Isolation and Characterization of Lytic Bacteriophages Active against Clinical Strains of E. coli and Development of a Phage Antimicrobial Cocktail
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Isolation of Bacteriophages
2.3. Determination of Bacteriophage Titer
2.4. Pure Bacteriophages Isolation
2.5. Bacteriophage Propagation
2.6. Concentration of Bacteriophages by Ultracentrifugation
2.7. Transmission Electron Microscopy Analysis
2.8. Phage DNA Isolation and Genome Sequencing
2.9. Bioinformatics Analysis
2.10. Assessment of the Lytic Activity Level of Bacteriophages
2.11. Host Range Determination
2.12. Development of a Bacteriophage Cocktail
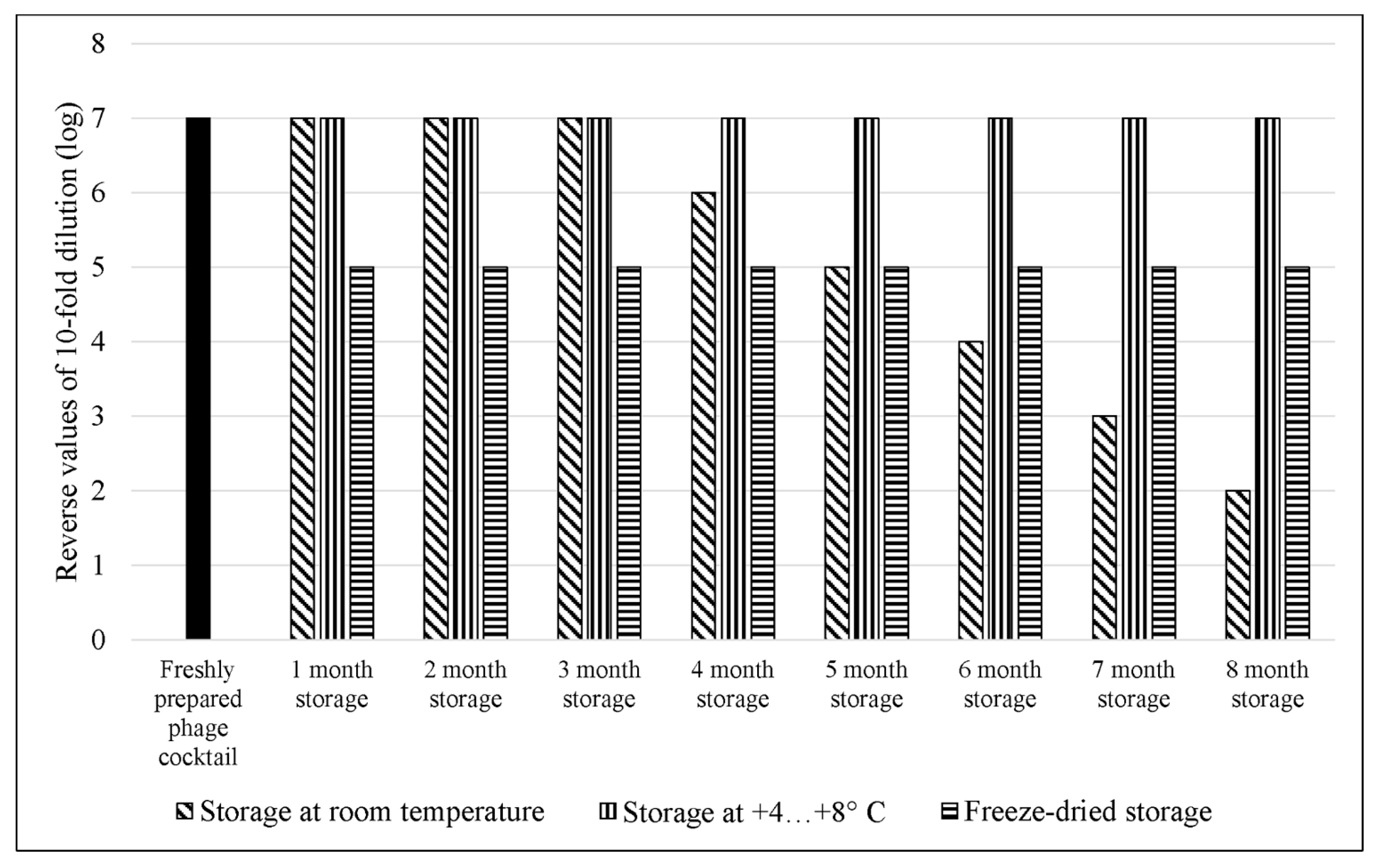
2.13. Investigation of the Bacteriophage Cocktail Stability after Storage
2.14. Statistics and Graphic Display of Results
3. Results
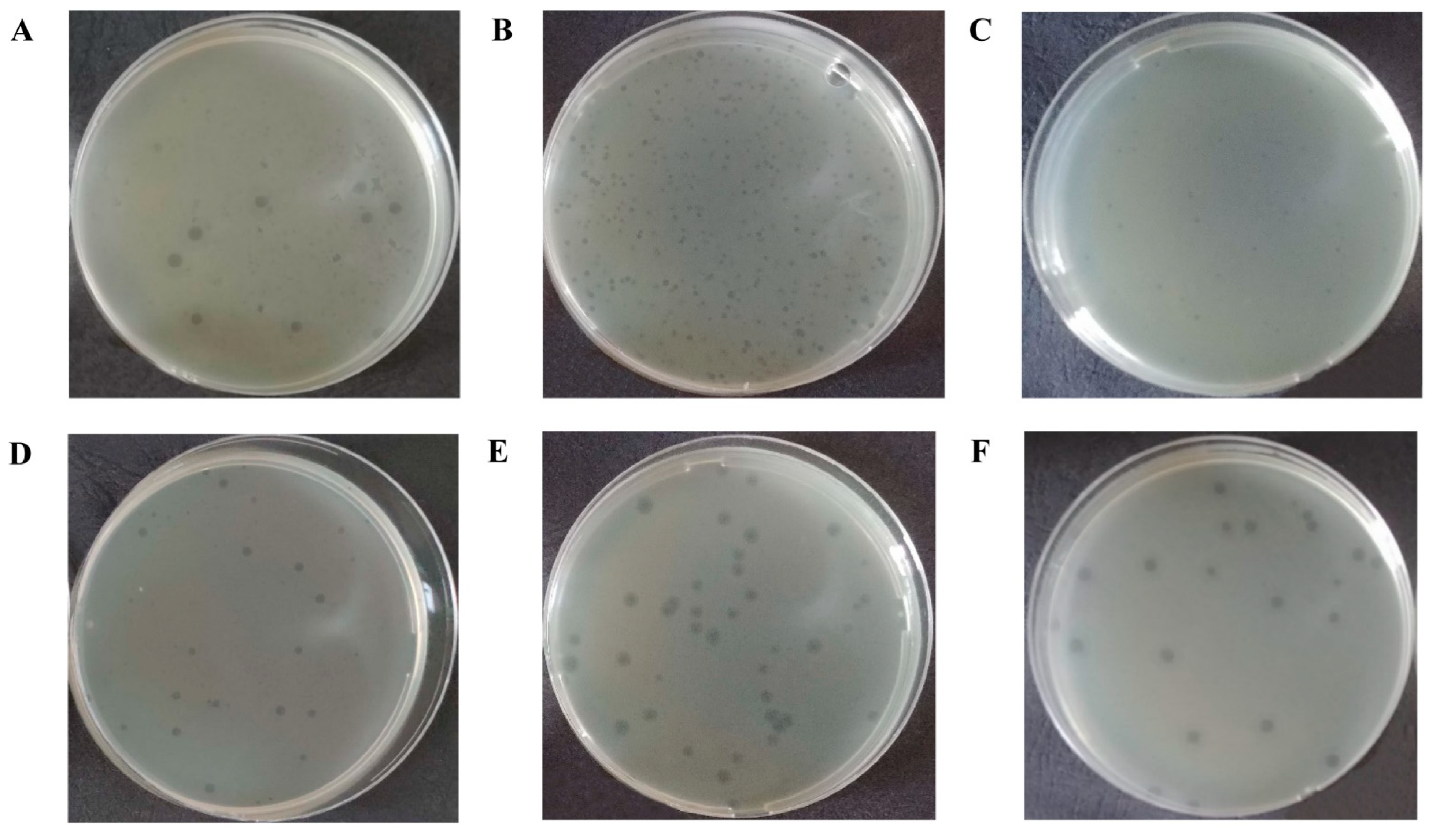
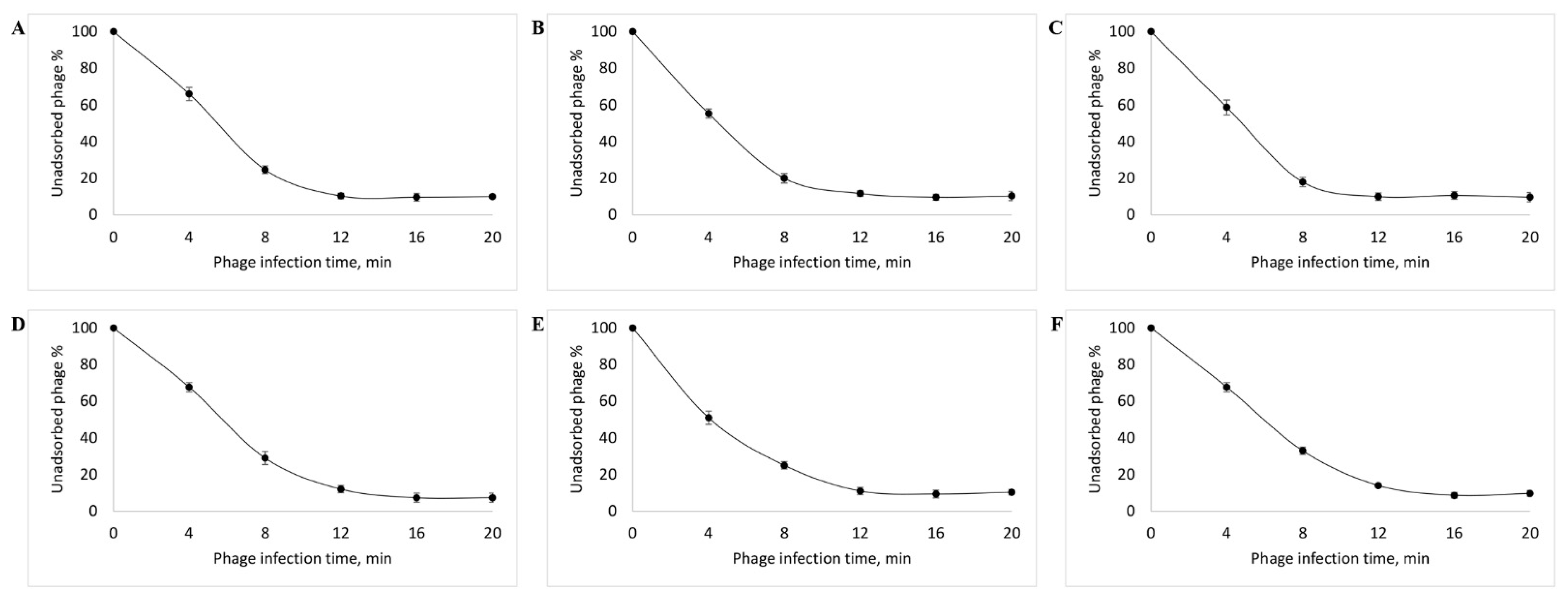
3.1. Isolation and Characterization of E. coli Bacteriophages
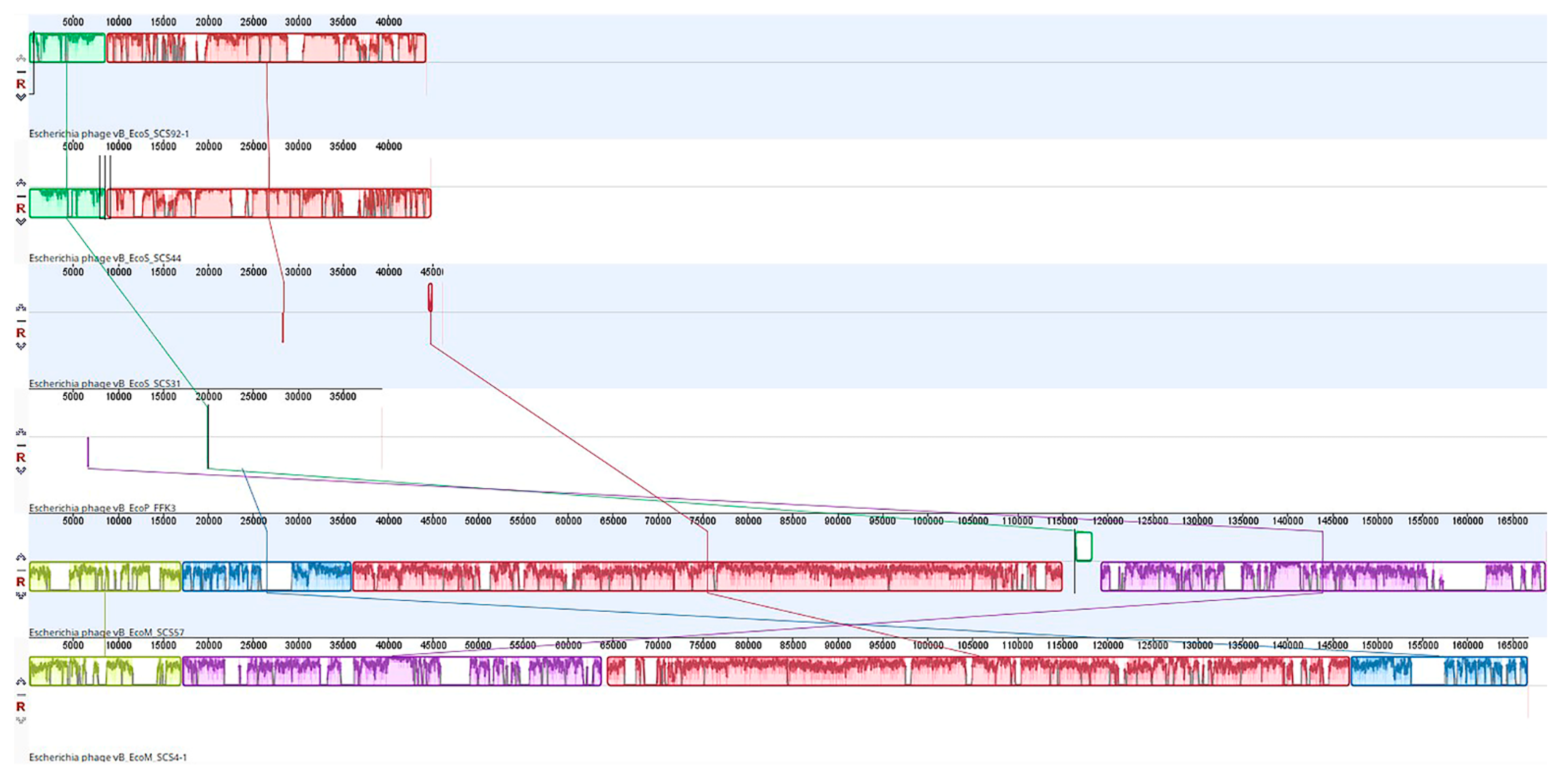
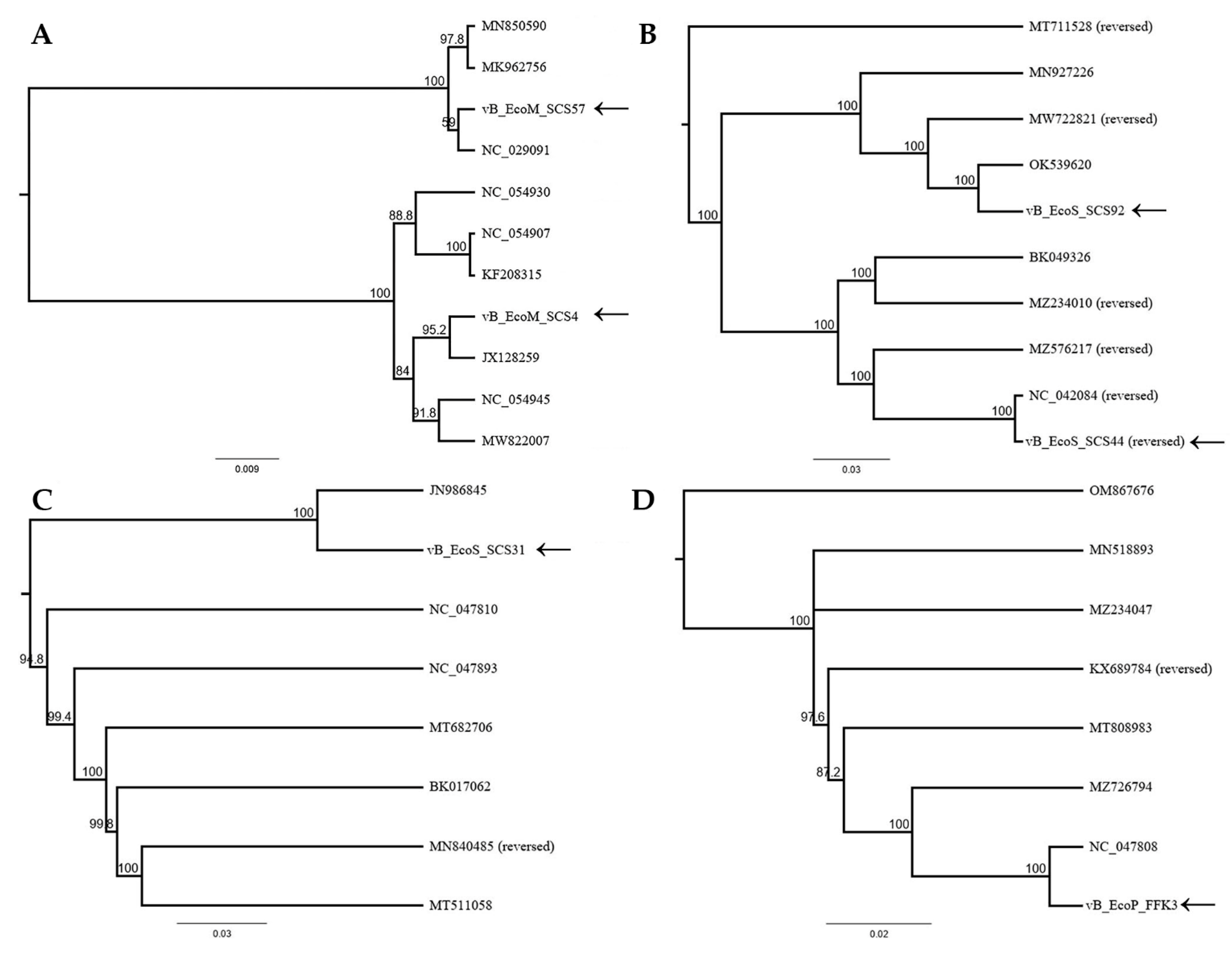
3.2. Bioinformatics Analysis and Characterization of the Bacteriophages’ Genome
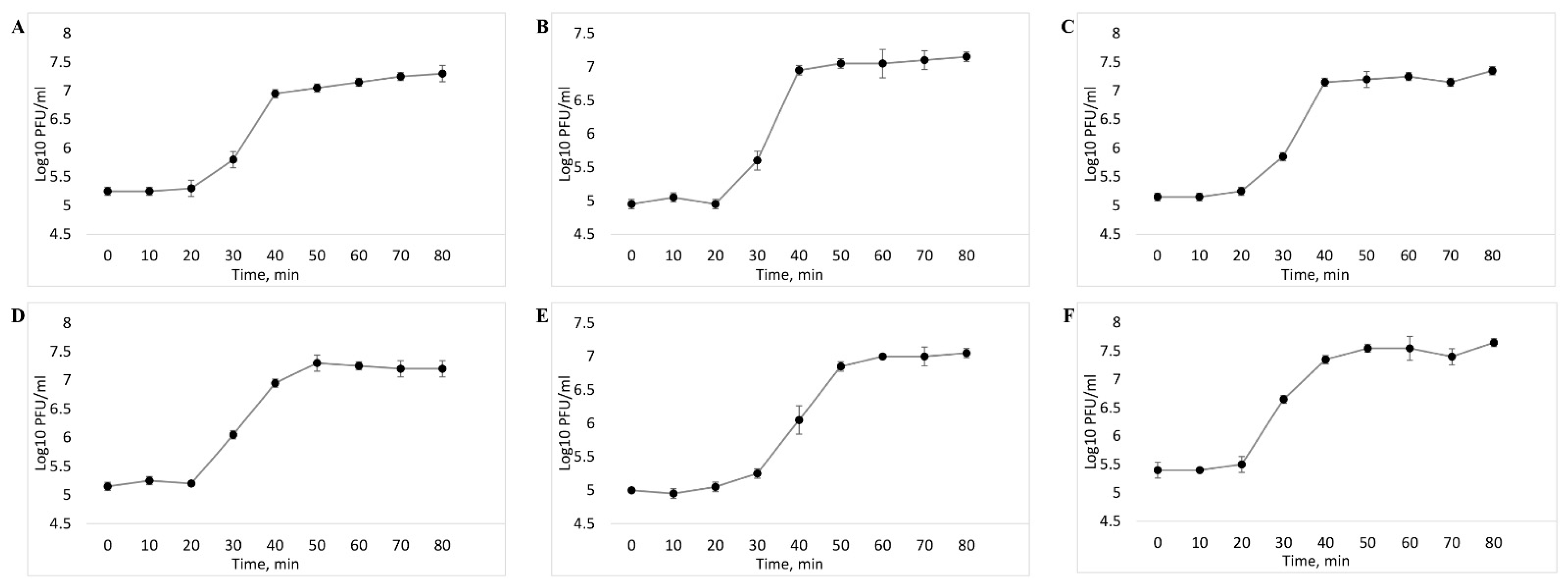
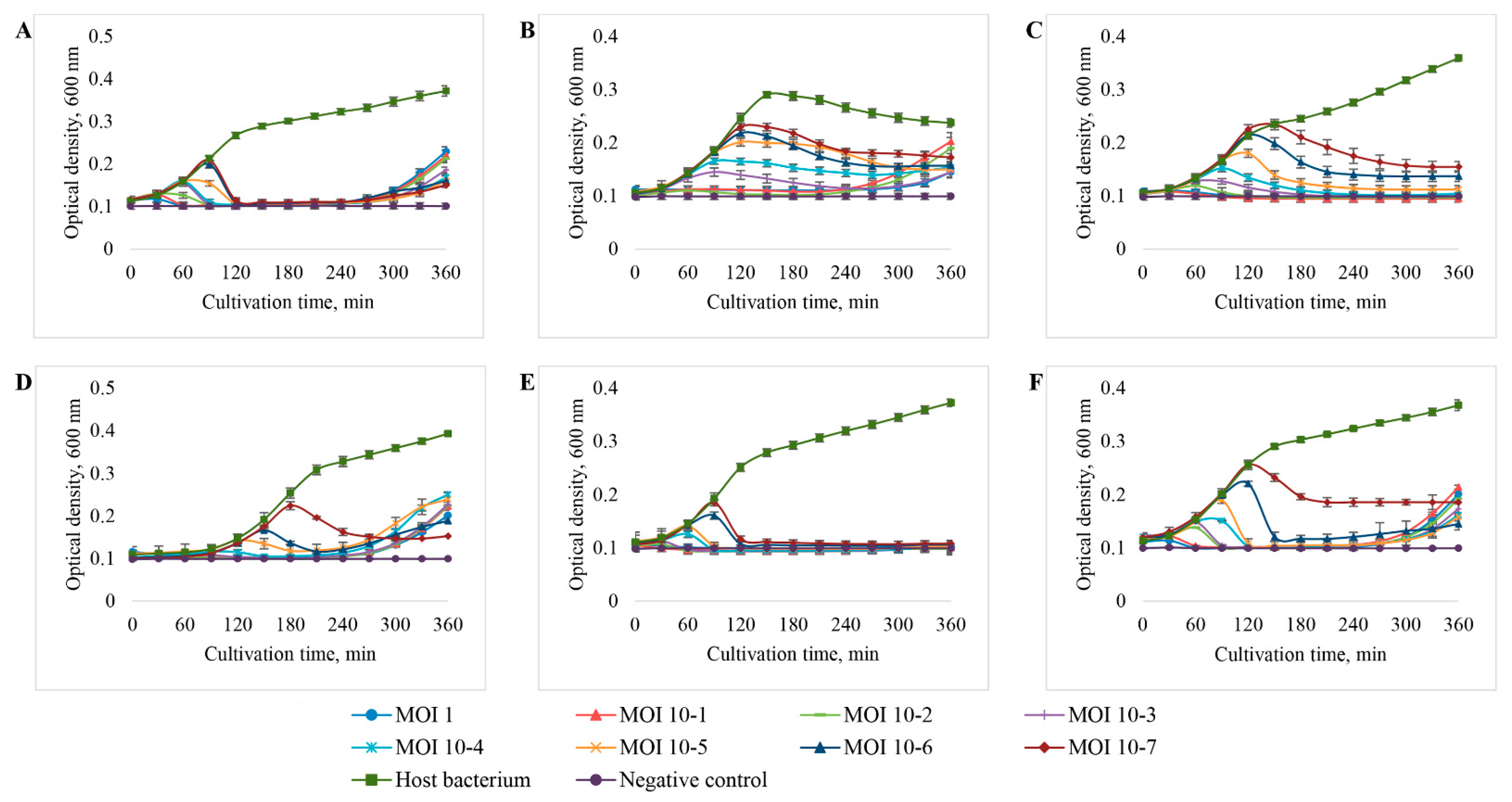
3.3. Determination of the Level of Lytic Activity
3.4. Host Range Analysis
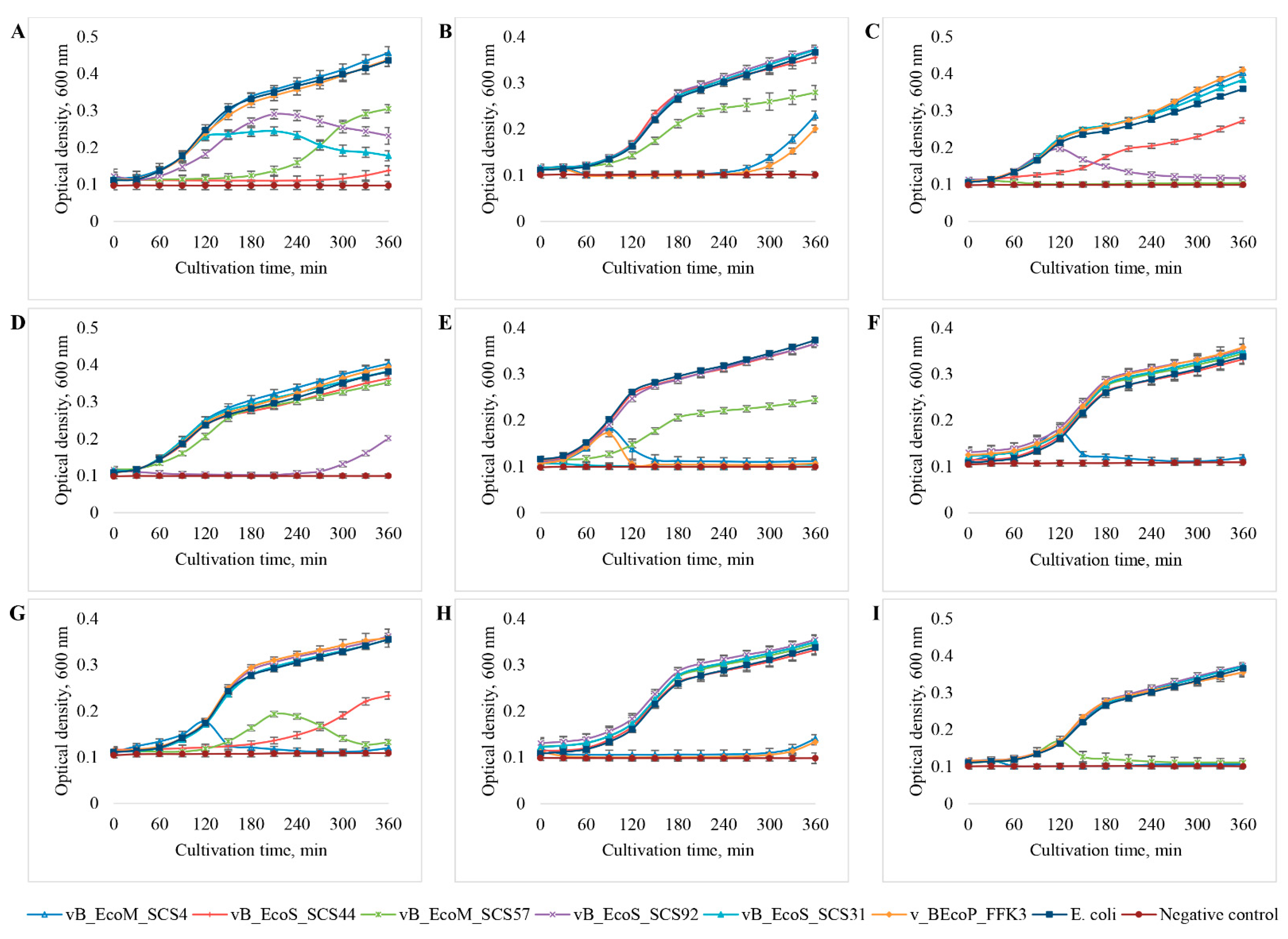
3.5. Development of the Bacteriophage Cocktail and Determination of Its Efficiency and Spectrum of Lytic Activity against E. coli
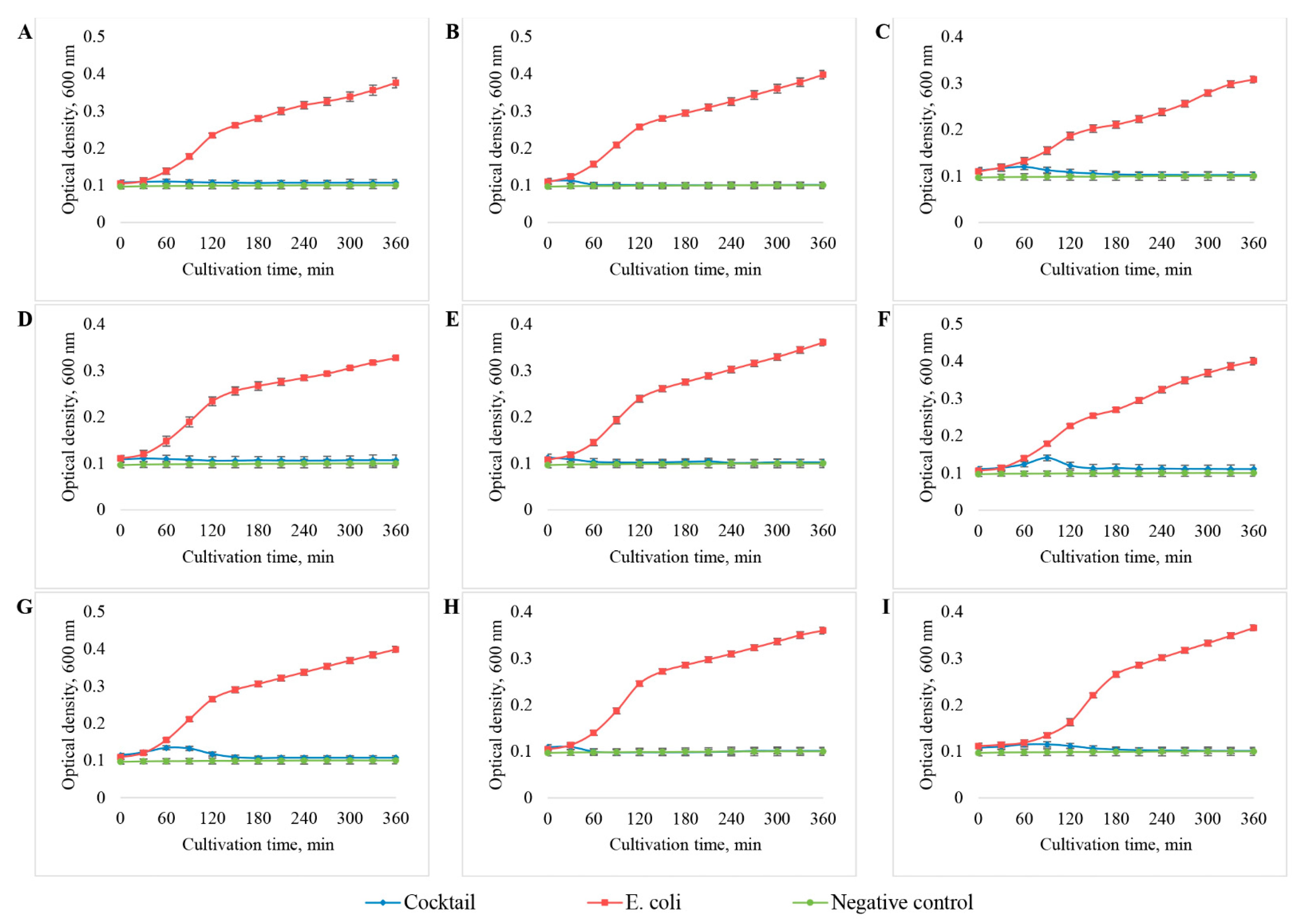
3.6. Comparative Study of the Efficiency of E. coli Bacteriophage Cocktail Storage Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Divatia, J.V.; Pulinilkunnathil, J.G.; Myatra, S.N. Nosocomial Infections and Ventilator-Associated Pneumonia in Cancer Patients. Oncol. Crit. Care 2019, 1419–1439. [Google Scholar] [CrossRef]
- Behnke, M.; Hansen, S.; Leistner, R.; Diaz, L.A.P.; Gropmann, A.; Sohr, D.; Gastmeier, P.; Piening, B. Nosocomial Infection and Antibiotic Use. Dtsch. Ärztebl. Int. 2013, 110, 627–633. [Google Scholar] [CrossRef] [Green Version]
- Leaper, D.J.; Van Goor, H.; Reilly, J.; Petrosillo, N.; Geiss, H.K.; Torres, A.J.; Berger, A. Surgical Site Infection–a European Perspective of Incidence and Economic Burden. Int. Wound J. 2004, 1, 247–273. [Google Scholar] [CrossRef]
- Khan, H.A.; Baig, F.K.; Mehboob, R. Nosocomial Infections: Epidemiology, Prevention, Control and Surveillance. Asian Pac. J. Trop. Biomed. 2017, 7, 478–482. [Google Scholar] [CrossRef]
- World Health Organization. Regional Office for the Eastern Mediterranean Eastern Mediterranean Health Journal. East. Mediterr. Health J. 2020, 26, 136–245. [Google Scholar]
- Vivas, R.; Barbosa, A.A.T.; Dolabela, S.S.; Jain, S. Multidrug-Resistant Bacteria and Alternative Methods to Control Them: An Overview. Microb. Drug Resist. 2019, 25, 890–908. [Google Scholar] [CrossRef]
- Iwu, C.D.; Korsten, L.; Okoh, A.I. The Incidence of Antibiotic Resistance within and beyond the Agricultural Ecosystem: A Concern for Public Health. MicrobiologyOpen 2020, 9, e1035. [Google Scholar] [CrossRef]
- Chang, R.Y.K.; Wallin, M.; Lin, Y.; Leung, S.S.Y.; Wang, H.; Morales, S.; Chan, H.-K. Phage Therapy for Respiratory Infections. Adv. Drug Deliv. Rev. 2018, 133, 76–86. [Google Scholar] [CrossRef]
- Cabal, A.; García-Castillo, M.; Cantón, R.; Gortázar, C.; Domínguez, L.; Álvarez, J. Prevalence of Escherichia coli Virulence Genes in Patients with Diarrhea and a Subpopulation of Healthy Volunteers in Madrid, Spain. Front. Microbiol. 2016, 7, 641. [Google Scholar] [CrossRef] [Green Version]
- Rahimzadeh, G.; Resch, G.; Rezai, M.S.; Nemati Hevelaee, E. Characterization and Lytic Activity of Isolated Escherichia coli Bacteriophages against Escherichia coli in Vitro. Iran. J. Med. Sci. 2020, 45, 298–303. [Google Scholar] [CrossRef]
- Barichello, T.; Fagundes, G.D.; Generoso, J.S.; Elias, S.G.; Simões, L.R.; Teixeira, A.L. Pathophysiology of Neonatal Acute Bacterial Meningitis. J. Med. Microbiol. 2013, 62, 1781–1789. [Google Scholar] [CrossRef]
- Hoffman, O.; Weber, R.J. Pathophysiology and Treatment of Bacterial Meningitis. Ther. Adv. Neurol. Disord. 2009, 2, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Chanishvili, N. Chapter 1-Phage Therapy—History from Twort and d’Herelle Through Soviet Experience to Current Approaches. In Advances in Virus Research; Łobocka, M., Szybalski, W., Eds.; Bacteriophages, Part B; Academic Press: Cambridge, MA, USA, 2012; Volume 83, pp. 3–40. [Google Scholar]
- Montso, P.K.; Mlambo, V.; Ateba, C.N. Characterization of Lytic Bacteriophages Infecting Multidrug-Resistant Shiga Toxigenic Atypical Escherichia coli O177 Strains Isolated From Cattle Feces. Front. Public Health 2019, 7, 355. [Google Scholar] [CrossRef]
- Chhibber, S.; Kaur, P.; Gondil, V.S. Simple Drop Cast Method for Enumeration of Bacteriophages. J. Virol. Methods 2018, 262, 1–5. [Google Scholar] [CrossRef]
- Manges, A.R.; Geum, H.M.; Guo, A.; Edens, T.J.; Fibke, C.D.; Pitout, J.D.D. Global Extraintestinal Pathogenic Escherichia coli (ExPEC) Lineages. Clin. Microbiol. Rev. 2019, 32, e00135-18. [Google Scholar] [CrossRef]
- Baldy-Chudzik, K.; Bok, E.; Mazurek, J. Well-known and new variants of pathogenic Escherichia coli as a consequence of the plastic genome. Postepy Hig. Med. Dosw. Online 2015, 69, 345–361. [Google Scholar] [CrossRef]
- Sarowska, J.; Futoma-Koloch, B.; Jama-Kmiecik, A.; Frej-Madrzak, M.; Ksiazczyk, M.; Bugla-PLoSkonska, G.; Choroszy-Krol, I. Virulence Factors, Prevalence and Potential Transmission of Extraintestinal Pathogenic Escherichia coli Isolated from Different Sources: Recent Reports. Gut Pathog. 2019, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- Pitout, J.D.D. Extraintestinal Pathogenic Escherichia coli: A Combination of Virulence with Antibiotic Resistance. Front. Microbiol. 2012, 3, 9. [Google Scholar] [CrossRef] [Green Version]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Russo, T.A.; Johnson, J.R. Medical and Economic Impact of Extraintestinal Infections Due to Escherichia coli: Focus on an Increasingly Important Endemic Problem. Microbes Infect. 2003, 5, 449–456. [Google Scholar] [CrossRef]
- Ramírez-Castillo, F.Y.; Moreno-Flores, A.C.; Avelar-González, F.J.; Márquez-Díaz, F.; Harel, J.; Guerrero-Barrera, A.L. An Evaluation of Multidrug-Resistant Escherichia coli Isolates in Urinary Tract Infections from Aguascalientes, Mexico: Cross-Sectional Study. Ann. Clin. Microbiol. Antimicrob. 2018, 17, 34. [Google Scholar] [CrossRef]
- Alkeskas, A.; Ogrodzki, P.; Saad, M.; Masood, N.; Rhoma, N.R.; Moore, K.; Farbos, A.; Paszkiewicz, K.; Forsythe, S. The Molecular Characterisation of Escherichia coli K1 Isolated from Neonatal Nasogastric Feeding Tubes. BMC Infect. Dis. 2015, 15, 449. [Google Scholar] [CrossRef] [Green Version]
- de Louvois, J.; Halket, S.; Harvey, D. Neonatal Meningitis in England and Wales: Sequelae at 5 Years of Age. Eur. J. Pediatr. 2005, 164, 730–734. [Google Scholar] [CrossRef]
- Mehall, J.R.; Kite, C.A.; Saltzman, D.A.; Wallett, T.; Jackson, R.J.; Smith, S.D. Prospective Study of the Incidence and Complications of Bacterial Contamination of Enteral Feeding in Neonates. J. Pediatr. Surg. 2002, 37, 1177–1182. [Google Scholar] [CrossRef]
- Hurrell, E.; Kucerova, E.; Loughlin, M.; Caubilla-Barron, J.; Hilton, A.; Armstrong, R.; Smith, C.; Grant, J.; Shoo, S.; Forsythe, S. Neonatal Enteral Feeding Tubes as Loci for Colonisation by Members of the Enterobacteriaceae. BMC Infect. Dis. 2009, 9, 146. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.-H.; Jong, A.Y. Cellular Mechanisms of Microbial Proteins Contributing to Invasion of the Blood–Brain Barrier. Cell. Microbiol. 2001, 3, 277–287. [Google Scholar] [CrossRef]
- El-Mokhtar, M.A.; Daef, E.; Mohamed Hussein, A.A.R.; Hashem, M.K.; Hassan, H.M. Emergence of Nosocomial Pneumonia Caused by Colistin-Resistant Escherichia coli in Patients Admitted to Chest Intensive Care Unit. Antibiotics 2021, 10, 226. [Google Scholar] [CrossRef]
- Torres, A.; Aznar, R.; Gatell, J.M.; Jiménez, P.; González, J.; Ferrer, A.; Celis, R.; Rodriguez-Roisin, R. Incidence, Risk, and Prognosis Factors of Nosocomial Pneumonia in Mechanically Ventilated Patients. Am. Rev. Respir. Dis. 1990, 142, 523–528. [Google Scholar] [CrossRef]
- Koulenti, D.; Tsigou, E.; Rello, J. Nosocomial Pneumonia in 27 ICUs in Europe: Perspectives from the EU-VAP/CAP Study. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2017, 36, 1999–2006. [Google Scholar] [CrossRef]
- La Combe, B.; Clermont, O.; Messika, J.; Eveillard, M.; Kouatchet, A.; Lasocki, S.; Corvec, S.; Lakhal, K.; Billard-Pomares, T.; Fernandes, R.; et al. Pneumonia-Specific Escherichia coli with Distinct Phylogenetic and Virulence Profiles, France, 2012–2014. Emerg. Infect. Dis. 2019, 25, 710–718. [Google Scholar] [CrossRef] [Green Version]
- Gould, L.H.; Mody, R.K.; Ong, K.L.; Clogher, P.; Cronquist, A.B.; Garman, K.N.; Lathrop, S.; Medus, C.; Spina, N.L.; Webb, T.H.; et al. Increased Recognition of Non-O157 Shiga Toxin–Producing Escherichia coli Infections in the United States During 2000–2010: Epidemiologic Features and Comparison with E. coli O157 Infections. Foodborne Pathog. Dis. 2013, 10, 453–460. [Google Scholar] [CrossRef] [PubMed]
- Barilli, E.; Vismarra, A.; Villa, Z.; Bonilauri, P.; Bacci, C. ESβL E. coli Isolated in Pig’s Chain: Genetic Analysis Associated to the Phenotype and Biofilm Synthesis Evaluation. Int. J. Food Microbiol. 2019, 289, 162–167. [Google Scholar] [CrossRef] [PubMed]
- Coque, T.M.; Baquero, F.; Cantón, R. Increasing Prevalence of ESBL-Producing Enterobacteriaceae in Europe. Eurosurveillance 2008, 13, 19044. [Google Scholar] [CrossRef] [PubMed]
- Cross, T.; Schoff, C.; Chudoff, D.; Graves, L.; Broomell, H.; Terry, K.; Farina, J.; Correa, A.; Shade, D.; Dunbar, D. An Optimized Enrichment Technique for the Isolation of Arthrobacter Bacteriophage Species from Soil Sample Isolates. J. Vis. Exp. JoVE 2015, 98, 52781. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Lee, J.Y.; Castillo, R.; Hixon, M.S.; Pujol, C.; Doppalapudi, V.R.; Shepard, H.M.; Wahl, G.M.; Lobl, T.J.; Chan, M.F. NB2001, a Novel Antibacterial Agent with Broad-Spectrum Activity and Enhanced Potency against β-Lactamase-Producing Strains. Antimicrob. Agents Chemother. 2002, 46, 1262–1268. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Liu, J.-H.; Lv, L.; Zong, Z.; Sun, Y.; Zheng, H.; Chen, Z.; Zeng, Z.-L. Characterization of Extended-Spectrum β-Lactamase Genes Found among Escherichia coli Isolates from Duck and Environmental Samples Obtained on a Duck Farm. Appl. Environ. Microbiol. 2012, 78, 3668–3673. [Google Scholar] [CrossRef] [Green Version]
- Croxen, M.A.; Finlay, B.B. Molecular Mechanisms of Escherichia coli Pathogenicity. Nat. Rev. Microbiol. 2010, 8, 26–38. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered Bacteriophages for Treatment of a Patient with a Disseminated Drug Resistant Mycobacterium Abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Gill, J.J.; Hyman, P. Phage Choice, Isolation, and Preparation for Phage Therapy. Curr. Pharm. Biotechnol. 2010, 11, 2–14. [Google Scholar] [CrossRef]
- Nobrega, F.L.; Costa, A.R.; Kluskens, L.D.; Azeredo, J. Revisiting Phage Therapy: New Applications for Old Resources. Trends Microbiol. 2015, 23, 185–191. [Google Scholar] [CrossRef] [Green Version]
- Keen, E.C. A Century of Phage Research: Bacteriophages and the Shaping of Modern Biology. BioEssays News Rev. Mol. Cell. Dev. Biol. 2015, 37, 6–9. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T.; García, P.; Mullany, P.; Aminov, R. Editorial: Phage Therapy: Past, Present and Future. Front. Microbiol. 2017, 8, 981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kortright, K.E.; Chan, B.K.; Koff, J.L.; Turner, P.E. Phage Therapy: A Renewed Approach to Combat Antibiotic-Resistant Bacteria. Cell Host Microbe 2019, 25, 219–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abedon, S.T.; Kuhl, S.J.; Blasdel, B.G.; Kutter, E.M. Phage Treatment of Human Infections. Bacteriophage 2011, 1, 66–85. [Google Scholar] [CrossRef] [Green Version]
- Korf, I.H.E.; Kittler, S.; Bierbrodt, A.; Mengden, R.; Rohde, C.; Rohde, M.; Kroj, A.; Lehnherr, T.; Fruth, A.; Flieger, A.; et al. In Vitro Evaluation of a Phage Cocktail Controlling Infections with Escherichia coli. Viruses 2020, 12, 1470. [Google Scholar] [CrossRef] [PubMed]
- Górski, A.; Międzybrodzki, R.; Łobocka, M.; Głowacka-Rutkowska, A.; Bednarek, A.; Borysowski, J.; Jończyk-Matysiak, E.; Łusiak-Szelachowska, M.; Weber-Dąbrowska, B.; Bagińska, N.; et al. Phage Therapy: What Have We Learned? Viruses 2018, 10, 288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chibeu, A.; Lingohr, E.J.; Masson, L.; Manges, A.; Harel, J.; Ackermann, H.-W.; Kropinski, A.M.; Boerlin, P. Bacteriophages with the Ability to Degrade Uropathogenic Escherichia coli Biofilms. Viruses 2012, 4, 471–487. [Google Scholar] [CrossRef] [Green Version]
- Endersen, L.; O’Mahony, J.; Hill, C.; Ross, R.P.; McAuliffe, O.; Coffey, A. Phage Therapy in the Food Industry. Annu. Rev. Food Sci. Technol. 2014, 5, 327–349. [Google Scholar] [CrossRef]
- Manohar, P.; Tamhankar, A.J.; Lundborg, C.S.; Ramesh, N. Isolation, Characterization and in Vivo Efficacy of Escherichia Phage MyPSH1131. PLoS ONE 2018, 13, e0206278. [Google Scholar] [CrossRef]
- Carvalho, C.; Costa, A.R.; Silva, F.; Oliveira, A. Bacteriophages and Their Derivatives for the Treatment and Control of Food-Producing Animal Infections. Crit. Rev. Microbiol. 2017, 43, 583–601. [Google Scholar] [CrossRef] [Green Version]
- Culot, A.; Grosset, N.; Gautier, M. Overcoming the Challenges of Phage Therapy for Industrial Aquaculture: A Review. Aquaculture 2019, 513, 734423. [Google Scholar] [CrossRef]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and Bacterial Plant Diseases. Front. Microbiol. 2017, 8, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordillo Altamirano, F.L.; Barr, J.J. Phage Therapy in the Postantibiotic Era. Clin. Microbiol. Rev. 2019, 32, e00066-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakasis, A.; Panitsa, G. Bacteriophage Therapy as an Alternative Treatment for Human Infections. A Comprehensive Review. Int. J. Antimicrob. Agents 2019, 53, 16–21. [Google Scholar] [CrossRef]
- Brüssow, H. Phage Therapy: The Escherichia coli Experience. Microbiology 2005, 151, 2133–2140. [Google Scholar] [CrossRef] [Green Version]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef] [Green Version]
- Merabishvili, M.; Pirnay, J.-P.; Verbeken, G.; Chanishvili, N.; Tediashvili, M.; Lashkhi, N.; Glonti, T.; Krylov, V.; Mast, J.; Van Parys, L.; et al. Quality-Controlled Small-Scale Production of a Well-Defined Bacteriophage Cocktail for Use in Human Clinical Trials. PLoS ONE 2009, 4, e4944. [Google Scholar] [CrossRef]
- Des Relations Numériques Entre Bactéries Lysogènes et Particules de Bactériophages. Available online: https://orbi.uliege.be/bitstream/2268/267857/1/Des%20relations%20num%C3%A9riques%20entre%20bact%C3%A9ries%20lysog%C3%A8nes%20et%20particules%20de%20bact%C3%A9riophages.pdf (accessed on 2 September 2022).
- Molecular Cloning: A Laboratory Manual. 2nd ed. Volumes. 1, 2 and 3|Sigma-Aldrich. Available online: http://www.sigmaaldrich.com/ (accessed on 2 September 2022).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Vilsker, M.; Moosa, Y.; Nooij, S.; Fonseca, V.; Ghysens, Y.; Dumon, K.; Pauwels, R.; Alcantara, L.C.; Vanden Eynden, E.; Vandamme, A.-M.; et al. Genome Detective: An Automated System for Virus Identification from High-Throughput Sequencing Data. Bioinformatics 2019, 35, 871–873. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, W.; Nickle, D.C.; Learn, G.H.; Maust, B.; Mullins, J.I. ViroBLAST: A Stand-Alone BLAST Web Server for Flexible Queries of Multiple Databases and User’s Datasets. Bioinformatics 2007, 23, 2334–2336. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuno, M.; Kasukawa, T.; Saito, R.; Adachi, J.; Suzuki, H.; Baldarelli, R.; Hayashizaki, Y.; Okazaki, Y. CDS Annotation in Full-Length CDNA Sequence. Genome Res. 2003, 13, 1478–1487. [Google Scholar] [CrossRef] [Green Version]
- Alves, D.R.; Perez-Esteban, P.; Kot, W.; Bean, J.E.; Arnot, T.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T.A. A Novel Bacteriophage Cocktail Reduces and Disperses P Seudomonas Aeruginosa Biofilms under Static and Flow Conditions. Microb. Biotechnol. 2015, 9, 61–74. [Google Scholar] [CrossRef] [Green Version]
- Wittebole, X.; De Roock, S.; Opal, S.M. A Historical Overview of Bacteriophage Therapy as an Alternative to Antibiotics for the Treatment of Bacterial Pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef]
- Chakraborty, S.; von Mentzer, A.; Begum, Y.A.; Manzur, M.; Hasan, M.; Ghosh, A.N.; Hossain, M.A.; Camilli, A.; Qadri, F. Phenotypic and Genomic Analyses of Bacteriophages Targeting Environmental and Clinical CS3-Expressing Enterotoxigenic Escherichia coli (ETEC) Strains. PLoS ONE 2018, 13, e0209357. [Google Scholar] [CrossRef] [Green Version]
- Korf, I.H.E.; Meier-Kolthoff, J.P.; Adriaenssens, E.M.; Kropinski, A.M.; Nimtz, M.; Rohde, M.; van Raaij, M.J.; Wittmann, J. Still Something to Discover: Novel Insights into Escherichia coli Phage Diversity and Taxonomy. Viruses 2019, 11, 454. [Google Scholar] [CrossRef] [Green Version]
- Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Grabski, M.; Topka, G.; Dydecka, A.; Kosznik-Kwaśnicka, K.; Grabowski, Ł.; Jurczak-Kurek, A.; Wołkowicz, T.; et al. Characterization of a Bacteriophage, VB_Eco4M-7, That Effectively Infects Many Escherichia coli O157 Strains. Sci. Rep. 2020, 10, 3743. [Google Scholar] [CrossRef] [Green Version]
- Alexyuk, M.S.; Bogoyavlenskiy, A.P.; Alexyuk, P.G.; Moldakhanov, Y.S.; Turmagambetova, A.S.; Anarkulova, E.I.; Berezin, V.E. Complete Genome Sequence of VB_EcoP_PR_Kaz2018, a T7-Like Bacteriophage. Microbiol. Resour. Announc. 2019, 8, e01323-19. [Google Scholar] [CrossRef] [PubMed]
- Maffei, E.; Shaidullina, A.; Burkolter, M.; Heyer, Y.; Estermann, F.; Druelle, V.; Sauer, P.; Willi, L.; Michaelis, S.; Hilbi, H.; et al. Systematic Exploration of Escherichia coli Phage–Host Interactions with the BASEL Phage Collection. PLoS Biol. 2021, 19, e3001424. [Google Scholar] [CrossRef]
- Badawy, S.; Baka, Z.A.M.; Abou-Dobara, M.I.; El-Sayed, A.K.A.; Skurnik, M. Biological and Molecular Characterization of FEg-Eco19, a Lytic Bacteriophage Active against an Antibiotic-Resistant Clinical Escherichia coli Isolate. Arch. Virol. 2022, 167, 1333–1341. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, F.; Rezk, N.; Fayez, M.S.; Abdelmoteleb, M.; Atteya, R.; Elhadidy, M.; El-Shibiny, A. Isolation, Characterization, and Genomic Analysis of Three Novel E. coli Bacteriophages That Effectively Infect E. coli O18. Microorganisms 2022, 10, 589. [Google Scholar] [CrossRef]
- Vera-Mansilla, J.; Sánchez, P.; Silva-Valenzuela, C.A.; Molina-Quiroz, R.C. Isolation and Characterization of Novel Lytic Phages Infecting Multidrug-Resistant Escherichia coli. Microbiol. Spectr. 2022, 10, e01678-21. [Google Scholar] [CrossRef] [PubMed]
- Olsen, N.S.; Forero-Junco, L.; Kot, W.; Hansen, L.H. Exploring the Remarkable Diversity of Culturable Escherichia coli Phages in the Danish Wastewater Environment. Viruses 2020, 12, 986. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Wang, I.-N. Bacteriophage Adsorption Rate and Optimal Lysis Time. Genetics 2008, 180, 471–482. [Google Scholar] [CrossRef] [Green Version]
- Pereira, C.; Duarte, J.; Costa, P.; Braz, M.; Almeida, A. Bacteriophages in the Control of Aeromonas Sp. in Aquaculture Systems: An Integrative View. Antibiotics 2022, 11, 163. [Google Scholar] [CrossRef]
- Dalmasso, M.; Strain, R.; Neve, H.; Franz, C.M.A.P.; Cousin, F.J.; Ross, R.P.; Hill, C. Three New Escherichia coli Phages from the Human Gut Show Promising Potential for Phage Therapy. PLoS ONE 2016, 11, e0156773. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Ryu, S. Characterization of a T5-Like Coliphage, SPC35, and Differential Development of Resistance to SPC35 in Salmonella Enterica Serovar Typhimurium and Escherichia coli. Appl. Environ. Microbiol. 2011, 77, 2042–2050. [Google Scholar] [CrossRef] [Green Version]
- Kocharunchitt, C.; Ross, T.; McNeil, D.L. Use of Bacteriophages as Biocontrol Agents to Control Salmonella Associated with Seed Sprouts. Int. J. Food Microbiol. 2009, 128, 453–459. [Google Scholar] [CrossRef]
- Fischer, S.; Kittler, S.; Klein, G.; Glünder, G. Impact of a Single Phage and a Phage Cocktail Application in Broilers on Reduction of Campylobacter Jejuni and Development of Resistance. PLoS ONE 2013, 8, e78543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsonos, J.; Vandenheuvel, D.; Briers, Y.; De Greve, H.; Hernalsteens, J.-P.; Lavigne, R. Hurdles in Bacteriophage Therapy: Deconstructing the Parameters. Vet. Microbiol. 2014, 171, 460–469. [Google Scholar] [CrossRef]
- Hong, Y.; Thimmapuram, J.; Zhang, J.; Collings, C.K.; Bhide, K.; Schmidt, K.; Ebner, P.D. The Impact of Orally Administered Phages on Host Immune Response and Surrounding Microbial Communities. Bacteriophage 2016, 6, e1211066. [Google Scholar] [CrossRef] [Green Version]
- Nobrega, F.L.; Vlot, M.; de Jonge, P.A.; Dreesens, L.L.; Beaumont, H.J.E.; Lavigne, R.; Dutilh, B.E.; Brouns, S.J.J. Targeting Mechanisms of Tailed Bacteriophages. Nat. Rev. Microbiol. 2018, 16, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Chan, B.K.; Abedon, S.T. Chapter 1-Phage Therapy Pharmacology: Phage Cocktails. In Advances in Applied Microbiology; Laskin, A.I., Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 78, pp. 1–23. [Google Scholar]
- O’Flynn, G.; Ross, R.P.; Fitzgerald, G.F.; Coffey, A. Evaluation of a Cocktail of Three Bacteriophages for Biocontrol of Escherichia coli O157:H7. Appl. Environ. Microbiol. 2004, 70, 3417–3424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, B.K.; Abedon, S.T.; Loc-Carrillo, C. Phage Cocktails and the Future of Phage Therapy. Future Microbiol. 2013, 8, 769–783. [Google Scholar] [CrossRef]
- Hall, A.R.; De Vos, D.; Friman, V.-P.; Pirnay, J.-P.; Buckling, A. Effects of Sequential and Simultaneous Applications of Bacteriophages on Populations of Pseudomonas Aeruginosa In Vitro and in Wax Moth Larvae. Appl. Environ. Microbiol. 2012, 78, 5646–5652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-Encoded Virion-Associated Enzymes to Overcome the Carbohydrate Barriers during the Infection Process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119. [Google Scholar] [CrossRef] [Green Version]
- Mangalea, M.R.; Duerkop, B.A. Fitness Trade-Offs Resulting from Bacteriophage Resistance Potentiate Synergistic Antibacterial Strategies. Infect. Immun. 2020, 88, e00926-19. [Google Scholar] [CrossRef] [Green Version]
- Kifelew, L.G.; Warner, M.S.; Morales, S.; Vaughan, L.; Woodman, R.; Fitridge, R.; Mitchell, J.G.; Speck, P. Efficacy of Phage Cocktail AB-SA01 Therapy in Diabetic Mouse Wound Infections Caused by Multidrug-Resistant Staphylococcus Aureus. BMC Microbiol. 2020, 20, 204. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hon, K.; Bouras, G.S.; Psaltis, A.J.; Shearwin, K.; Wormald, P.-J.; Vreugde, S. APTC-C-SA01: A Novel Bacteriophage Cocktail Targeting Staphylococcus Aureus and MRSA Biofilms. Int. J. Mol. Sci. 2022, 23, 6116. [Google Scholar] [CrossRef]
- Naghizadeh, M.; Karimi Torshizi, M.A.; Rahimi, S.; Dalgaard, T.S. Synergistic Effect of Phage Therapy Using a Cocktail Rather than a Single Phage in the Control of Severe Colibacillosis in Quails. Poult. Sci. 2019, 98, 653–663. [Google Scholar] [CrossRef] [PubMed]
- Rosner, D.; Clark, J. Formulations for Bacteriophage Therapy and the Potential Uses of Immobilization. Pharmaceuticals 2021, 14, 359. [Google Scholar] [CrossRef] [PubMed]
- Ly, A.; Carrigy, N.B.; Wang, H.; Harrison, M.; Sauvageau, D.; Martin, A.R.; Vehring, R.; Finlay, W.H. Atmospheric Spray Freeze Drying of Sugar Solution With Phage D29. Front. Microbiol. 2019, 10, 488. [Google Scholar] [CrossRef] [PubMed]
- Clark, W.A. Comparison of Several Methods for Preserving Bacteriophages. Appl. Microbiol. 1962, 10, 466–471. [Google Scholar] [CrossRef]
- González-Menéndez, E.; Fernández, L.; Gutiérrez, D.; Rodríguez, A.; Martínez, B.; García, P. Comparative Analysis of Different Preservation Techniques for the Storage of Staphylococcus Phages Aimed for the Industrial Development of Phage-Based Antimicrobial Products. PLoS ONE 2018, 13, e0205728. [Google Scholar] [CrossRef]

| E. coli Strains | Antibiotic Resistance | Organization Providing Bacterial Strains |
|---|---|---|
| 444 | CFZ, CAZ, LVX, CIP, AZM, PIP | JSC “Central Clinical Hospital”, Almaty |
| 753 | PEN, fluoroquinolones I-III generation, TMP | |
| 3957 | AMP, PIP, GEN, SXT | |
| 3992 | CIP, LVX | |
| 4231 | Resistance to the antibiotics used has not been identified | |
| 4238 | AMP, CXM, CTX, CAZ, FEP, GEN, CIP, LVX, SXT | |
| 4320 | Resistance to the antibiotics used has not been identified | |
| 4325 | AMP, CIP, LVX | |
| 15597 | − | ATCC |
| Bacteriophage Strain | Bacteriophage Isolation Place | Host Bacterium Strain | Plaques |
|---|---|---|---|
| vB_EcoM_SCS4 | Sewage collector of Shymkent, Kazakhstan | E. coli 753 | Limpid plaque ≈4 mm in diameter with a slight halo up to 8 mm |
| vB_EcoS_SCS44 | E. coli 444 | Limpid plaque with a diameter of ≈2 mm without halo | |
| vB_EcoM_SCS57 | E. coli 3957 | Limpid plaque with a diameter of ≈1.5 mm without halo | |
| vB_EcoS_SCS92 | E. coli 3992 | Limpid plaque with a diameter of ≈2 mm without halo | |
| vB_EcoD_SCS31 | E. coli 4231 | Limpid plaque with a diameter of ≈4 mm with a pronounced halo up to 6 mm | |
| vB_EcoP_FFK3 | Sewage disposal fields of Konaev, Kazakhstan | E. coli 753 | A limpid plaque ≈4 mm in diameter with a slight halo up to 7 mm. |
| Phages | Length (bp) | No. of ORFs | No. of tRNAs | GC Content (%) |
|---|---|---|---|---|
| vB_EcoM_SCS57 | 168,771 | 272 | 2 | 37.6 |
| vB_EcoM_SCS4 | 166,783 | 269 | 0 | 35.4 |
| vB_EcoS_SCS92 | 44,240 | 72 | 0 | 51.0 |
| vB_EcoS_SCS44 | 44,829 | 78 | 0 | 50.6 |
| vB_EcoS_SCS31 | 46,054 | 78 | 1 | 43.5 |
| vB_EcoP_FFK3 | 49,300 | 48 | 0 | 49.8 |
| E. coli Bacteriophage Strain | MOI | |||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 10−1 | 10−2 | 10−3 | 10−4 | 10−5 | 10−6 | 10−7 | |
| vB_EcoM_SCS4 | ++ * | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| vB_EcoS_SCS44 | ++ | ++ | ++ | ++ | + | + | + | + |
| vB_EcoM_SCS57 | ++ | ++ | ++ | ++ | ++ | ++ | + | + |
| vB_EcoS_SCS92 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + |
| vB_EcoS_SCS31 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | ++ |
| v_BEcoP_FFK3 | ++ | ++ | ++ | ++ | ++ | ++ | ++ | + |
| Bacteriophage Strain | Clinical Strains of E. coli | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 444 | 753 | 3957 | 3992 | 4231 | 4238 | 4320 | 4325 | 15597 | |
| vB_EcoM_SCS4 | − * | +++ | − | − | +++ | +++ | +++ | +++ | +++ |
| vB_EcoS_SCS44 | +++ | − | ++ | − | − | − | ++ | − | − |
| vB_EcoM_SCS57 | +++ | + | +++ | − | ++ | − | +++ | − | +++ |
| vB_EcoS_SCS92 | ++ | − | +++ | +++ | − | − | − | − | − |
| vB_EcoS_SCS31 | ++ | − | − | − | +++ | − | − | − | − |
| v_BEcoP_FFK3 | − | +++ | − | − | +++ | − | − | +++ | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexyuk, P.; Bogoyavlenskiy, A.; Alexyuk, M.; Akanova, K.; Moldakhanov, Y.; Berezin, V. Isolation and Characterization of Lytic Bacteriophages Active against Clinical Strains of E. coli and Development of a Phage Antimicrobial Cocktail. Viruses 2022, 14, 2381. https://doi.org/10.3390/v14112381
Alexyuk P, Bogoyavlenskiy A, Alexyuk M, Akanova K, Moldakhanov Y, Berezin V. Isolation and Characterization of Lytic Bacteriophages Active against Clinical Strains of E. coli and Development of a Phage Antimicrobial Cocktail. Viruses. 2022; 14(11):2381. https://doi.org/10.3390/v14112381
Chicago/Turabian StyleAlexyuk, Pavel, Andrey Bogoyavlenskiy, Madina Alexyuk, Kuralay Akanova, Yergali Moldakhanov, and Vladimir Berezin. 2022. "Isolation and Characterization of Lytic Bacteriophages Active against Clinical Strains of E. coli and Development of a Phage Antimicrobial Cocktail" Viruses 14, no. 11: 2381. https://doi.org/10.3390/v14112381
APA StyleAlexyuk, P., Bogoyavlenskiy, A., Alexyuk, M., Akanova, K., Moldakhanov, Y., & Berezin, V. (2022). Isolation and Characterization of Lytic Bacteriophages Active against Clinical Strains of E. coli and Development of a Phage Antimicrobial Cocktail. Viruses, 14(11), 2381. https://doi.org/10.3390/v14112381







