1. Introduction and Background
In this study, we concern ourselves with the week-by-week chronology of evolution of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) genomes as an illustration of emergence of variants of concern (VOC) of the virus and other elements of virus evolution. For this purpose, we downloaded almost 5 million genomic sequences from the GISAID database, collected from week 1 until week 97 of the pandemic. Using the original Wuhan consensus genome as a reference, we aligned all the sequences and split these into subsets, each including the sequences registered in a 1-week-long window. In each of the 97 time points, we created a list of variant sites at which the genomes differed from the Wuhan genome sequence, be it nucleotide substitutions, deleted nucleotides or non-sequenceable sites or site runs.
We categorized the genomes into disjoint subsets: non-variant of concern (non-VOC) mostly present in the early period of the pandemic, the Alpha (“British”) VOC, the Beta (“South African”) VOC and the Delta (“Indian”) VOC. In our data series, we observe the early stages of the Omicron VOC but not the latter’s divergence into substrains.
We decided not to include Omicron variant data in our analysis. One reason is the staggering count of genomes and the very rich diversification of Omicron variants. Therefore, we focused on the relatively simple “traveling wave” pattern of the pre-Omicron period. Simulations in the end section of Results qualitatively reproduce the pattern of the pre-Omicron era but would not be helpful to understand the Omicron data.
In our considerations, the benchmark is the hypothesis of the strongly asexual evolution of the virus, which implies that all VOC are clonal and share the same ancestral sequence. Recombination or repeated instances of variant emergence may contradict this hypothesis in its simple form. Recombination may occur, for example, if a mixture of more than 1 strain infects a host cell where they may trade portions of their genomes.
The SARS-CoV-2, which caused the current COVID-19 pandemic is a single-stranded RNA virus and it is expected to mutate at a pace of
nucleotide substitutions per site per year [
1,
2]. Although most of these mutations are either deleterious or neutral, some of them may impact the transmissibility and infectivity of the emerging strain. In addition, the accumulation of mutations may lead to immune escape, leading to an increased likelihood of reinfection. These features are observed in some of the VOC [
3,
4]. We turn now to some background information. It has to be noted that many recent papers discuss adaptation and purifying selection to variants evolution. A helpful introduction and recent literature review are provided by Neher [
5], and we return to the subject in the Discussion.
1.1. B.1.1.7 (Alpha) Variant
The B.1.1.7 variant, later recognized as a variant of concern, was first detected in November 2020 in a sample taken on 20 September 2020 in the United Kingdom. On 14 December 2020, Public Health authorities in England reported a new SARS-CoV-2 variant referred to as Variant Under Investigation (later recognized as VOC) [
6]. The B.1.1.7 variant is characterized by 15 non-synonymous mutations and 3 deletions [
7,
8] (
Table A1). Several amino acid mutations are observed in the S protein of the Alpha variant, including D614G, N501Y and deletions H69-V70. It was reported that the S-receptor-binding domain (RBD) N501Y mutation increases the binding affinity to the ACE-2 receptor, facilitating transmission [
9]. With transmissibility increased by 43–90% [
10,
11], and about a twofold replicative advantage [
12], the Alpha variant began to spread, quickly outnumbering the original Wuhan strain.
1.2. B.1.351 (Beta) Variant
Another of several SARS-CoV-2 variants believed to be of particular importance was announced for the first time on 18 December 2020 by South Africa’s health department. The first sample was detected in the Nelson Mandela Bay metropolitan area of the Eastern Cape province of South Africa in October 2020. The B.1.351 variant is characterized by 17 mutations, with 9 of them in the Spike protein coding region [
13] (
Table A2), including three critical mutations in the RBD (K417N, E484K and N501Y) that impact viral fitness, transmissibility and survival adaptations [
9].
1.3. B.1.617.2 (Delta) Variant
The B.1.617.2 variant appeared in Maharashtra state in India in October 2020 [
14,
15] and quickly became dominant in most countries. This variant is characterized by rapid transmission and spread, which is indicative of selective advantages against other VOC such as B.1.1.7 or B.1.351. Studies suggest a high risk of hospitalization compared with the Wuhan strain or the B.1.1.7 variant [
16,
17] and higher potential of immune evasion [
15,
18,
19]. The B.1.617.2 variant is characterized by 2 deletions and 18 mutations, with 9 of them in the Spike protein coding region [
19] (
Table A3). Some of the most important Delta variant mutations are the P681R mutation present in the Spike insertion region, which distinguishes SARS-CoV-2 from, among others, bat coronaviruses [
20] and T478K. Spike mutation, which has impact on infectivity and pathogenesis, facilitates viral replication and is potentially responsible for antibody escape [
19,
21].
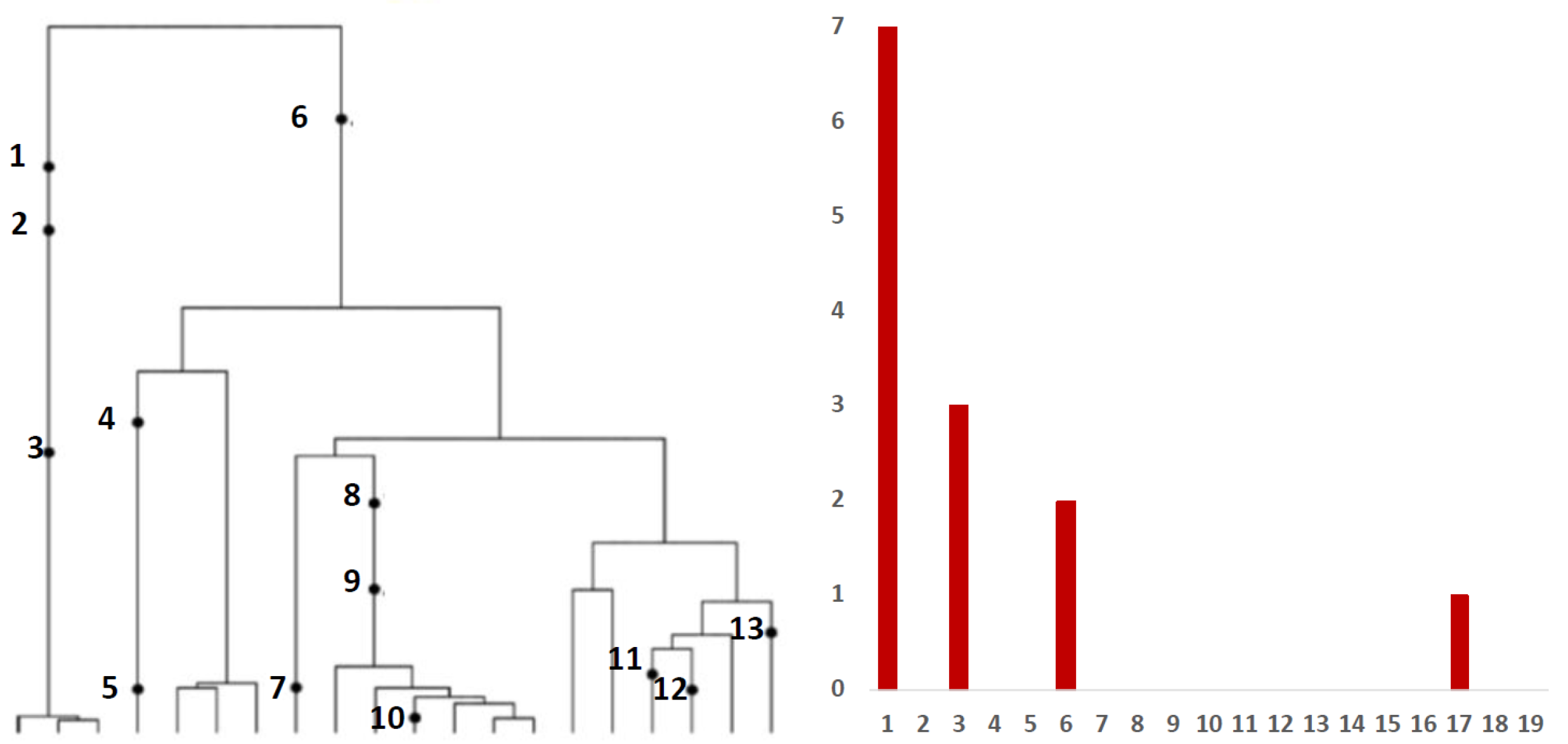
We are exploring the history of each of the segregating sites present in Alpha, Beta and Delta VOC. We are trying to answer the question of whether defining mutations were accumulating gradually until they formed a sequence characteristic of the Alpha, Beta and Delta variants, or whether this phenomena can be explained by the recombination of two genomes with subsets of mutations.
We then use the longitudinal data of evolution of mutation frequencies to classify the genetic forces active at different ranges of the mutational spectrum. We investigate neutrality of the mutations at the lowest frequencies with the Griffiths–Tavaré theory [
22]. At the mid-frequency range, we look for negative selection using the Tung–Durrett model [
23] assuming clone competition. These results add up to a model of genomic evolution. Certain observations, such as mutation “bands” persistent over the epidemic history, suggest contribution of genetic forces different from mutation, drift and selection, including recombination or other genome transformations. In addition, we investigate a“toy” mathematical model based on the Tug-of-War concept [
24] to verify if it may qualitatively reproduce how new variants (clones) stem from rare advantageous driver mutations, and then acquire neutral or disadvantageous passenger mutations which gradually reduce their fitness.
4. Discussion
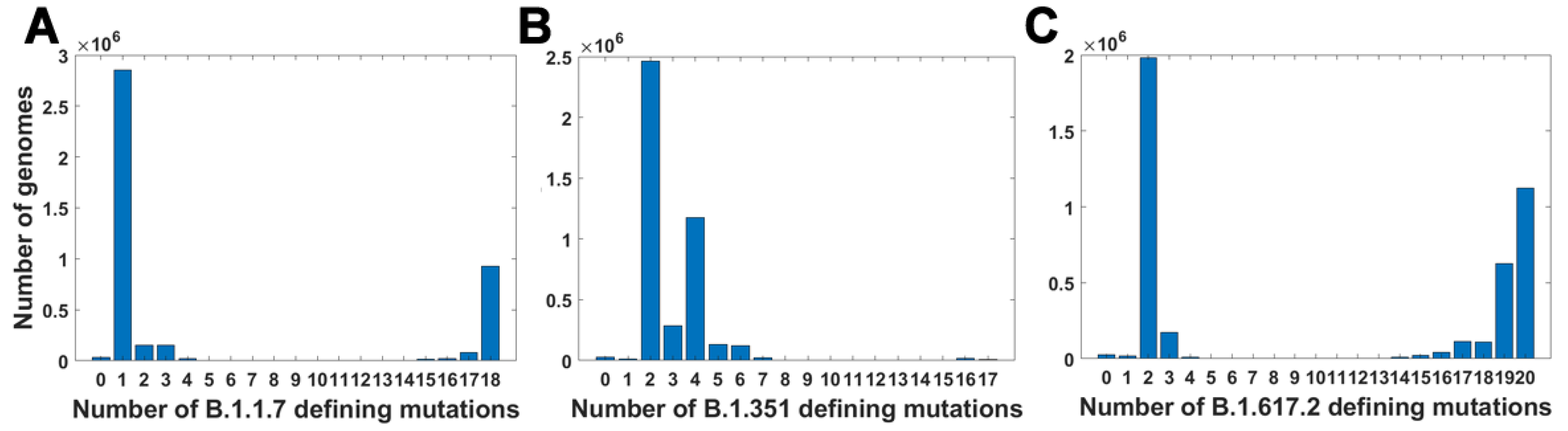
In this study, we accumulated and aligned 4.7 million SARS-CoV-2 genomes from the GISAID database and carried out a comprehensive set of analyses. This collection covers the period until the end of October 2021, i.e., the beginnings of the Omicron variant. First, we explored combinatorial complexity of the genomic variants emerging and their timing, indicating very strong, albeit hidden, selection forces. To this end, we analyzed SARS-CoV-2 genomes to determine how individual mutations that define the Alpha, Beta and Delta variants were appearing over time and how these were interfering with neutral and mildly deleterious mutations in different ranges of mutation frequency. Our analyses showed that the VOC-defining mutations did not arise gradually but rather co-evolved rapidly, leading to the emergence of the full VOC strain (
Figure 3). We did not observe transient states, which would be expected under neutral evolution. In addition, the recorded assortment of haplotypes involving the VOC-defining mutations demonstrated that maybe around 1% of combinatorially feasible variants appeared in the known viral strains (
Table A4,
Table A5 and
Table A6). These results seem to indicate that segregating sites in the Alpha, Beta and Delta variants evolved under strong positive selection, with a possible contribution of recombinations among viruses carrying subsets of VOC-defining mutations. Research has shown that the latter is common in bat coronaviruses [
44] and might indeed also be affecting the evolution of SARS-CoV-2 [
45]. Observed mutation patterns may also be due to mutation hotspots, which were detected in the region encoding the Spike protein [
46].
As noted in Neher [
5], recently, Hill et al. [
47] and Tay et al. [
48] investigated the dichotomous pattern of SARS-CoV-2 evolution with step-wise evolution within clades or variants and atypical bursts of evolution leading to new variants and showed that the rate of evolution along branches giving rise to new variants is up to four-fold higher than the background rate. However, this does not seem to exclude selection as the underlying mechanism; please see further on.
In addition, we cannot rule out the possibility that genomes carrying subsets of VOC-defining mutations avoided collection and sequencing. In the data gathered by GISAID, we can clearly see temporal differences in the number of sequenced genomes (as shown in
Figure A1A), but more importantly, most of the collected genomes come from Europe and the United States. The under-representation of sequences from other parts of the world might alter our conclusions.
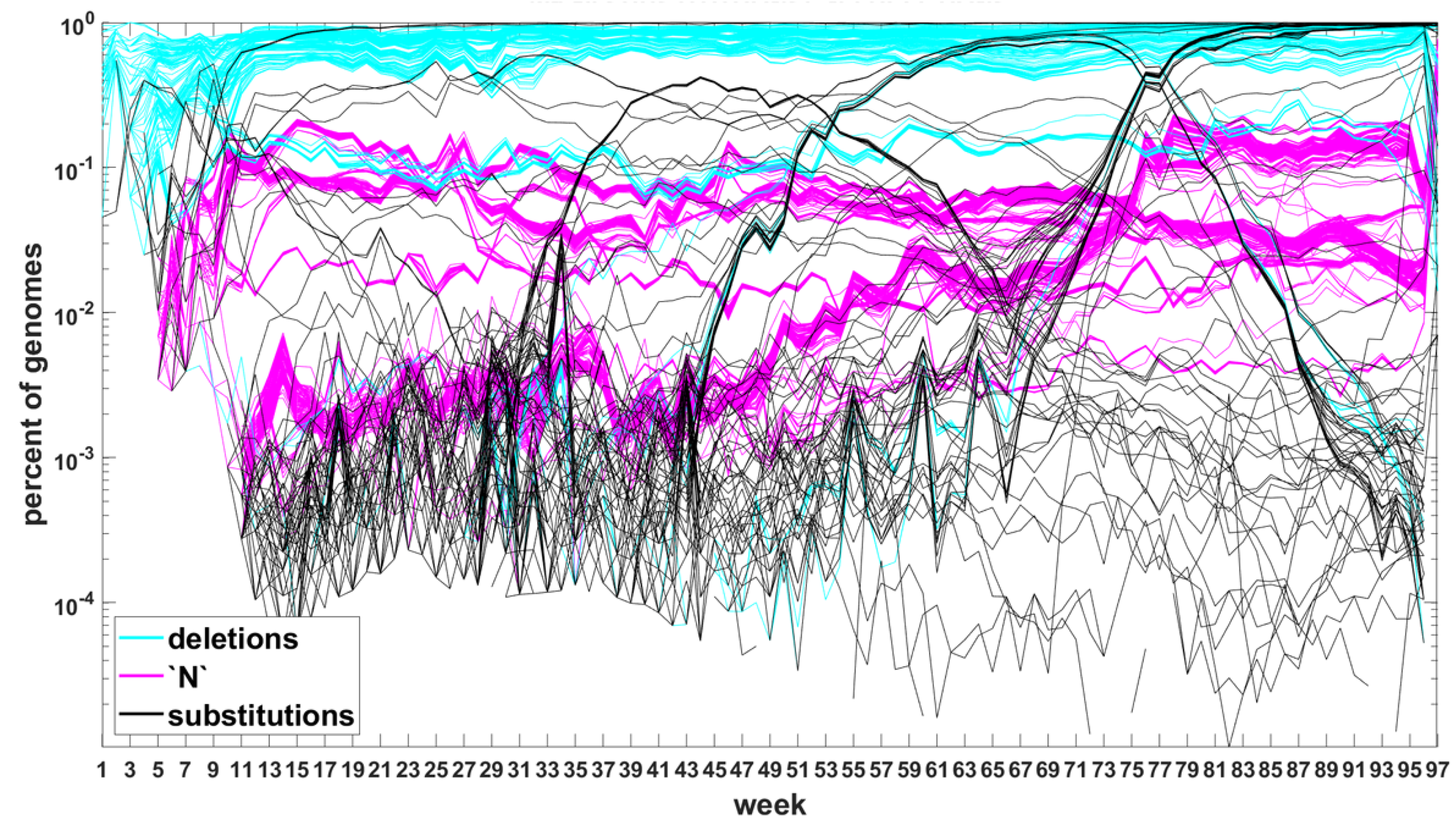
To explore in some detail the evolutionary forces at work, we developed time trajectories of mutations at all 29,903 sites of the SARS-CoV-2 genome, week by week, and stratified them into trends related to (i) point substitutions, (ii) deletions and (iii) non-sequenceable regions (
Figure 7,
Figure 8 and
Figure A5,
Figure A6 and
Figure A7). Among others, as mentioned earlier on, this allowed us to track the non-standard variant-defining mutations, left out in the original definitions of the variants of concern.
We focused on classifying the genetic forces active at different ranges of the mutational spectrum. A “reasonable” presumption might be that at the lower end of the mutational spectrum, there exists a “neutral foam” that is affected by mutation and drift, counteracting each other and creating a barrier, prohibiting the evolutionary process from dying out (see further on). Moving further up the frequency spectrum, one might expect forces related to competition and selection show their presence, with negative selection increasing with the size of the VOC genome population and accumulation of deleterious mutations.
As evident from
Figure 9, we observe the agreement of the lowest-frequency mutation SFS with the Griffiths–Tavaré theory [
22] under the Infinite Sites Model (ISM) and neutrality. This is consistent with the results of IAM testing; the numbers of single-copy haplotypes agree with two models under neutrality, though further terms diverge (
Figure 14). If we widen the frequency range, we observe the SFS to be much more consistent with the Tung–Durrett model (
Figure 11 and
Figure 12), assuming clone competition and selection [
23]. The coefficients of the fitting model indicate the possibility of selection acting to promote the gradual growth slowdown, as observed in the history of the VOC.
These results add up to a model of genomic evolution, which partly fits into the classical drift barrier ideas. Classically, drift barrier prevents the mutations from dominating fitness change too easily, as explained in a body of theoretical work in the field of evolutionary genetics, such as [
49,
50,
51]. These papers concern the interplay among mutation, drift and selection, in the absence of recombination (asexual reproduction), where epistasis plays a major role. In our case, a somewhat different barrier, arguably present at the bottom of the mutation frequency spectrum, contributes to injecting mutants, which becomes successful, but then their growth rate decays and they are replaced by others. Certain observations, such as mutations “bands” persistent over the epidemic history, suggest the contribution of genetic forces different from mutation, drift and selection, including recombinations and other genome transformations.
As already mentioned, Neher [
5] reviewed the mechanisms of new strain formation in influenza A and HIV-1 viruses and emphasized the exceptional nature of the dichotomous pattern of SARS-CoV-2 evolution with step-wise evolution within clades or variants and atypical bursts of evolution leading to new VOC [
47,
48]. Furthermore, [
5] concluded that a difference in evolutionary rate is only seen for non-synonymous changes, while the rate of synonymous evolution within variants was compatible with that seen between variants. The paper also systematized the knowledge regarding substitution types, leading to new adaptations. These conclusions do not contradict our finding of neutrality at the lowest frequencies of the SFS and gradually picking up negative selection at the mid-range frequencies, as documented in
Figure 9,
Figure 10,
Figure 11 and
Figure 12. To synthesize our findings and contribute to the discussion regarding mechanisms of adaptation leading to wave-form succession of the VOC, we proposed a Tug-of-War-type model (see [
34] and
Section 2.6 for details) in which new variants (clones) stem from rare advantageous driver mutations, and then acquire neutral or disadvantageous passenger mutations which gradually reduce the fitness of the variant, which can be then outcompeted by a new variant due to other driver mutations. Although the current version is a “toy” model, and lacks the resolution necessary for predictive power, it reproduces the succession of clones resembling the Alpha, Beta and Delta pattern (
Figure 15) and provides a mathematically consistent mechanism of VOC emergence and replacement.