Life after Cleavage: The Story of a β-Retroviral (MMTV) Signal Peptide—From Murine Lymphoma to Human Breast Cancer
Abstract
1. MMTV and Cancer
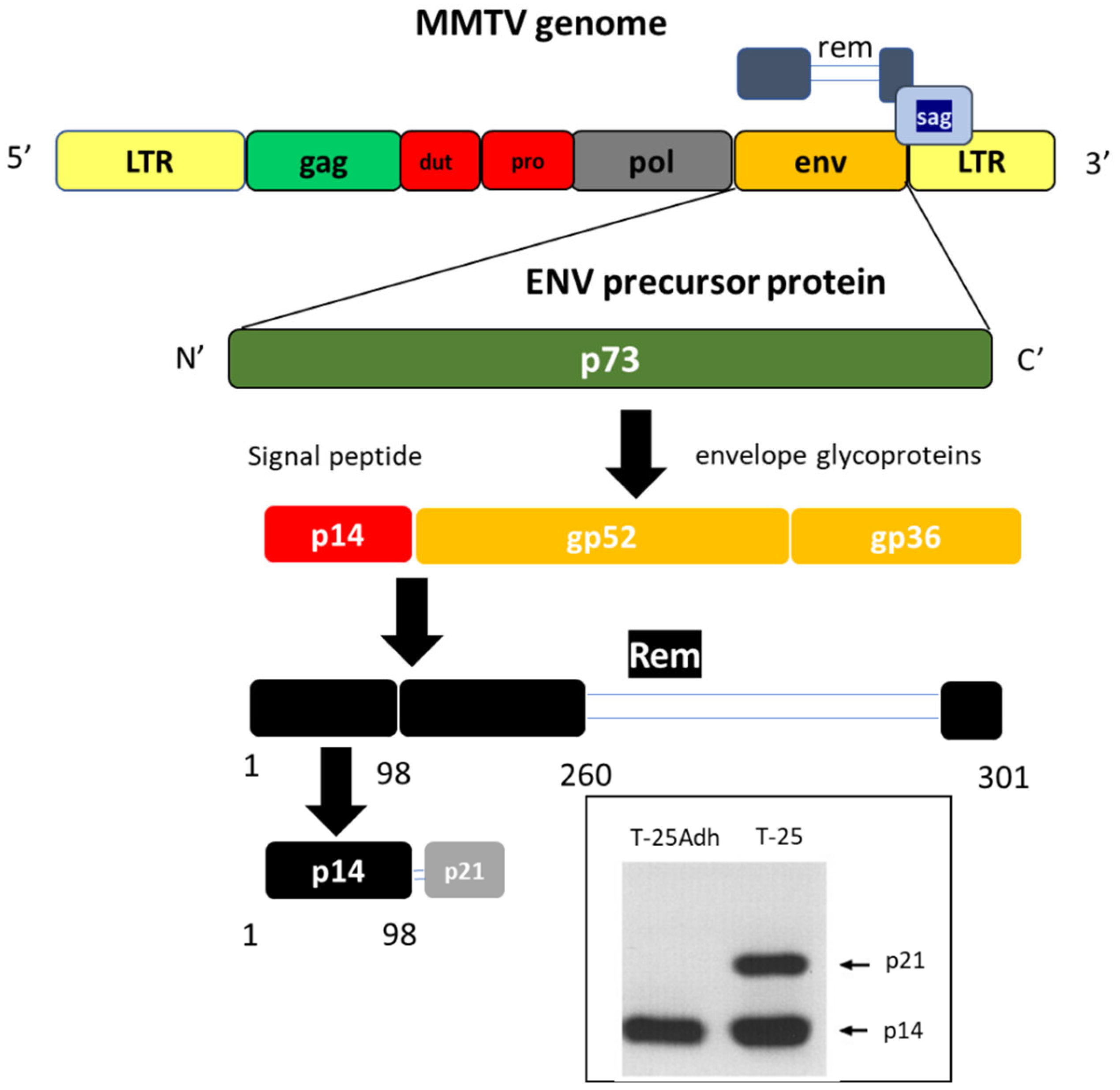
1.1. MMTV Genome
1.2. MMTV Infection and Life Cycle
1.3. MMTV in Breast Cancer
2. The Lymphoma Connection
3. Enter the Signal Peptide, p14
4. A Word on Signal Peptides
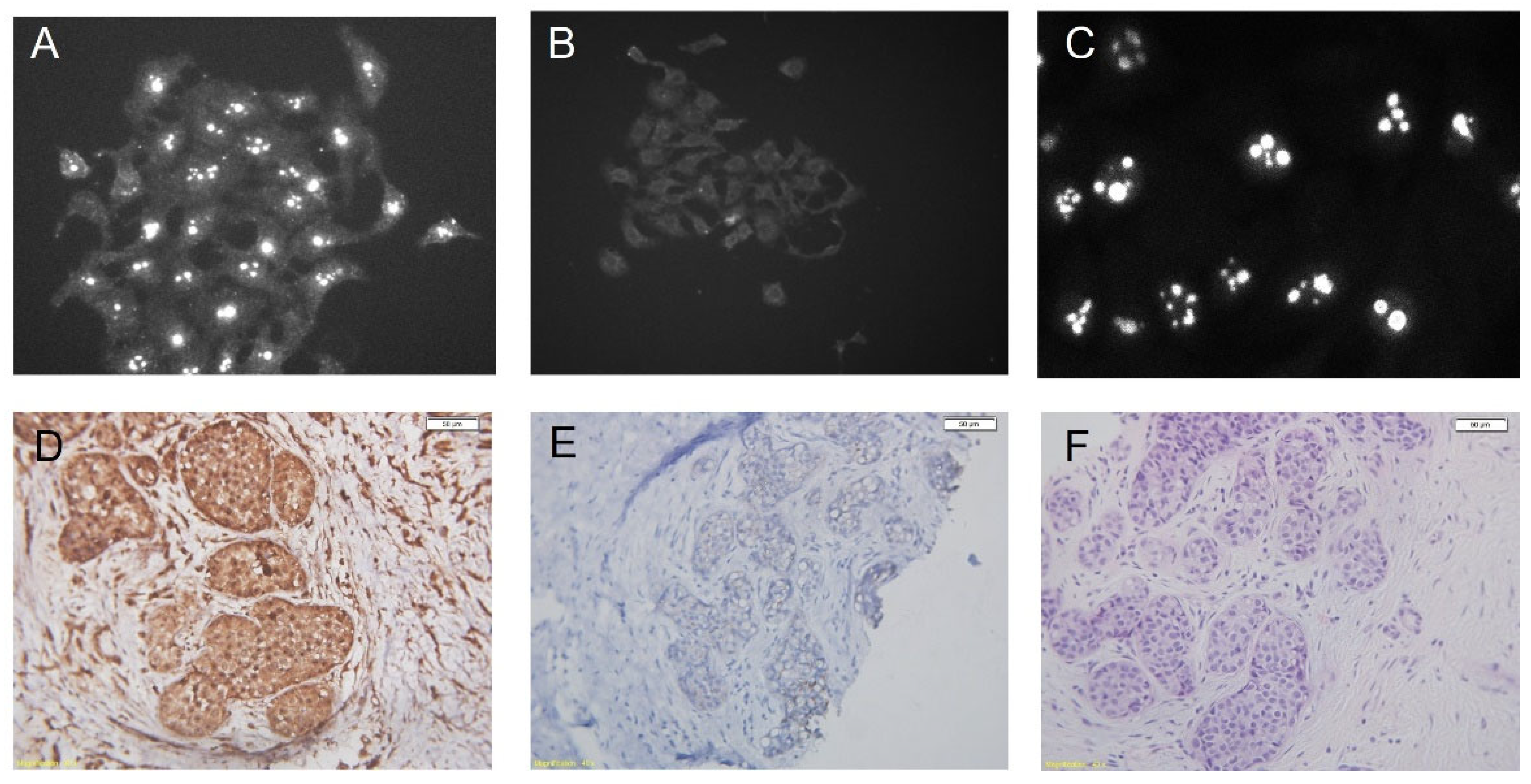
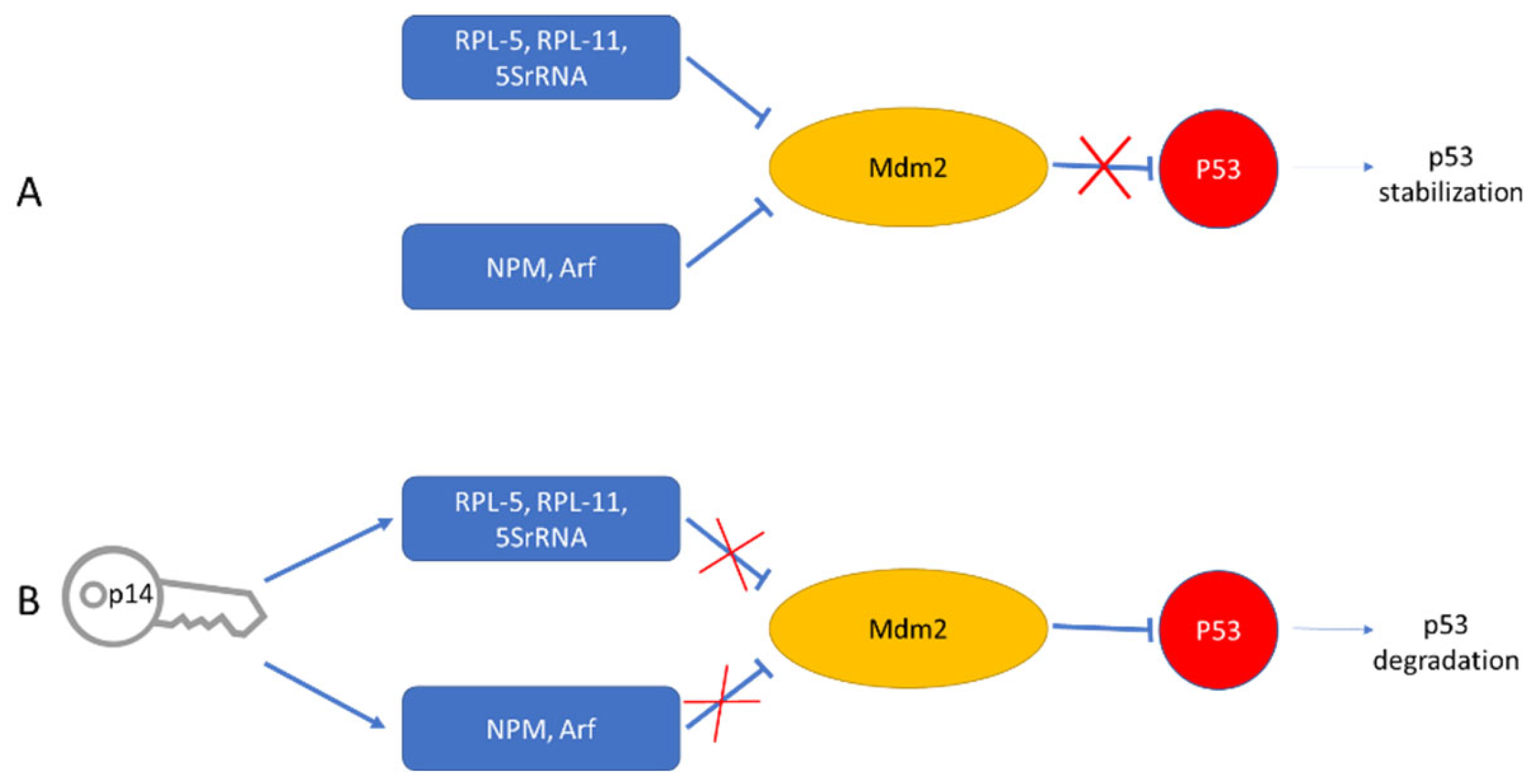
5. p14 Is Not an Orphan Nucleolar Signal Peptide
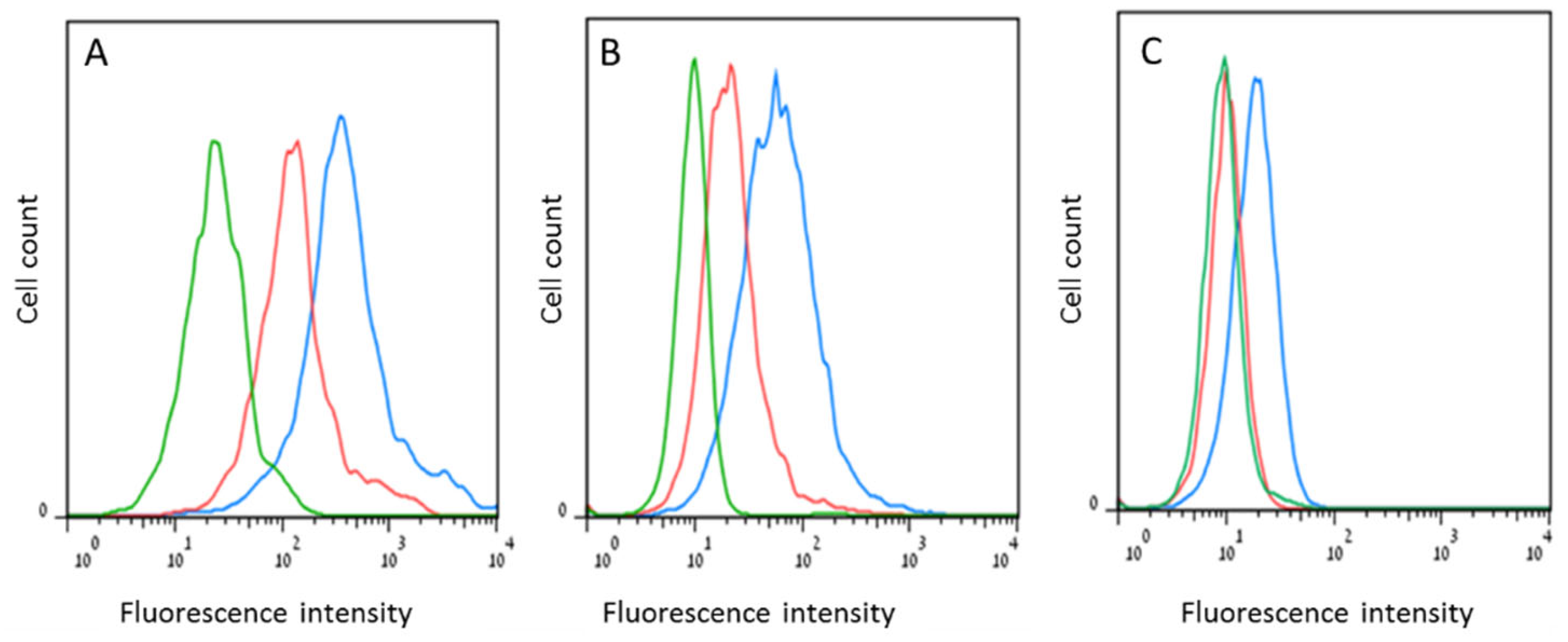
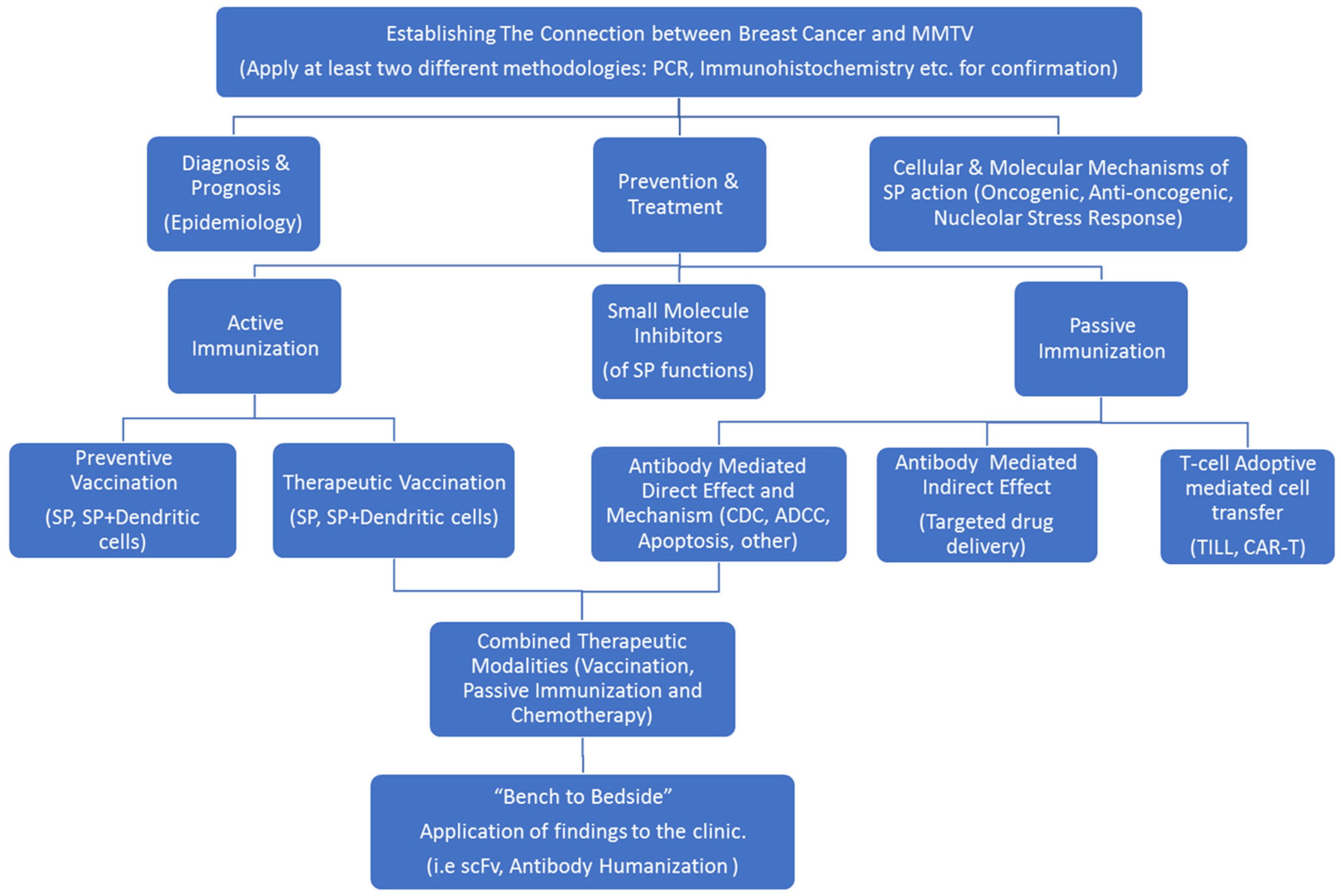
6. Strategies for p14 Investigation in Human Breast Cancer
- Diagnosis, of human breast cancer.
- Translational research involving the targeting, prevention and treatment of MMTV-associated murine lymphomas and mammary carcinomas.
- Assessing post-ER targeting functions of p14 and putative mechanisms involved.
6.1. p14 as a Diagnostic Marker for MMTV-Associated Breast Cancers
6.2. Signal Peptide-Mediated Targeting, Prevention and Treatment of MMTV-Associated Murine Lymphoma and Mammary Carcinoma (Translational Research)
6.3. Post ER Targeting Functions of p14
7. Treatment Modalities Based on Target Localization
7.1. With Regards to Cell Surface Expression of p14
7.2. With Regards to Intracellular Expression of p14
8. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sarid, R.; Gao, S.-J. Viruses and human cancer: From detection to causality. Cancer Lett. 2011, 305, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Bitnner, J.J. Mammary cancer in mice of different ages and stocks following the administration of the mammary tumor agent. Cancer Res. 1956, 16, 1038–1042. [Google Scholar]
- Michalides, R.; Wagenaar, E.; Hilkens, J.; Hilgers, J.; Groner, B.; Hynes, N.E. Acquisition of proviral DNA of mouse mammary tumor virus in thymic leukemia cells from GR mice. J. Virol. 1982, 43, 819–829. [Google Scholar] [CrossRef] [PubMed]
- Racevskis, J. Altered mouse mammary tumor virus transcript synthesis in T-cell lymphoma cells. J. Virol. 1990, 64, 4043–4050. [Google Scholar] [CrossRef] [PubMed]
- Yanagawa, S.; Kakimi, K.; Tanaka, H.; Murakami, A.; Nakagawa, Y.; Kubo, Y.; Yamada, Y.; Hiai, H.; Kuribayashi, K.; Masuda, T. Mouse mammary tumor virus with rearranged long terminal repeats causes murine lymphomas. J. Virol. 1993, 67, 112–118. [Google Scholar] [CrossRef]
- Wang, Y.; Holland, J.F.; Bleiweiss, I.J.; Melana, S.; Liu, X.; Pelisson, I.; Cantarella, A.; Stellrecht, K.; Mani, S.; Pogo, B.G. Detection of mammary tumor virus env gene-like sequences in human breast cancer. Cancer Res. 1995, 55, 5173–5179. [Google Scholar]
- Holland, J.F.; Pogo, B.G.T. Mouse Mammary Tumor Virus-Like Viral Infection and Human Breast Cancer. Clin. Cancer Res. 2004, 10, 5647–5649. [Google Scholar] [CrossRef]
- Lawson, J.S.; Glenn, W.K.; Salmons, B.; Ye, Y.; Heng, B.; Moody, P.; Johal, H.; Rawlinson, W.D.; Delprado, W.; Lutze-Mann, L.; et al. Mouse Mammary Tumor Virus–like Sequences in Human Breast Cancer. Cancer Res. 2010, 70, 3576–3585. [Google Scholar] [CrossRef]
- Mazzanti, C.M.; Al Hamad, M.; Fanelli, G.; Scatena, C.; Zammarchi, F.; Zavaglia, K.; Lessi, F.; Pistello, M.; Naccarato, A.G.; Bevilacqua, G. A Mouse Mammary Tumor Virus env-Like Exogenous Sequence Is Strictly Related to Progression of Human Sporadic Breast Carcinoma. Am. J. Pathol. 2011, 179, 2083–2090. [Google Scholar] [CrossRef]
- Moore, R.; Dixon, M.; Smith, R.; Peters, G.; Dickson, C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: Two frameshift suppression events are required for translation of gag and pol. J. Virol. 1987, 61, 480–490. [Google Scholar] [CrossRef]
- Hizi, A.; Henderson, L.E.; Copeland, T.D.; Sowder, R.C.; Hixson, C.V.; Oroszlan, S. Characterization of mouse mammary tumor virus gag-pro gene products and the ribosomal frameshift site by protein sequencing. Proc. Natl. Acad. Sci. USA 1987, 84, 7041–7045. [Google Scholar] [CrossRef] [PubMed]
- Jacks, T.; Townsley, K.; Varmus, H.E.; Majors, J. Two efficient ribosomal frameshifting events are required for synthesis of mouse mammary tumor virus gag-related polyproteins. Proc. Natl. Acad. Sci. USA 1987, 84, 4298–4302. [Google Scholar] [CrossRef] [PubMed]
- Hook, L.M.; Agafonova, Y.; Ross, S.R.; Turner, S.J.; Golovkina, T.V. Genetics of Mouse Mammary Tumor Virus-Induced Mammary Tumors: Linkage of Tumor Induction to the gag Gene. J. Virol. 2000, 74, 8876–8883. [Google Scholar] [CrossRef] [PubMed]
- Burzyn, D.; Rassa, J.C.; Kim, D.; Nepomnaschy, I.; Ross, S.R.; Piazzon, I. Toll-Like Receptor 4-Dependent Activation of Dendritic Cells by a Retrovirus. J. Virol. 2004, 78, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Cardiff, R.D.; Munn, R.J. Comparative pathology of mammary tumorigenesis in transgenic mice. Cancer Lett. 1995, 90, 13–19. [Google Scholar] [CrossRef]
- Held, W.; Acha-Orbea, H.; MacDonald, H.; Waanders, G.A. Superantigens and retroviral infection: Insights from mouse mammary tumor virus. Immunol. Today 1994, 15, 184–190. [Google Scholar] [CrossRef]
- Matsuzawa, A.; Nakano, H.; Yoshimoto, T.; Sayama, K. Biology of mouse mammary tumor virus (MMTV). Cancer Lett. 1995, 90, 3–11. [Google Scholar] [CrossRef]
- Parisi, F.; Freer, G.; Mazzanti, C.M.; Pistello, M.; Poli, A. Mouse Mammary Tumor Virus (MMTV) and MMTV-like Viruses: An In-depth Look at a Controversial Issue. Viruses 2022, 14, 977. [Google Scholar] [CrossRef]
- Ross, S.R. Mouse Mammary Tumor Virus Molecular Biology and Oncogenesis. Viruses 2010, 2, 2000–2012. [Google Scholar] [CrossRef]
- Al Ahmad, M.; Akhlaq, S.; Rizvi, T.A.; Mustafa, F. Detection of Mouse Mammary Tumor Virus (MMTV) Particles in an Immortalized T Cell Line Based on Electrical Parameters. IEEE Access 2018, 6, 63597–63605. [Google Scholar] [CrossRef]
- Schulman, H.M.; Ponka, P.; Wilczynska, A.; Gauthier, Y.; Shyamala, G. Transferrin receptor and ferritin levels during murine mammary gland development. Biochim. Biophys. Acta 1989, 1010, 1–6. [Google Scholar] [CrossRef]
- Futran, J.; Kemp, J.D.; Field, E.; Vora, A.; Ashman, R.F. Transferrin receptor synthesis is an early event in B cell activation. J. Immunol. 1989, 143, 787–792. [Google Scholar] [PubMed]
- Brekelmans, P.; Van Soest, P.; Voerman, J.; Platenburg, P.P.; Leenen, P.J.; Van Ewijk, W. Transferrin Receptor Expression as a Marker of Immature Cycling Thymocytes in the Mouse. Cell. Immunol. 1994, 159, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, F.; Al Amri, D.; Al Ali, F.; Al Sari, N.; Al Suwaidi, S.; Jayanth, P.; Philips, P.S.; Rizvi, T.A. Sequences within Both the 5′ UTR and Gag Are Required for Optimal In Vivo Packaging and Propagation of Mouse Mammary Tumor Virus (MMTV) Genomic RNA. PLoS ONE 2012, 7, e47088. [Google Scholar] [CrossRef] [PubMed]
- Aktar, S.J.; Vivet-Boudou, V.; Ali, L.M.; Jabeen, A.; Kalloush, R.M.; Richer, D.; Mustafa, F.; Marquet, R.; Rizvi, T.A. Structural basis of genomic RNA (gRNA) dimerization and packaging determinants of mouse mammary tumor virus (MMTV). Retrovirology 2014, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Mustafa, F.; Vivet-Boudou, V.; Jabeen, A.; Ali, L.M.; Kalloush, R.M.; Marquet, R.; Rizvi, T.A. The bifurcated stem loop 4 (SL4) is crucial for efficient packaging of mouse mammary tumor virus (MMTV) genomic RNA. RNA Biol. 2018, 15, 1047–1059. [Google Scholar] [CrossRef]
- Chameettachal, A.; Vivet-Boudou, V.; Pitchai, F.N.N.; Pillai, V.N.; Ali, L.M.; Krishnan, A.; Bernacchi, S.; Mustafa, F.; Marquet, R.; Rizvi, T.A. A purine loop and the primer binding site are critical for the selective encapsidation of mouse mammary tumor virus genomic RNA by Pr77Gag. Nucleic Acids Res. 2021, 49, 4668–4688. [Google Scholar] [CrossRef]
- Melana, S.M.; Nepomnaschy, I.; Sakalian, M.; Abbott, A.; Hasa, J.; Holland, J.F.; Pogo, B.G. Characterization of Viral Particles Isolated from Primary Cultures of Human Breast Cancer Cells. Cancer Res. 2007, 67, 8960–8965. [Google Scholar] [CrossRef]
- Melana, S.M.; Nepomnaschy, I.; Hasa, J.; Djougarian, A.; Djougarian, A.; Holland, J.F.; Pogo, B.G. Detection of human mammary tumor virus proteins in human breast cancer cells. J. Virol. Methods 2010, 163, 157–161. [Google Scholar] [CrossRef]
- Etkind, P.; Du, J.; Khan, A.; Pillitteri, J.; Wiernik, P.H. Mouse mammary tumor virus-like ENV gene sequences in human breast tumors and in a lymphoma of a breast cancer patient. Clin. Cancer Res. 2000, 6, 1273–1278. [Google Scholar]
- Etkind, P.R.; Stewart, A.F.R.; Dorai, T.; Purcell, D.J.; Wiernik, P.H. Clonal Isolation of Different Strains of Mouse Mammary Tumor Virus-Like DNA Sequences from Both the Breast Tumors and Non-Hodgkin’s Lymphomas of Individual Patients Diagnosed with Both Malignancies. Clin. Cancer Res. 2004, 10, 5656–5664. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Indik, S.; Günzburg, W.H.; Salmons, B.; Rouault, F.; de Cicco, R.L.; Bassi, D.E.; Zucker, S.; Seidah, N.G.; Klein-Szanto, A.J. Mouse Mammary Tumor Virus Infects Human Cells. Cancer Res. 2005, 65, 6651–6659. [Google Scholar] [CrossRef]
- Indik, S.; Günzburg, W.H.; Kulich, P.; Salmons, B.; Rouault, F. Rapid spread of mouse mammary tumor virus in cultured human breast cells. Retrovirology 2007, 4, 73. [Google Scholar] [CrossRef] [PubMed]
- Konstantoulas, C.J.; Lamp, B.; Rumenapf, T.H.; Indik, S. Single amino acid substitution (G42E) in the receptor binding domain of mouse mammary tumour virus envelope protein facilitates infection of non-murine cells in a transferrin receptor 1-independent manner. Retrovirology 2015, 12, 43. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mazzanti, C.M.; Lessi, F.; Armogida, I.; Zavaglia, K.; Franceschi, S.; AI Hamad, M.; Roncella, M.; Ghilli, M.; Boldrini, A.; Aretini, P.; et al. Human saliva as route of inter-human infection for mouse mammary tumor virus. Oncotarget 2015, 6, 18355–18363. [Google Scholar] [CrossRef]
- Levine, P.H.; Pogo, B.G.-T.; Klouj, A.; Coronel, S.; Woodson, K.; Melana, S.M.; Mourali, N.; Holland, J.F. Increasing evidence for a human breast carcinoma virus with geographic differences. Cancer 2004, 101, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.-L.; Zhang, X.-L.; Yang, M.; Lin, J.; Yue, Y.-F.; Li, Y.-D.; Wang, X.; Shu, Q.; Jin, H.-C. Prevalence and characteristics of mouse mammary tumor virus-like virus associated breast cancer in China. Infect. Agents Cancer 2021, 16, 47. [Google Scholar] [CrossRef]
- Stewart, T.H.M.; Sage, R.D.; Stewart, A.; Cameron, D.W. Breast cancer incidence highest in the range of one species of house mouse, Mus domesticus. Br. J. Cancer 2000, 82, 446–451. [Google Scholar] [CrossRef]
- Stewart, A.F.R.; Chen, H.-H. Revisiting the MMTV Zoonotic Hypothesis to Account for Geographic Variation in Breast Cancer Incidence. Viruses 2022, 14, 559. [Google Scholar] [CrossRef]
- Tang, K.-W.; Larsson, E. Tumour virology in the era of high-throughput genomics. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160265. [Google Scholar] [CrossRef]
- Amarante, M.K.; Pereira, N.D.S.; Vitiello, G.A.F.; Watanabe, M.A.E. Involvement of a mouse mammary tumor virus (MMTV) homologue in human breast cancer: Evidence for, against and possible causes of controversies. Microb. Pathog. 2019, 130, 283–294. [Google Scholar] [CrossRef]
- Zapatka, M.; Pathogens, P.; Borozan, I.; Brewer, D.S.; Iskar, M.; Grundhoff, A.; Alawi, M.; Desai, N.; Sültmann, H.; Moch, H.; et al. The landscape of viral associations in human cancers. Nat. Genet. 2020, 52, 320–330. [Google Scholar] [CrossRef] [PubMed]
- Mason, A.; Zhang, G. Linking human beta retrovirus infection with primary biliary cirrhosis. Gastroenterol. Clin. Biol. 2010, 34, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Sharon, D.; Mason, A.L. Role of novel retroviruses in chronic liver disease: Assessing the link of human betaretrovirus with primary biliary cirrhosis. Curr. Infect. Dis. Rep. 2015, 17, 460. [Google Scholar] [CrossRef]
- Mason, A.L. Is PBC a viral infectious disease? Best Pract. Res. Clin. Gastroenterol. 2018, 34–35, 27–39. [Google Scholar] [CrossRef]
- Bevilacqua, G. The Viral Origin of Human Breast Cancer: From the Mouse Mammary Tumor Virus (MMTV) to the Human Betaretrovirus (HBRV). Viruses 2022, 14, 1704. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M. Viral Xenogenization of Intact Tumor Cells. Adv. Cancer Res. 1979, 30, 279–299. [Google Scholar] [CrossRef]
- Hochman, J.; Katz, A.; Levy, E.; Eshel, S. Substrate-adhering lymphoid cells show impaired tumorigenicity and increased immunogenicity. Nature 1981, 290, 248–249. [Google Scholar] [CrossRef]
- Hochman, J.; Levy, E.; Mador, N.; Gottesman, M.M.; Shearer, G.M.; Okon, E. Cell adhesiveness is related to tumorigenicity in malignant lymphoid cells. J. Cell Biol. 1984, 99, 1282–1288. [Google Scholar] [CrossRef]
- Hochman, J.; Mador, N.; Panet, A. Tubular structures in S49 mouse lymphoma are regulated through in vivo host-cell interaction and in vitro interferon treatment. J. Cell Biol. 1985, 100, 1351–1356. [Google Scholar] [CrossRef]
- Mador, N.; Falk, H.; Bergel, M.; Panet, A.; Hochman, J. Variant mouse lymphoma cells with modified response to interferon demonstrate enhanced immunogenicity. Cancer Immunol. Immunother. 1997, 44, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Hochman, J.; Park, S.S.; Lazarovici, P.; Bergel, M.; Gottesman, M.M. Monoclonal Antibodies to Immunogenic Lymphoma Cell Variants Displaying Impaired Neoplastic Properties: Characterization and Applications. J. Natl. Cancer Inst. 1990, 82, 1821–1826. [Google Scholar] [CrossRef] [PubMed]
- Hoch-Marchaim, H.; Hasson, T.; Rorman, E.; Cohen, S.; Hochman, J. Nucleolar Localization of Mouse Mammary Tumor Virus Proteins in T-Cell Lymphomas. Virology 1998, 242, 246–254. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hoch-Marchaim, H.; Weiss, A.M.; Bar-Sinai, A.; Fromer, M.; Adermann, K.; Hochman, J. The leader peptide of MMTV Env precursor localizes to the nucleoli in MMTV-derived T cell lymphomas and interacts with nucleolar protein B23. Virology 2003, 313, 22–32. [Google Scholar] [CrossRef]
- Bar-Sinai, A.; Bassa, N.; Fischette, M.; Gottesman, M.M.; Love, D.C.; Hanover, J.A.; Hochman, J. Mouse Mammary Tumor Virus Env–Derived Peptide Associates with Nucleolar Targets in Lymphoma, Mammary Carcinoma, and Human Breast Cancer. Cancer Res. 2005, 65, 7223–7230. [Google Scholar] [CrossRef][Green Version]
- Mertz, J.A.; Simper, M.S.; Lozano, M.M.; Payne, S.M.; Dudley, J.P. Mouse Mammary Tumor Virus Encodes a Self-Regulatory RNA Export Protein and Is a Complex Retrovirus. J. Virol. 2005, 79, 14737–14747. [Google Scholar] [CrossRef]
- Indik, S.; Günzburg, W.H.; Salmons, B.; Rouault, F. A novel, mouse mammary tumor virus encoded protein with Rev-like properties. Virology 2005, 337, 1–6. [Google Scholar] [CrossRef]
- Redmond, S.; Dickson, C. Sequence and expression of the mouse mammary tumour virus env gene. EMBO J. 1983, 2, 125–131. [Google Scholar] [CrossRef]
- Byun, H.; Halani, N.; Mertz, J.A.; Ali, A.F.; Lozano, M.M.; Dudley, J.P. Retroviral Rem protein requires processing by signal peptidase and retrotranslocation for nuclear function. Proc. Natl. Acad. Sci. USA 2010, 107, 12287–12292. [Google Scholar] [CrossRef]
- Byun, H.; Halani, N.; Gou, Y.; Nash, A.K.; Lozano, M.M.; Dudley, J.P. Requirements for Mouse Mammary Tumor Virus Rem Signal Peptide Processing and Function. J. Virol. 2012, 86, 214–225. [Google Scholar] [CrossRef][Green Version]
- Byun, H.; Das, P.; Yu, H.; Aleman, A.; Lozano, M.M.; Matouschek, A.; Dudley, J.P. Mouse Mammary Tumor Virus Signal Peptide Uses a Novel p97-Dependent and Derlin-Independent Retrotranslocation Mechanism to Escape Proteasomal Degradation. mBio 2017, 8, e00328-17. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.K.; Gou, Y.; Lozano, M.M.; Dudley, J.P. Unconventional p97/VCP-Mediated Endoplasmic Reticulum-to-Endosome Trafficking of a Retroviral Protein. J. Virol. 2021, 95, e00531-21. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Xu, W.K.; Gautam, A.K.S.; Lozano, M.M.; Dudley, J.P. A Retrotranslocation Assay That Predicts Defective VCP/p97-Mediated Trafficking of a Retroviral Signal Peptide. mBio 2022, 13, e02953-21. [Google Scholar] [CrossRef] [PubMed]
- Dultz, E.; Hildenbeutel, M.; Martoglio, B.; Hochman, J.; Dobberstein, B.; Kapp, K. The Signal Peptide of the Mouse Mammary Tumor Virus Rem Protein Is Released from the Endoplasmic Reticulum Membrane and Accumulates in Nucleoli. J. Biol. Chem. 2008, 283, 9966–9976. [Google Scholar] [CrossRef]
- Blobel, G. Intracellular protein topogenesis. Proc. Natl. Acad. Sci. USA 1980, 77, 1496–1500. [Google Scholar] [CrossRef]
- Walter, P.; Blobel, G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 1980, 77, 7112–7116. [Google Scholar] [CrossRef]
- Meyer, D.I.; Dobberstein, B. Identification and characterization of a membrane component essential for the translocation of nascent proteins across the membrane of the endoplasmic reticulum. J. Cell Biol. 1980, 87, 503–508. [Google Scholar] [CrossRef]
- Martoglio, B.; Dobberstein, B. Signal sequences: More than just greasy peptides. Trends Cell Biol. 1998, 8, 410–415. [Google Scholar] [CrossRef]
- Eichler, R.; Lenz, O.; Strecker, T.; Eickmann, M.; Klenk, H.-D.; Garten, W. Identification of Lassa virus glycoprotein signal peptide as a trans-acting maturation factor. EMBO Rep. 2003, 4, 1084–1088. [Google Scholar] [CrossRef]
- Eichler, R.; Lenz, O.; Strecker, T.; Garten, W. Signal peptide of Lassa virus glycoprotein GP-C exhibits an unusual length. FEBS Lett. 2003, 538, 203–206. [Google Scholar] [CrossRef]
- Agnihothram, S.S.; York, J.; Nunberg, J.H. Role of the Stable Signal Peptide and Cytoplasmic Domain of G2 in Regulating Intracellular Transport of the Junín Virus Envelope Glycoprotein Complex. J. Virol. 2006, 80, 5189–5198. [Google Scholar] [CrossRef] [PubMed]
- York, J.; Nunberg, J.H. Role of the Stable Signal Peptide of Junín Arenavirus Envelope Glycoprotein in pH-Dependent Membrane Fusion. J. Virol. 2006, 80, 7775–7780. [Google Scholar] [CrossRef] [PubMed]
- Froeschke, M.; Basler, M.; Groettrup, M.; Dobberstein, B. Long-lived Signal Peptide of Lymphocytic Choriomeningitis Virus Glycoprotein pGP-C. J. Biol. Chem. 2003, 278, 41914–41920. [Google Scholar] [CrossRef] [PubMed]
- Mclauchlan, J.; Lemberg, M.K.; Hope, G.; Martoglio, B. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J. 2002, 21, 3980–3988. [Google Scholar] [CrossRef]
- Ait-Goughoulte, M.; Hourioux, C.; Patient, R.; Trassard, S.; Brand, D.; Roingeard, P. Core protein cleavage by signal peptide peptidase is required for hepatitis C virus-like particle assembly. J. Gen. Virol. 2006, 87, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Hourioux, C.; Ait-Goughoulte, M.; Patient, R.; Fouquenet, D.; Arcanger-Doudet, F.; Brand, D.; Martin, A.; Roingeard, P. Core protein domains involved in hepatitis C virus-like particle assembly and budding at the endoplasmic reticulum membrane. Cell. Microbiol. 2006, 9, 1014–1027. [Google Scholar] [CrossRef]
- Lemberg, M.K.; Bland, F.A.; Weihofen, A.; Braud, V.M.; Martoglio, B. Intramembrane Proteolysis of Signal Peptides: An Essential Step in the Generation of HLA-E Epitopes. J. Immunol. 2001, 167, 6441–6446. [Google Scholar] [CrossRef]
- Prod’Homme, V.; Tomasec, P.; Cunningham, C.; Lemberg, M.K.; Stanton, R.; McSharry, B.; Wang, E.; Cuff, S.; Martoglio, B.; Davison, A.J.; et al. Human Cytomegalovirus UL40 Signal Peptide Regulates Cell Surface Expression of the NK Cell Ligands HLA-E and gpUL18. J. Immunol. 2012, 188, 2794–2804. [Google Scholar] [CrossRef]
- Quinan, B.R.; Flesch, I.E.; Pinho, T.M.; Coelho, F.M.; Tscharke, D.C.; da Fonseca, F.G. An intact signal peptide on dengue virus E protein enhances immunogenicity for CD8+ T cells and antibody when expressed from modified vaccinia Ankara. Vaccine 2014, 32, 2972–2979. [Google Scholar] [CrossRef]
- Subramanian, R.P.; Wildschutte, J.H.; Russo, C.; Coffin, J.M. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology 2011, 8, 90. [Google Scholar] [CrossRef]
- Ruggieri, A.; Maldener, E.; Sauter, M.; Mueller-Lantzsch, N.; Meese, E.; Fackler, O.T.; Mayer, J. Human endogenous retrovirus HERV-K(HML-2) encodes a stable signal peptide with biological properties distinct from Rec. Retrovirology 2009, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Armezzani, A.; Arnaud, F.; Caporale, M.; di Meo, G.; Iannuzzi, L.; Murgia, C.; Palmarini, M. The Signal Peptide of a Recently Integrated Endogenous Sheep Betaretrovirus Envelope Plays a Major Role in Eluding Gag-Mediated Late Restriction. J. Virol. 2011, 85, 7118–7128. [Google Scholar] [CrossRef] [PubMed]
- Caporale, M.; Arnaud, F.; Mura, M.; Golder, M.; Murgia, C.; Palmarini, M. The Signal Peptide of a Simple Retrovirus Envelope Functions as a Posttranscriptional Regulator of Viral Gene Expression. J. Virol. 2009, 83, 4591–4604. [Google Scholar] [CrossRef]
- Civita, P.; Menicagli, M.; Scopelliti, C.; Lessi, F.; Millanta, F.; Borsacchi, S.; Parisi, F.; Freer, G.; Pistello, M.; Mazzanti, C.M.; et al. Mouse mammary tumour virus-like env nucleotide and p14 signal peptide are present in feline mammary carcinomas, but not in neoplastic or dysplastic canine mammary lesions. PLoS ONE 2018, 13, e0200839. [Google Scholar] [CrossRef]
- Braitbard, O.; Roniger, M.; Bar-Sinai, A.; Rajchman, D.; Gross, T.; Abramovitch, H.; La Ferla, M.; Franceschi, S.; Lessi, F.; Naccarato, A.G.; et al. A new immunization and treatment strategy for mouse mammary tumor virus (MMTV) associated cancers. Oncotarget 2016, 7, 21168–21180. [Google Scholar] [CrossRef]
- Lawson, J.S.; Mazzanti, C.; Civita, P.; Menicagli, M.; Ngan, C.C.; Whitaker, N.J.; Hochman, J.; Braitbard, O.; Yosufi, B.; Glenn, W.K. Association of Mouse Mammary Tumor Virus with Human Breast Cancer: Histology, Immunohistochemistry and Polymerase Chain Reaction Analyses. Front. Oncol. 2018, 8, 141. [Google Scholar] [CrossRef] [PubMed]
- Feldman, D.; Roniger, M.; Bar-Sinai, A.; Braitbard, O.; Natan, C.; Love, D.C.; Hanover, J.A.; Hochman, J. The Signal Peptide of Mouse Mammary Tumor Virus-Env: A Phosphoprotein Tumor Modulator. Mol. Cancer Res. 2012, 10, 1077–1086. [Google Scholar] [CrossRef]
- Katz, E.; Lareef, M.H.; Rassa, J.C.; Grande, S.M.; King, L.B.; Russo, J.; Ross, S.R.; Monroe, J.G. MMTV Env encodes an ITAM responsible for transformation of mammary epithelial cells in three-dimensional culture. J. Exp. Med. 2005, 201, 431–439. [Google Scholar] [CrossRef]
- Naccarato, A.G.; Lessi, F.; Zavaglia, K.; Scatena, C.; Al Hamad, M.A.; Aretini, P.; Menicagli, M.; Roncella, M.; Ghilli, M.; Caligo, M.A.; et al. Mouse mammary tumor virus (MMTV)-like exogenous sequences are associated with sporadic but not hereditary human breast carcinoma. Aging 2019, 11, 7236–7241. [Google Scholar] [CrossRef]
- Parisi, F.; Muscatello, L.V.; Civita, P.; Lessi, F.; Menicagli, M.; Millanta, F.; Brunetti, B.; Benazzi, C.; Sarli, G.; Freer, G.; et al. Pathological Features and Molecular Phenotype of MMTV Like-Positive Feline Mammary Carcinomas. Animals 2021, 11, 2821. [Google Scholar] [CrossRef]
- Parisi, F.; Lessi, F.; Menicagli, M.; Civita, P.; Liotti, R.; Millanta, F.; Freer, G.; Pistello, M.; Mazzanti, C.M.; Poli, A. Presence of a mouse mammary tumour virus-like in feline lymphomas: A preliminary study. Infect. Agents Cancer 2022, 17, 35. [Google Scholar] [CrossRef] [PubMed]
- Roniger, M.; Braitbard, O.; Bar-Sinai, A.; Hochman, N.; Hochman, J. Abstract #367: Use of MMTV-p14 as a vaccine against MMTV-bearing tumors. Cancer Res. 2009, 69, 367. [Google Scholar]
- Stein, S.; Weiss, A.; Adermann, K.; Lazarovici, P.; Hochman, J.; Wellhöner, H. A disulfide conjugate between anti-tetanus antibodies and HIV (37-72)Tat neutralizes tetanus toxin inside chromaffin cells. FEBS Lett. 1999, 458, 383–386. [Google Scholar] [CrossRef]
- Wellhöner, H.; Weiss, A.; Schulz, A.; Adermann, K.; Braitbard, O.; Bar-Sinai, A.; Hochman, J. Reversing ABCB1-mediated multi-drug resistance from within cells using translocating immune conjugates. J. Drug Target. 2012, 20, 445–452. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hochman, J.; Braitbard, O. Life after Cleavage: The Story of a β-Retroviral (MMTV) Signal Peptide—From Murine Lymphoma to Human Breast Cancer. Viruses 2022, 14, 2435. https://doi.org/10.3390/v14112435
Hochman J, Braitbard O. Life after Cleavage: The Story of a β-Retroviral (MMTV) Signal Peptide—From Murine Lymphoma to Human Breast Cancer. Viruses. 2022; 14(11):2435. https://doi.org/10.3390/v14112435
Chicago/Turabian StyleHochman, Jacob, and Ori Braitbard. 2022. "Life after Cleavage: The Story of a β-Retroviral (MMTV) Signal Peptide—From Murine Lymphoma to Human Breast Cancer" Viruses 14, no. 11: 2435. https://doi.org/10.3390/v14112435
APA StyleHochman, J., & Braitbard, O. (2022). Life after Cleavage: The Story of a β-Retroviral (MMTV) Signal Peptide—From Murine Lymphoma to Human Breast Cancer. Viruses, 14(11), 2435. https://doi.org/10.3390/v14112435





