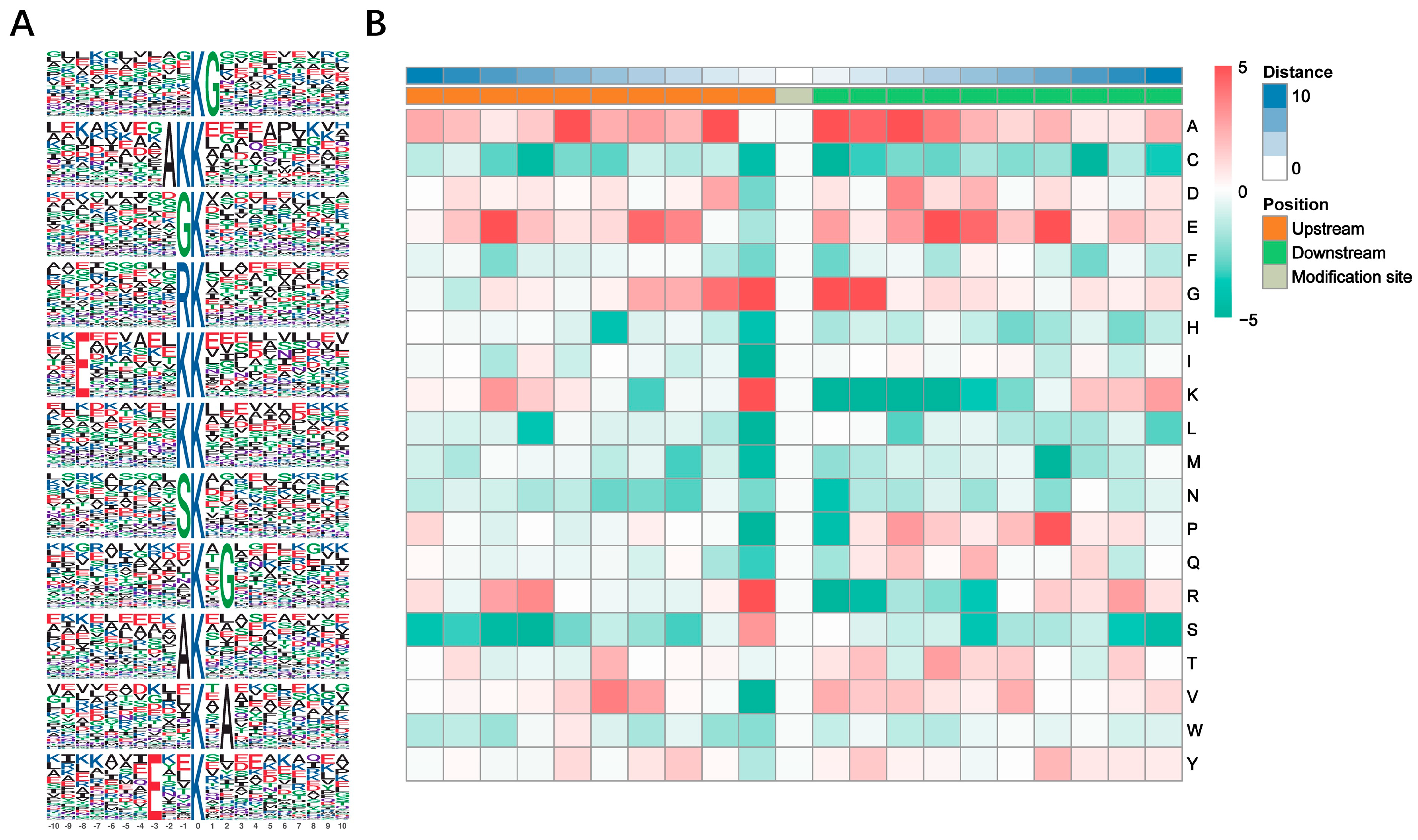
3.2. Motif Analysis of Kub Sites
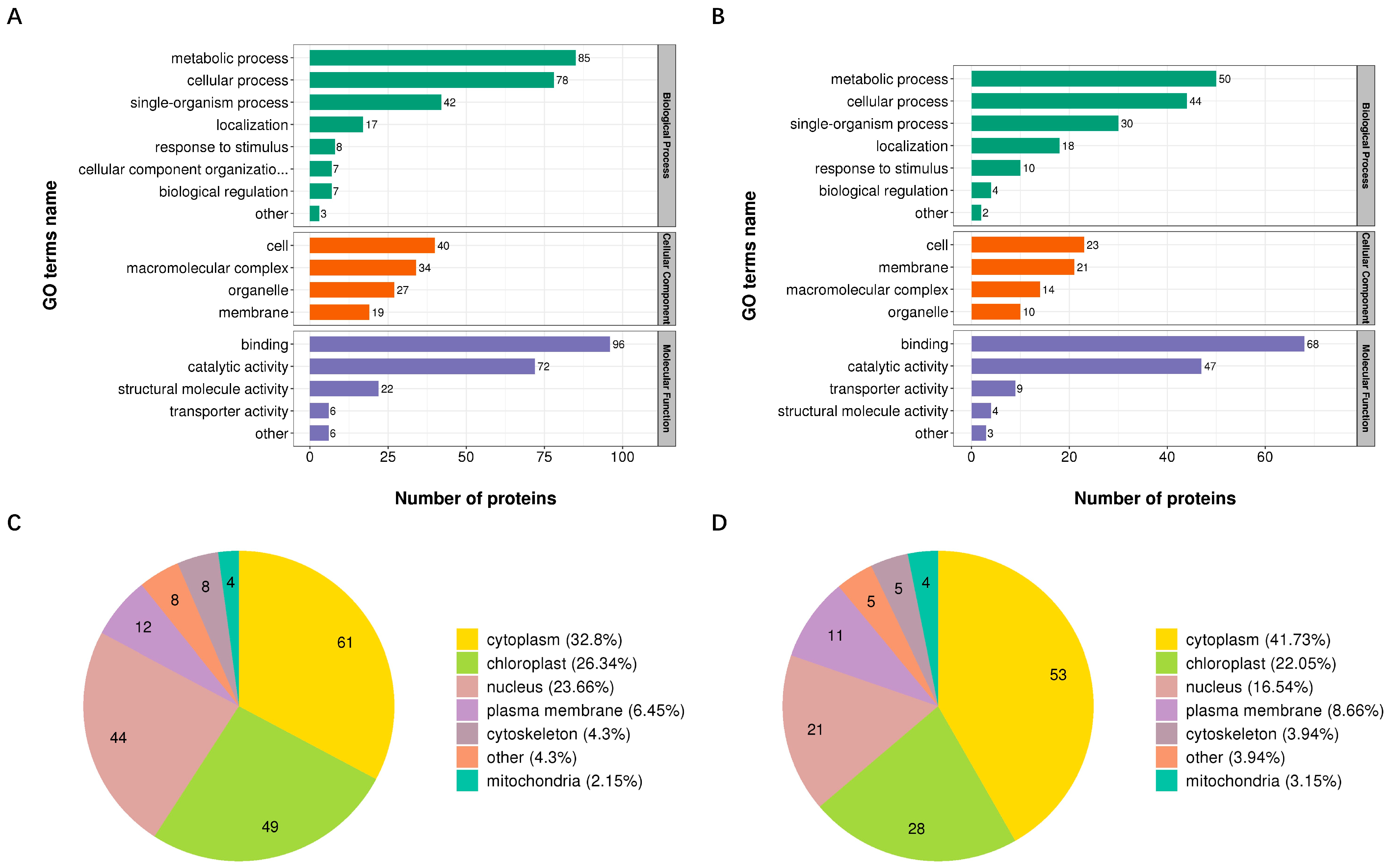
3.3. Functional Classification of Proteins with DUSs
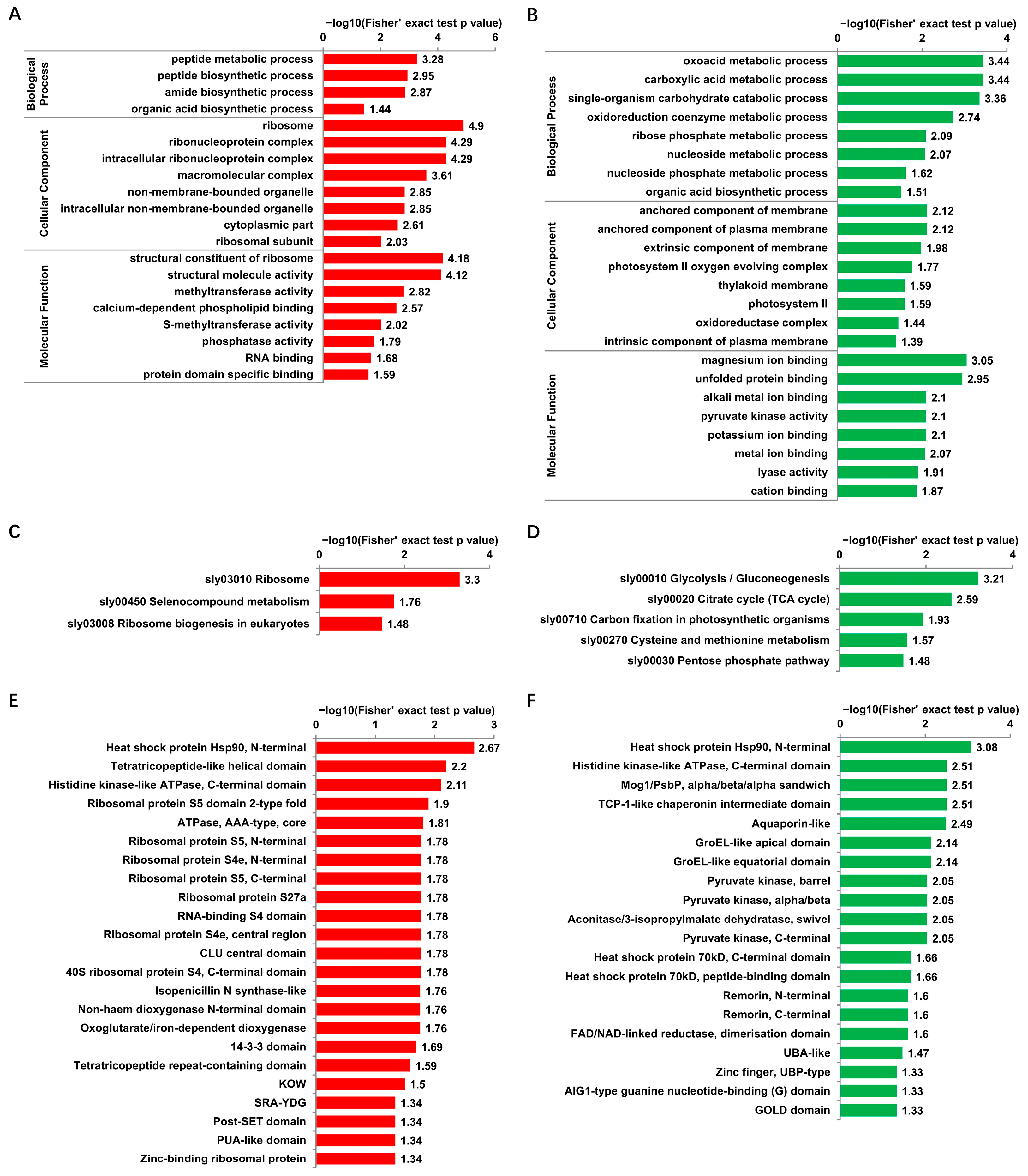
3.4. Functional Enrichment of Proteins with DUSs
3.5. Influence of Host Proteins with DUSs on RSV Infections
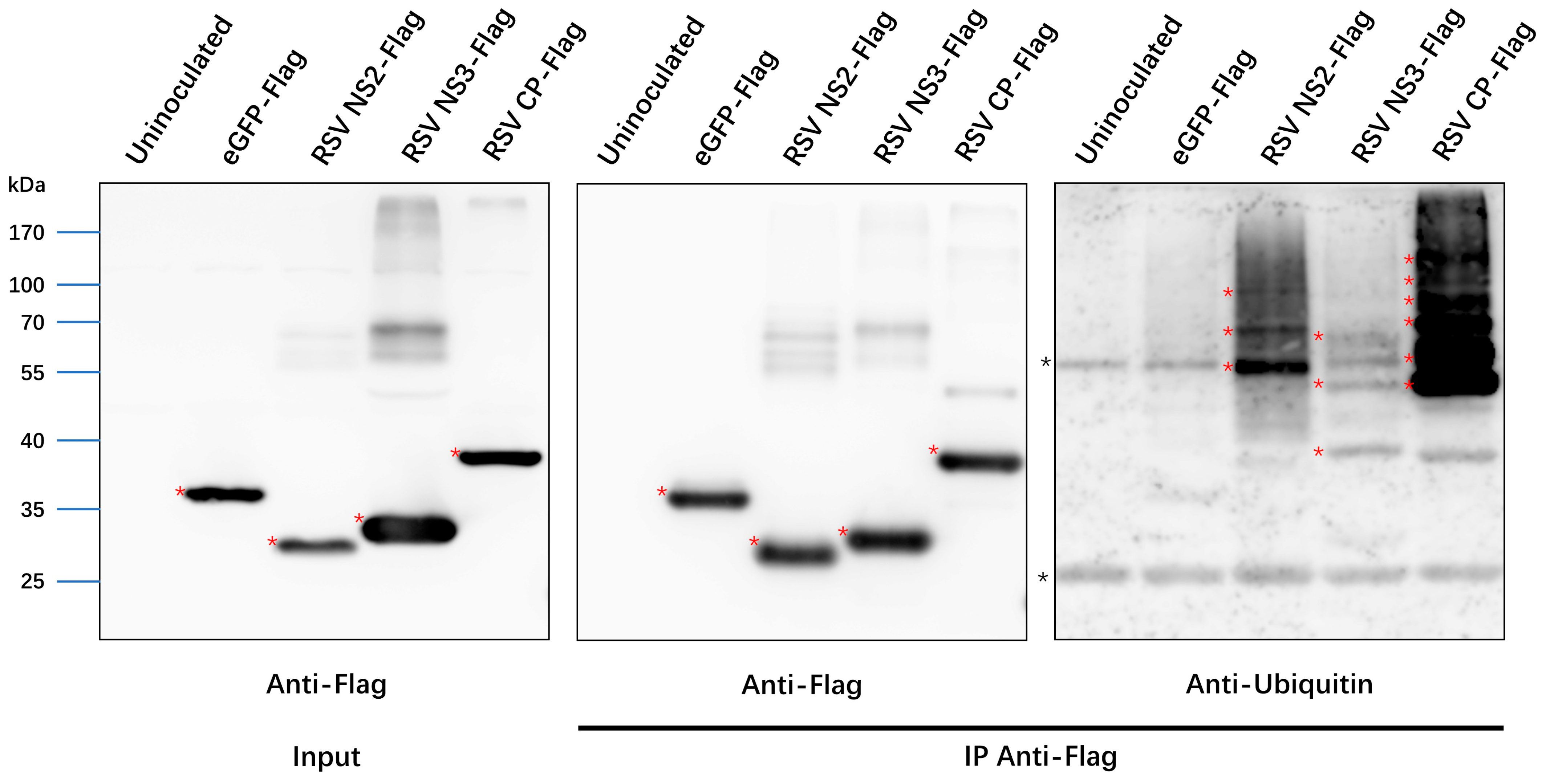
3.6. Ubiquitination of RSV Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu, Y.; Fu, S.; Tao, X.R.; Zhou, X.P. Rice stripe virus: Exploring Molecular Weapons in the Arsenal of a Negative-Sense RNA Virus. Annu. Rev. Phytopathol. 2021, 59, 351–371. [Google Scholar] [CrossRef] [PubMed]
- Hibino, H. Biology and epidemiology of rice viruses. Annu. Rev. Phytopathol. 1996, 34, 249–274. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Huang, L.Z.; Fu, S.; Wu, J.X.; Zhou, X.P. Population Diversity of Rice Stripe Virus-Derived siRNAs in Three Different Hosts and RNAi-Based Antiviral Immunity in Laodelphgax striatellus. PLoS ONE 2012, 7, e46238. [Google Scholar] [CrossRef]
- Komander, D. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 2009, 37, 937–953. [Google Scholar] [CrossRef] [Green Version]
- Varshavsky, A. The Ubiquitin System, Autophagy, and Regulated Protein Degradation. Annu. Rev. Biochem. 2017, 86, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J.J.; Sun, L.J.J. Nonproteolytic Functions of Ubiquitin in Cell Signaling. Mol. Cell 2009, 33, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Grabbe, C.; Husnjak, K.; Dikic, I. The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 2011, 12, 295–307. [Google Scholar] [CrossRef] [Green Version]
- Shen, Q.T.; Hu, T.; Bao, M.; Cao, L.G.; Zhang, H.W.; Song, F.M.; Xie, Q.; Zhou, X.P. Tobacco RING E3 Ligase NtRFP1 Mediates Ubiquitination and Proteasomal Degradation of a Geminivirus-Encoded beta C1. Mol. Plant 2016, 9, 911–925. [Google Scholar] [CrossRef] [Green Version]
- Derrien, B.; Baumberger, N.; Schepetilnikov, M.; Viotti, C.; De Cillia, J.; Ziegler-Graff, V.; Isono, E.; Schumacher, K.; Genschik, P. Degradation of the antiviral component ARGONAUTE1 by the autophagy pathway. Proc. Natl. Acad. Sci. USA 2012, 109, 15942–15946. [Google Scholar] [CrossRef] [Green Version]
- Chiu, M.H.; Chen, I.H.; Baulcombe, D.C.; Tsai, C.H. The silencing suppressor P25 of Potato virus X interacts with Argonaute1 and mediates its degradation through the proteasome pathway. Mol. Plant Pathol. 2010, 11, 641–649. [Google Scholar] [CrossRef]
- Xu, G.Q.; Paige, J.S.; Jaffrey, S.R. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 2010, 28, 868–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, X.; Kang, H.X.; Liu, W.D.; Wang, G.L. Comprehensive Profiling of the Rice Ubiquitome Reveals the Significance of Lysine Ubiquitination in Young Leaves. J. Proteome Res. 2015, 14, 2017–2025. [Google Scholar] [CrossRef] [PubMed]
- Zhan, H.X.; Song, L.Y.; Kamran, A.; Han, F.; Li, B.; Zhou, Z.C.; Liu, T.B.; Shen, L.L.; Li, Y.; Wang, F.L.; et al. Comprehensive Proteomic Analysis of Lysine Ubiquitination in Seedling Leaves of Nicotiana tabacum. ACS Omega 2020, 5, 20122–20133. [Google Scholar] [CrossRef]
- He, D.L.; Li, M.; Damaris, R.N.; Bu, C.; Xue, J.Y.; Yang, P.F. Quantitative ubiquitylomics approach for characterizing the dynamic change and extensive modulation of ubiquitylation in rice seed germination. Plant J. 2020, 101, 1430–1447. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.D.; Hou, Y.; Shen, J.Y.; Mehra, R.; Kallianpur, A.; Culver, D.A.; Gack, M.U.; Farha, S.; Zein, J.; Comhair, S.; et al. A network medicine approach to investigation and population-based validation of disease manifestations and drug repurposing for COVID-19. PLoS Biol. 2020, 18, e3000970. [Google Scholar] [CrossRef]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.Q.; Oubraham, L.; Wang, A.Q.; et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef]
- Kong, L.F.; Wu, J.X.; Lu, L.N.; Xu, Y.; Zhou, X.P. Interaction between Rice stripe virus Disease-Specific Protein and Host PsbP Enhances Virus Symptoms. Mol. Plant 2014, 7, 691–708. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhou, X.P. Role of rice stripe virus NSvc4 in cell-to-cell movement and symptom development in Nicotiana benthamiana. Front. Plant Sci. 2012, 3, 269. [Google Scholar] [CrossRef] [Green Version]
- Xiong, R.Y.; Wu, J.X.; Zhou, Y.J.; Zhou, X.P. Characterization and subcellular localization of an RNA silencing suppressor encoded by Rice stripe tenuivirus. Virology 2009, 387, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Li, C.Y.; Xu, Y.; Fu, S.; Liu, Y.; Li, Z.D.; Zhang, T.Z.; Wu, J.X.; Zhou, X.P. The unfolded protein response plays dual roles in rice stripe virus infection through fine-tuning the movement protein accumulation. PLoS Pathog. 2021, 17, e1009370. [Google Scholar] [CrossRef]
- Ping, L.; Chao, C.; Ping, L.; Yibo, D. A comprehensive examination of the lysine acetylation targets in paper mulberry based on proteomics analyses. PLoS ONE 2021, 16, e0240947. [Google Scholar]
- Yu, Y.; Wang, H.Y.; Rao, X.C.; Liu, L.X.; Zheng, P.; Li, W.X.; Zhou, W.; Chai, T.J.; Ji, P.; Song, J.L.; et al. Proteomic Profiling of Lysine Acetylation Indicates Mitochondrial Dysfunction in the Hippocampus of Gut Microbiota-Absent Mice. Front. Mol. Neurosci. 2021, 14, 594332. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.C.; Su, J.Y.; Wang, N.N.; Zhao, W.; Zhu, M.Y.; Su, S. Comprehensive analysis of the ubiquitome in rabies virus-infected brain tissue of Mus musculus. Vet. Microbiol. 2020, 241, 108552. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Wang, Y.; Ding, Y.Q.; Qiu, C.; Sun, L.T.; Gai, Z.S.; Gu, H.L.; Ding, Z.T. Global Ubiquitome Profiling Revealed the Roles of Ubiquitinated Proteins in Metabolic Pathways of Tea Leaves in Responding to Drought Stress. Sci. Rep. 2019, 9, 4286. [Google Scholar] [CrossRef] [Green Version]
- Ma, T.T.; Fu, S.; Wang, K.; Wang, Y.Q.; Wu, J.X.; Zhou, X.P. Palmitoylation Is Indispensable for Remorin to Restrict Tobacco Mosaic Virus Cell-to-Cell Movement in Nicotiana benthamiana. Viruses 2022, 14, 1324. [Google Scholar] [CrossRef]
- Van Aalderen, M.C.; van den Biggelaar, M.; Remmerswaal, E.B.M.; van Alphen, F.P.J.; Meijer, A.B.; ten Berge, I.J.M.; van Lier, R.A.W. Label-free Analysis of CD8+ T Cell Subset Proteomes Supports a Progressive Differentiation Model of Human-Virus-Specific T Cells. Cell Rep. 2017, 19, 1068–1079. [Google Scholar] [CrossRef] [Green Version]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, Z.; Shen, M.; Ge, L.Q.; Liu, F. Ubiquitin-Conjugating Enzyme E2 E Inhibits the Accumulation of Rice Stripe Virus in Laodelphax striatellus (Fallen). Viruses 2020, 12, 908. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, L.F.; Gao, J.Q.; Ma, H.R.; Li, C.Y.; Song, Y.Z.; Zhu, X.P.; Zhu, C.X. Silencing suppressors of rice black-streaked dwarf virus and rice stripe virus hijack the 26S proteasome of Laodelphax striatellus to facilitate virus accumulation and transmission. Pest Manag. Sci. 2022, 78, 2940–2951. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, J.X.; Fu, S.; Li, C.Y.; Zhu, Z.R.; Zhou, X.P. Rice Stripe Tenuivirus Nonstructural Protein 3 Hijacks the 26S Proteasome of the Small Brown Planthopper via Direct Interaction with Regulatory Particle Non-ATPase Subunit 3. J. Virol. 2015, 89, 4296–4310. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.L.; Gu, X.M.; Li, J.; Liang, C.Y. The N-terminal cysteine protease domain of rice stripe tenuivirus Pc1 possesses deubiquitinating enzyme activity. Virus Genes 2021, 57, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.H.; Lin, L.; Lu, Y.W.; Peng, J.J.; Zheng, H.Y.; Yang, Q.K.; Rao, S.F.; Wu, G.W.; Li, J.M.; Chen, Z.; et al. Ubiquitin-Like protein 5 interacts with the silencing suppressor p3 of rice stripe virus and mediates its degradation through the 26S proteasome pathway. PLoS Pathog. 2020, 16, e1008780. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.; Wang, K.; Ma, T.T.; Liang, Y.; Ma, Z.H.; Wu, J.X.; Xu, Y.; Zhou, X.P. An evolutionarily conserved C4HC3-type E3 ligase regulates plant broad-spectrum resistance against pathogens. Plant Cell 2022, 34, 1822–1843. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.M.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.M.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef]
- Chen, X.J.; Li, P.F.; Zhang, G.W.; Kang, L.H.; Qin, B.; Mao, X.M.; Qin, M.M.; Cao, Y.; Wang, Y.; Guan, H.J. Comprehensive Profiling of Proteome and Ubiquitome Changes in Human Lens Epithelial Cell Line after Ultraviolet-B Irradiation. ACS Omega 2020, 5, 32171–32182. [Google Scholar] [CrossRef] [PubMed]
- Vere, G.; Kealy, R.; Kessler, B.M.; Pinto-Fernandez, A. Ubiquitomics: An Overview and Future. Biomolecules 2020, 10, 1453. [Google Scholar] [CrossRef]
- Theurillat, J.P.P.; Udeshi, N.D.; Errington, W.J.; Svinkina, T.; Baca, S.C.; Pop, M.; Wild, P.J.; Blattner, M.; Groner, A.C.; Rubin, M.A.; et al. Ubiquitylome analysis identifies dysregulation of effector substrates in SPOP-mutant prostate cancer. Science 2014, 346, 85–89. [Google Scholar] [CrossRef] [Green Version]
- Sureshkumar, S.; Todesco, M.; Schneeberger, K.; Harilal, R.; Balasubramanian, S.; Weigel, D. A Genetic Defect Caused by a Triplet Repeat Expansion in Arabidopsis thaliana. Science 2009, 323, 1060–1063. [Google Scholar] [CrossRef]
- Decker, M.; Adamska, M.; Cronin, A.; Di Giallonardo, F.; Burgener, J.; Marowsky, A.; Falck, J.R.; Morisseau, C.; Hammock, B.D.; Gruzdev, A.; et al. EH3 (ABHD9): The first member of a new epoxide hydrolase family with high activity for fatty acid epoxides. J. Lipid Res. 2012, 53, 2038–2045. [Google Scholar] [CrossRef] [Green Version]
- Guo, A.; Durner, J.; Klessig, D.F. Characterization of a tobacco epoxide hydrolase gene induced during the resistance response to TMV. Plant J. 1998, 15, 647–656. [Google Scholar] [CrossRef]
- Iwasaki, S.; Takeda, A.; Motose, H.; Watanabe, Y. Characterization of Arabidopsis decapping proteins AtDCPI and AtDCP2, which are essential for post-embryonic development. FEBS Lett. 2007, 581, 2455–2459. [Google Scholar] [CrossRef] [PubMed]
- Li, F.F.; Wang, A.M. RNA decay is an antiviral defense in plants that is counteracted by viral RNA silencing suppressors. PLoS Pathog. 2018, 14, e1007228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.J.; Jin, J.; Qiu, P.; Gao, F.L.; Lin, W.Z.; Xie, G.H.; He, S.M.; Liu, S.M.; Du, Z.G.; Wu, Z.J. Rice Stripe Tenuivirus Has a Greater Tendency to Use the Prime-and-Realign Mechanism in Transcription of Genomic than in Transcription of Antigenomic Template RNAs. J. Virol. 2018, 92, e0141417. [Google Scholar] [CrossRef] [PubMed]


| Identified | Quantified | ||
|---|---|---|---|
| Proteome | Proteins | 5519 | 3549 |
| Ubiquitome | Sites | 2530 | 1345 |
| Proteins | 1469 | 862 |
| Regulated Type | Fold Change > 1.5 | |
|---|---|---|
| Proteome RSV/Mock | Up-regulated | 323 proteins |
| Down-regulated | 276 proteins | |
| Normalized ubiquitome RSV/Mock | Up-regulated | 244 sites |
| Down-regulated | 155 sites |
| Protein | Position | Ubiquitome RSV/Mock Ratio | Ubiquitome RSV/Mock p Value | Proteome RSV/Mock Ratio | Proteome RSV/Mock p Value | Normalized Ubiquitome RSV/Mock Ratio | Normalized Ubiquitome RSV/Mock p Value |
|---|---|---|---|---|---|---|---|
| LeuC | K701 | 0.096 | 1.82 × 10−5 | 1.51 | 5.89 × 10−5 | 0.086 | 6.09 × 10−6 |
| EPHX3 | K222 | 0.637 | 1.08 × 10−1 | 3.288 | 7.81 × 10−3 | 0.129 | 7.03 × 10−4 |
| DCP1 | K138 | 1.998 | 1.41 × 10−4 | 0.512 | 1.25 × 10−2 | 3.896 | 6.3 × 10−5 |
| RSV Proteins | Sequence Coverage | Identified Kub Sites |
|---|---|---|
| RdRp | 2.5% | 1270 |
| NS2 | 67.8% | 16, 18, 87 |
| NSvc2 | 24.7% | 42, 243, 300, 351, 353, 575 |
| NS3 | 89.1% | 87, 112, 127, 138, 157, 177, 184, 193 |
| CP | 78.3% | 5, 21, 88, 90, 214, 231, 246, 256, 289, 292, 300 |
| SP | 52.8% | 51, 52, 136, 144, 147, 158, 161, 162, 177 |
| MP | 37.8% | 13, 25, 33, 39, 243, 252, 279 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Li, C.; Wang, Y.; Xu, Y.; Wu, J.; Zhou, X. Comprehensive Analysis of Ubiquitome Changes in Nicotiana benthamiana after Rice Stripe Virus Infection. Viruses 2022, 14, 2349. https://doi.org/10.3390/v14112349
Liu Y, Li C, Wang Y, Xu Y, Wu J, Zhou X. Comprehensive Analysis of Ubiquitome Changes in Nicotiana benthamiana after Rice Stripe Virus Infection. Viruses. 2022; 14(11):2349. https://doi.org/10.3390/v14112349
Chicago/Turabian StyleLiu, Yu, Chenyang Li, Yaqin Wang, Yi Xu, Jianxiang Wu, and Xueping Zhou. 2022. "Comprehensive Analysis of Ubiquitome Changes in Nicotiana benthamiana after Rice Stripe Virus Infection" Viruses 14, no. 11: 2349. https://doi.org/10.3390/v14112349
APA StyleLiu, Y., Li, C., Wang, Y., Xu, Y., Wu, J., & Zhou, X. (2022). Comprehensive Analysis of Ubiquitome Changes in Nicotiana benthamiana after Rice Stripe Virus Infection. Viruses, 14(11), 2349. https://doi.org/10.3390/v14112349






