Canine Coronavirus Activates Aryl Hydrocarbon Receptor during In Vitro Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Virus Infection
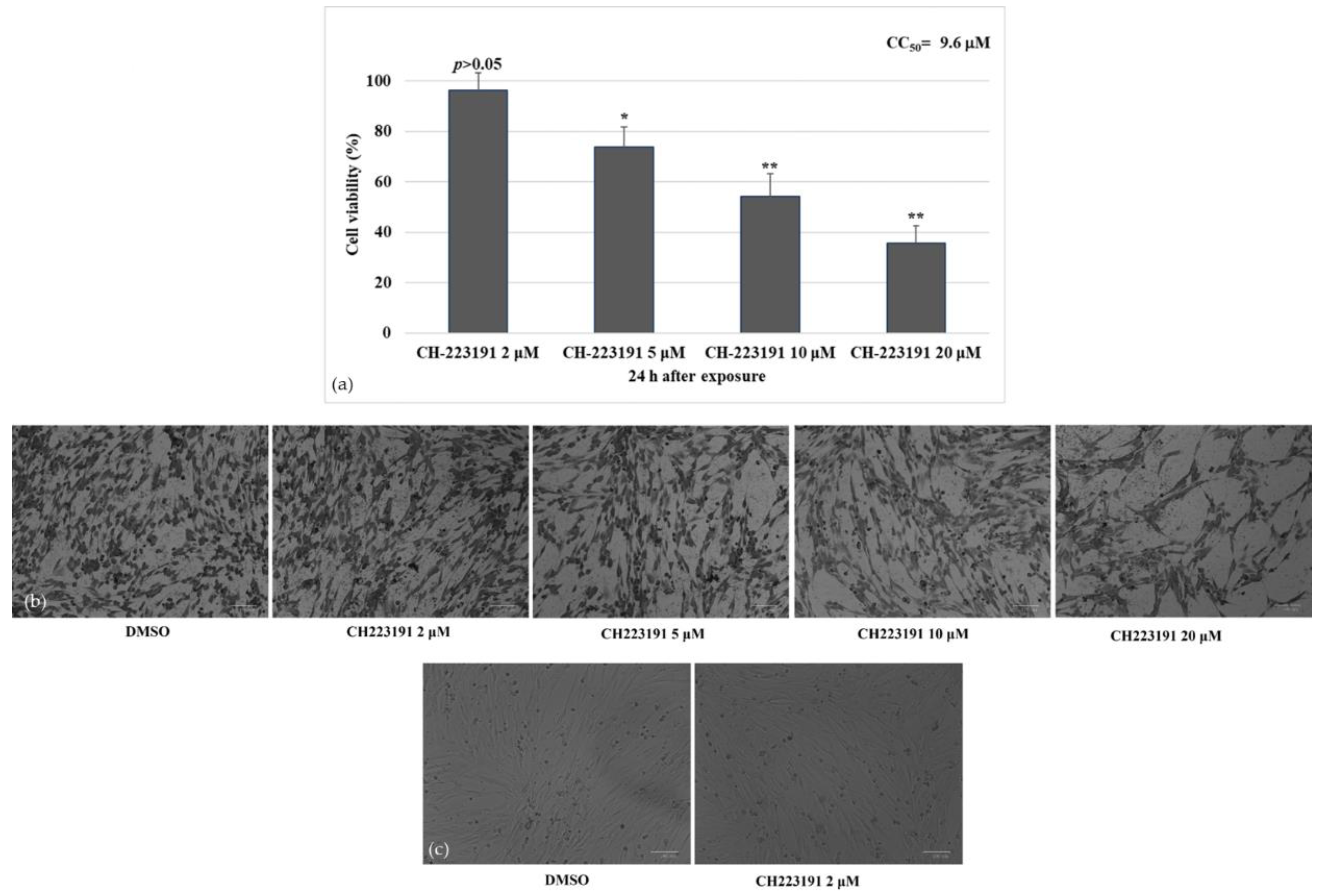
2.2. Cell Viability
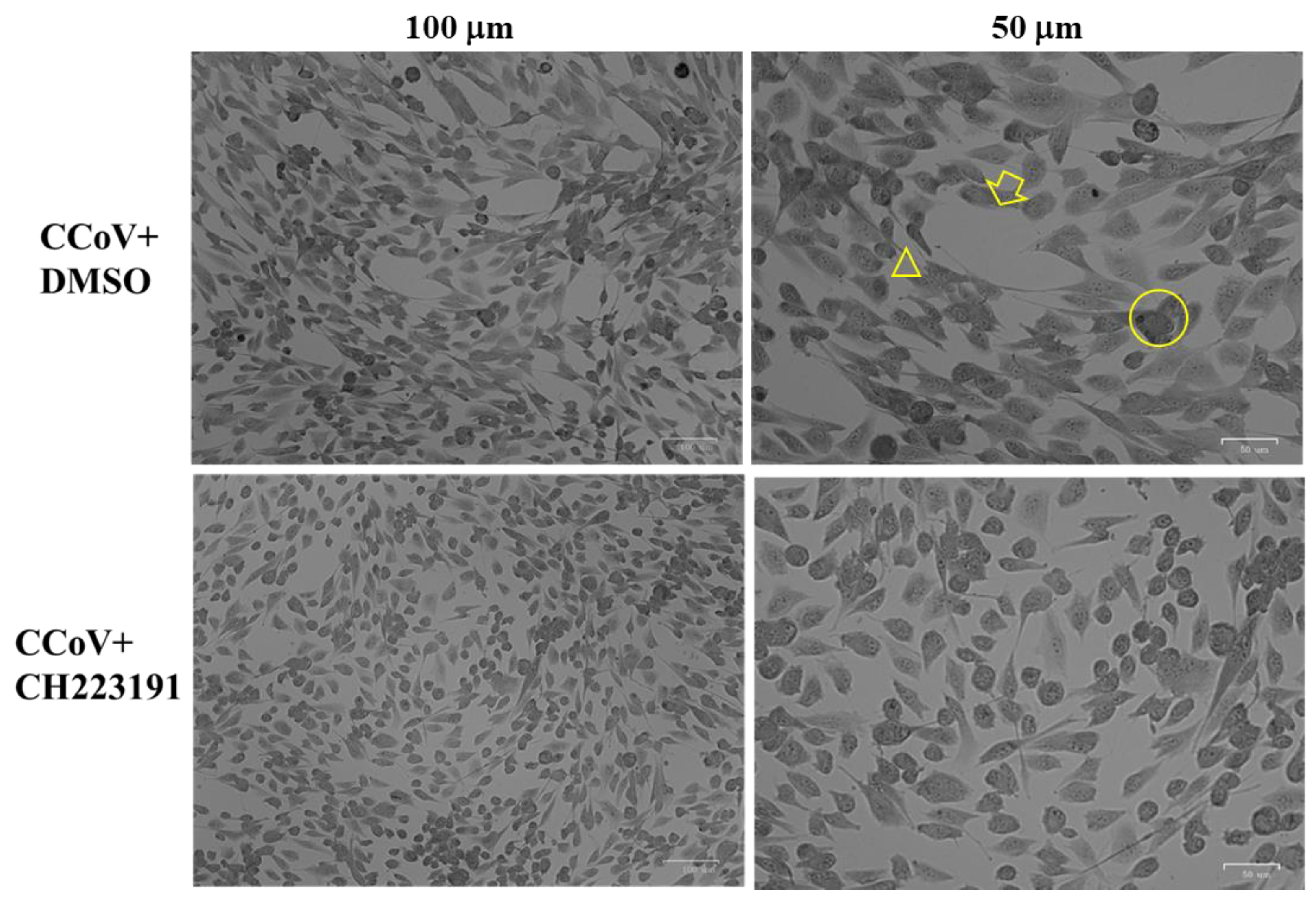
2.3. Examination of Cell Morphology
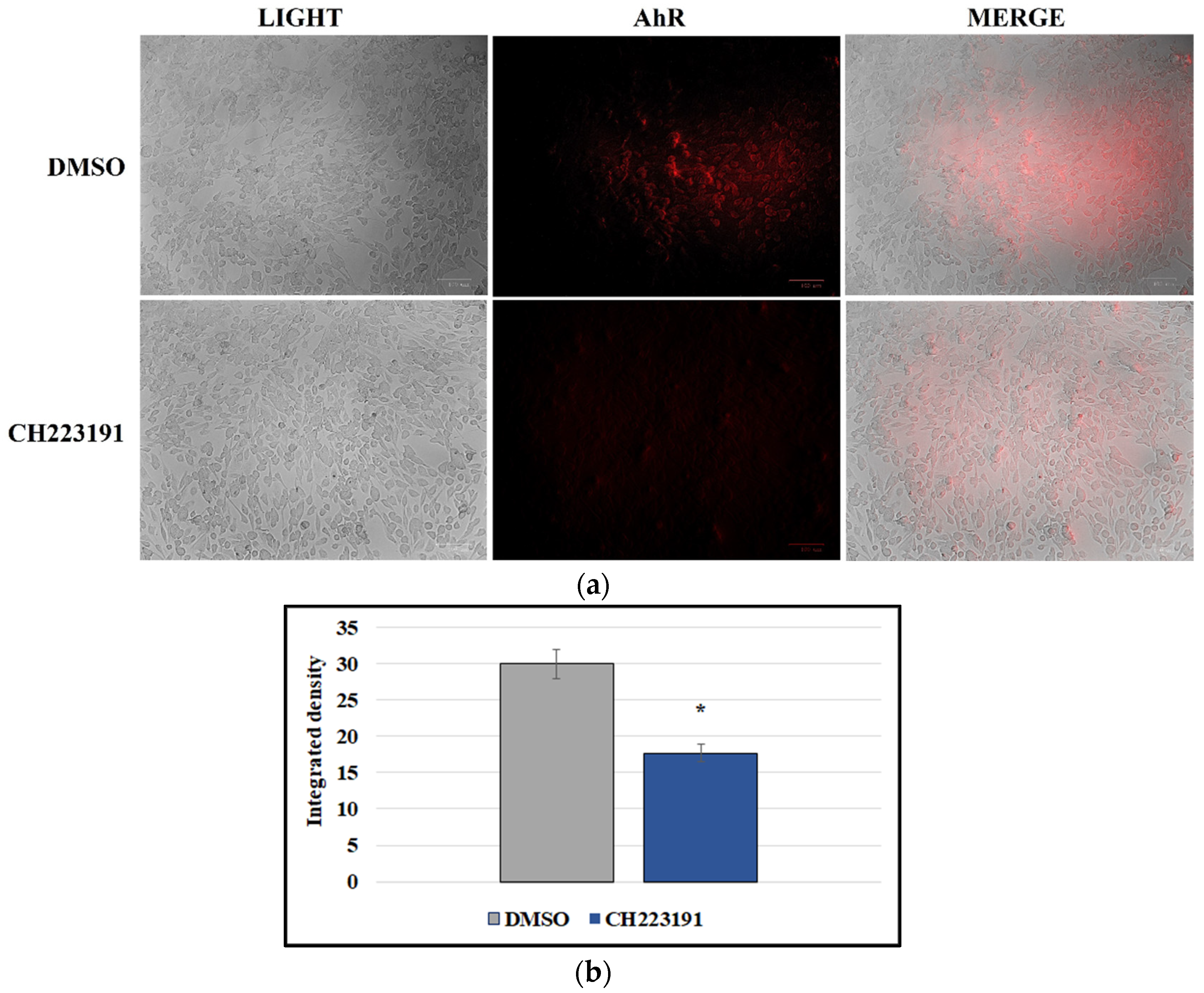
2.4. Immunofluorescence Staining
2.5. Virus Infection
2.6. Viral Nucleic Acids Extraction Procedures
2.7. CCoV Viral Load Quantification by Real-Time Reverse Transcription PCR (RT-qPCR)
2.8. Statistical Analysis
3. Results
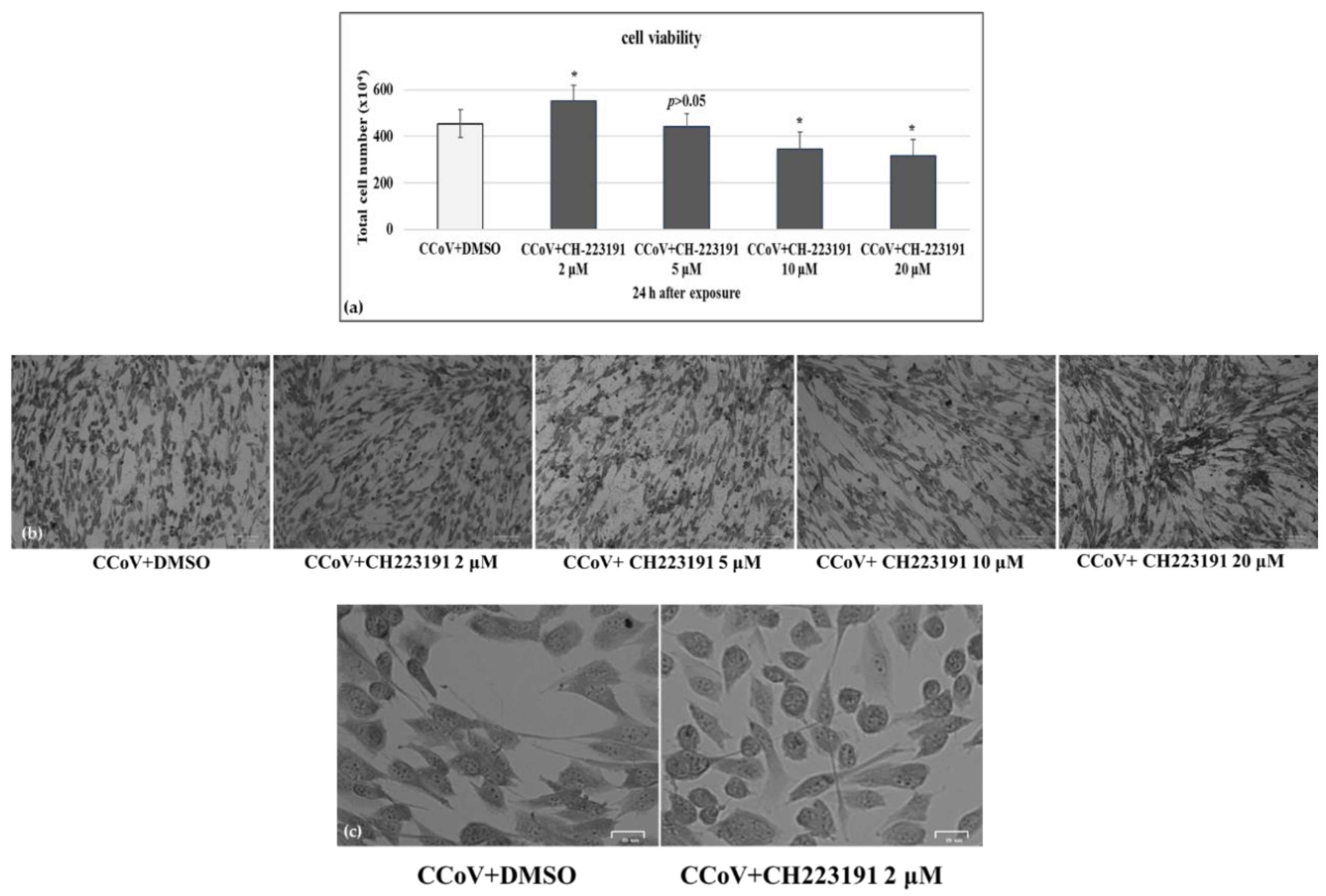
3.1. AhR Inhibitor Increases Cell Viability during CCoV Infection
3.2. AhR Inhibitor Reduces Morphological Cell Death Features during CCoV Infection in A72 Cells
3.3. A72 Cells Express AhR
3.4. CCoV Infection Activates the Expression of AhR in A72 Cells
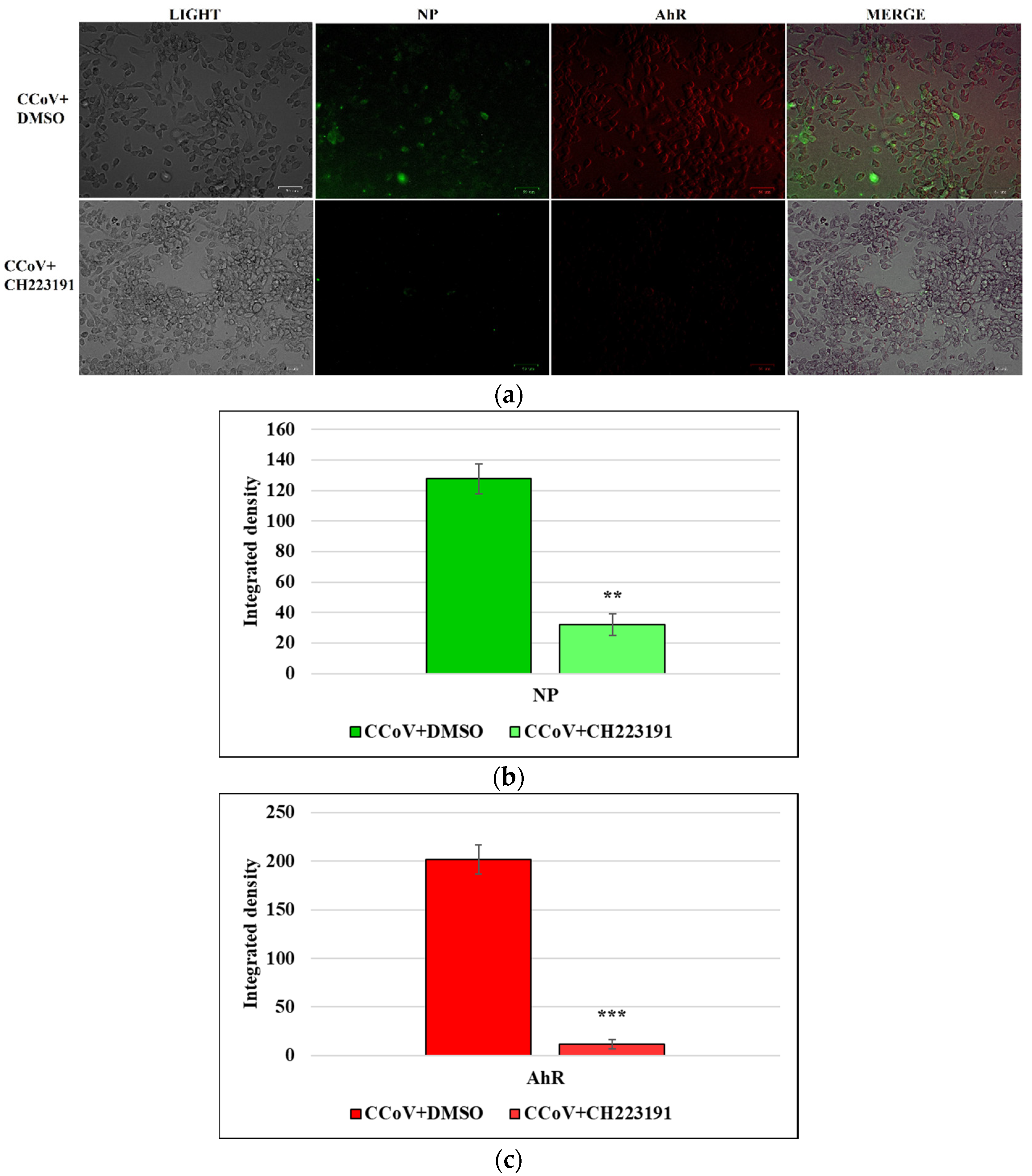
3.5. AhR Inhibitor Inhibits Both AhR and NP Expression during CCoV Infection in A72 Cells
3.6. AhR Inhibitor Decreases Virus Yield during CCoV Infection in A72 Cells
3.6.1. CPE Evaluation
3.6.2. Standard Curve and Virus Quantification
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kenney, S.P.; Wang, Q.; Vlasova, A.; Jung, K.; Saif, L. Naturally Occurring Animal Coronaviruses as Models for Studying Highly Pathogenic Human Coronaviral Disease. Vet. Pathol. 2021, 58, 438–452. [Google Scholar] [CrossRef] [PubMed]
- Timurkan, M.O.; Aydin, K.; Dincer, E.; Coskun, N. Molecular characterization of canine coronaviruses: An enteric and pantropic approach. Arch. Virol. 2021, 166, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Pratelli, A.; Buonavoglia, A.; Lanave, G.; Tempesta, M.; Camero, M.; Martella, V.; Decaro, N. One world, one health, one virology of the mysterious labyrinth of coronaviruses: The canine coronavirus affair. Lancet Microbe 2021, 2, e646–e647. [Google Scholar]
- Pratelli, A.; Tempesta, M.; Elia, G.; Martella, V.; Decaro, N.; Buonavoglia, C. The knotty biology of canine coronavirus: A worrying model of coronaviruses’ danger. Vet. Sci. 2022, 144, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Buonavoglia, C.; Decaro, N.; Martella, V.; Elia, G.; Campolo, M.; Desario, C.; Castagnaro, M.; Tempesta, M. Canine Coronavirus Highly Pathogenic for Dogs. Emerg. Infect. Dis. 2006, 12, 492. [Google Scholar] [CrossRef] [PubMed]
- Decaro, N.; Martella, V.; Elia, G.; Campolo, M.; Desario, C.; Cirone, F.; Tempesta, M.; Buonavoglia, C. Molecular characterisation of the virulent canine coronavirus CB/05 strain. Virus Res. 2007, 125, 54–60. [Google Scholar] [CrossRef]
- Decaro, N.; Mari, V.; Campolo, M.; Lorusso, A.; Camero, M.; Elia, G.; Martella, V.; Cordioli, P.; Enjuanes, L.; Buonavoglia, C. Recombinant canine coronaviruses related to transmissible gastroenteritis virus of swine are circulating in dogs. J. Virol. 2009, 83, 1532–1537. [Google Scholar]
- Decaro, N.; Elia, G.; Martella, V.; Campolo, M.; Mari, V.; Desario, C.; Lucente, M.S.; Lorusso, E.; Kanellos, T.; Gibbons, R.H.; et al. Immunity after natural exposure to enteric canine coronavirus does not provide complete protection against infection with the new pantropic CB/05 strain. Vaccine 2010, 28, 724–729. [Google Scholar]
- Vlasova, A.N.; Diaz, A.; Damtie, D.; Xiu, L.; Toh, T.H.; Lee, J.S.; Saif, L.J.; Gray, G.C. Novel canine coronavirus isolated from a hospitalized pneumonia patient, East Malaysia. Clin. Infect. Dis. 2022, 74, 446–454. [Google Scholar]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution. MedRxiv 2021, 75. [Google Scholar] [CrossRef]
- Vlasova, A.N.; Toh, T.H.; Lee, J.S.Y.; Poovorawan, Y.; Davis, P.; Azevedo, M.S.P.; Lednicky, J.A.; Saif, L.J.; Gray, G.C. Animal alphacoronaviruses found in human patients with acute respiratory illness in different countries. Emerg. Microbes Infect. 2022, 11, 699–702. [Google Scholar] [CrossRef] [PubMed]
- Theamboonlers, A.; Samransamruajkit, R.; Thongme, C.; Amonsin, A.; Chongsrisawat, V.; Poovorawan, Y. Human Coronavirus Infection among Children with Acute Lower Respiratory Tract Infection in Thailand. Intervirology 2007, 50, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.S.; Mullis, L.B.; Pereira, O., Jr.; Saif, L.J.; Vlasova, A.; Zhang, X.; Owens, R.J.; Paulson, D.; Yaylor, D.; Haynes, L.M.; et al. Human respiratory coronaviruses detected in patients with influenza-like illness in Arkansas, USA. Virol. Mycol. Infect. Dis. 2014, 2014 (Suppl. S2), 004. [Google Scholar]
- Yang, T.; Feng, Y.L.; Chen, L.; Vaziri, N.D.; Zhao, Y.Y. Dietary natural flavonoids treating cancer by targeting aryl hydrocarbon receptor. Crit. Rev. Toxicol. 2019, 49, 445–460. [Google Scholar] [CrossRef] [PubMed]
- Torti, M.F.; Giovannoni, F.; Quintana, F.J.; Garcia, C.C. The Aryl Hydrocarbon Receptor as a Modulator of Anti-viral Immunity. Front. Immunol. 2021, 12, 624293. [Google Scholar] [CrossRef]
- Lawrence, B.P.; Vorderstrasse, B.A. New insights into the aryl hydrocarbon receptor as a modulator of host responses to infection. Semin. Immunopathol. 2013, 35, 615–626. [Google Scholar] [CrossRef]
- Gutierrez-Vazquez, C.; Quintana, F.J. Regulation of the immune response by the aryl hydrocarbon receptor. Immunity 2018, 48, 19–33. [Google Scholar] [CrossRef]
- Rothhammer, V.; Quintana, F.J. The aryl hydrocarbon receptor: An environmental sensor integrating immune responses in health and disease. Nat. Rev. Immunol. 2019, 19, 184–197. [Google Scholar] [CrossRef]
- Yamada, T.; Horimoto, H.; Kameyama, T.; Hayakawa, S.; Yamato, H.; Dazai, M.; Takada, A.; Kida, H.; Bott, D.; Zhou, A.C.; et al. Constitutive aryl hydrocarbon receptor signaling constrains type I interferon-mediated antiviral innate defense. Nat. Immunol. 2016, 17, 687–694. [Google Scholar] [CrossRef]
- Giovannoni, F.; Bosch, I.; Polonio, C.M.; Torti, M.F.; Wheeler, M.A.; Li, Z.; Romorini, L.; Rodriguez Varela, M.S.; Rothhammer, V.; Barroso, A.; et al. AHR is a Zika virus host factor and a candidate target for antiviral therapy. Nat. Neurosci. 2020, 23, 939–951. [Google Scholar] [CrossRef]
- Fiorito, F.; Cerracchio, C.; Salvatore, M.M.; Serra, F.; Pucciarelli, A.; Amoroso, M.G.; Nicoletti, R.; Andolfi, A. Antiviral Property of the Fungal Metabolite 3-O-Methylfunicone in Bovine Herpesvirus 1 Infection. Microorganisms 2022, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.S.F.; Chan, K.H.; Cheng, V.C.C.; Woo, P.C.Y.; Lau, S.K.P.; Lam, C.C.K.; Chan, T.L.; Wu, A.K.L.; Hung, I.F.N.; Leung, S.Y.; et al. Comparative Host Gene Transcription by Microarray Analysis Early after Infection of the Huh7 Cell Line by Severe Acute Respiratory Syndrome Coronavirus and Human coronavirus 229E. J. Virol. 2005, 79, 6180–6193. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, M.E.; Shaban, M.G.; Mackin, S.R.; Fehr, A.R.; Perlman, S. Murine coronavirus infection activates the aryl hydrocarbon receptor in an indoleamine 2,3-dioxygenase-independent manner, contributing to cytokine modulation and proviral TCDD-inducible-PARP expression. J. Virol. 2020, 94, 319–335. [Google Scholar] [CrossRef] [PubMed]
- Giovannoni, F.; Li, Z.; Remes-Lenicov, F.; Dávola, M.E.; Elizalde, M.; Paletta, A.; Ashkar, A.A.; Mossman, K.L.; Dugour, A.V.; Figueroa, J.M.; et al. AHR signaling is induced by infection with coronaviruses. Nat. Commun. 2021, 12, 5148. [Google Scholar] [CrossRef]
- Guarnieri, T. Hypothesis: Emerging Roles for Aryl Hydrocarbon Receptor in Orchestrating CoV-2-Related Inflammation. Cells 2022, 11, 648. [Google Scholar] [CrossRef]
- Binn, L.N.; Marchwicki, R.H.; Stephenson, E.H. Establishment of a canine cell line: Derivation, characterization, and viral spectrum. Am. J. Vet. Res. 1980, 41, 855–860. [Google Scholar]
- De Martino, L.; Marfè, G.; Longo, M.; Fiorito, F.; Montagnaro, S.; Iovane, V.; Decaro, N.; Pagnini, U. Bid cleavage, cytochrome c release and caspase activation in canine coronavirus-induced apoptosis. Vet. Microbiol. 2010, 141, 36–45. [Google Scholar] [CrossRef]
- Marfè, G.; Tafani, M.; Fiorito, F.; Pagnini, U.; Iovane, G.; De Martino, L. Involvement of FOXO transcription factors, TRAIL-FasL/Fas, and sirtuin proteins family in canine coronavirus type II-induced apoptosis. PLoS ONE 2011, 6, e27313. [Google Scholar] [CrossRef]
- Kim, S.H.; Henry, E.C.; Kim, D.K.; Kim, Y.H.; Shin, K.J.; Han, M.S.; Lee, T.G.; Kang, J.K.; Gasiewicz, T.A.; Ryu, S.H.; et al. Novel Compound 2-Methyl-2H-pyrazole-3-carboxylic Acid (2-methyl-4-o-tolylazo-phenyl)-amide (CH-223191) Prevents 2,3,7,8-TCDD-Induced Toxicity by Antagonizing the Aryl Hydrocarbon Receptor. Mol. Pharmacol. 2006, 69, 1871–1878. [Google Scholar] [CrossRef]
- Fiorito, F.; Marfè, G.; De Blasio, E.; Granato, G.E.; Tafani, M.; De Martino, L.; Montagnaro, S.; Florio, S.; Pagnini, U. 2,3,7,8-Tetrachlorodibenzo-p-dioxin regulates Bovine Herpesvirus type 1 induced apoptosis by modulating Bcl-2 family members. Apoptosis 2008, 13, 1243–1252. [Google Scholar] [CrossRef]
- Chowanadisai, W.; Graham, D.M.; Keen, C.L.; Rucker, R.B.; Messerli, M.A. Neurulation and neurite extension require the zinc transporter ZIP12 (slc39a12). Proc. Natl. Acad. Sci. USA 2013, 110, 9903–9908. [Google Scholar] [CrossRef] [PubMed]
- Fiorito, F.; Nocera, F.P.; Cantiello, A.; Iovane, V.; Lambiase, S.; Piccolo, M.; Ferraro, M.G.; Santamaria, R.; De Martino, L. Bovine herpesvirus-1 infection in mouse neuroblastoma (Neuro-2A) cells. Vet. Microbiol. 2020, 247, 108762. [Google Scholar] [CrossRef] [PubMed]
- Leite, M.; Quinta-Costa, M.; Leite, P.S.; Guimarães, J.E. Critical evaluation of techniques to detect and measure cell death-Study in a model of UV radiation of the leukaemic cell line HL60. Anal. Cell. Pathol. 1999, 19, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Kroemer, G.; Levine, B. Autophagic cell death: The story of a misnomer. Nat. Rev. Mol. Cell Biol. 2008, 9, 1004–1010. [Google Scholar] [CrossRef]
- Zakeri, Z.; Lockshin, R.A. Cell death: History and future. Adv. Exp. Med. Biol. 2008, 615, 1–11. [Google Scholar]
- Banfalvi, G. Methods to detect apoptotic cell death. Apoptosis 2017, 22, 306–323. [Google Scholar] [CrossRef]
- Altamura, G.; Power, K.; Martano, M.; degli Uberti, B.; Galiero, G.; De Luca, G.; Maiolino, P.; Borzacchiello, G. Felis catus papillomavirus type-2 E6 binds to E6AP, promotes E6AP/p53 binding and enhances p53 proteasomal degradation. Sci. Rep. 2018, 8, 17529. [Google Scholar] [CrossRef]
- Decaro, N.; Mari, V.; Elia, G.; Addie, D.D.; Camero, M.; Lucente, M.S.; Martella, V.; Buonavoglia, C. Recombinant canine coronaviruses in dogs, Europe. Emerg. Infect. Dis. 2010, 16, 41–47. [Google Scholar] [CrossRef]
- Amici, C.; Di Caro, A.; Ciucci, A.; Chiappa, L.; Castilletti, C.; Martella, V.; Decaro, N.; Buonavoglia, C.; Capobianchi, M.R.; Santoro, G.M. Indomethacin has a potent antiviral activity against SARS coronavirus. Antivir. Ther. 2006, 11, 1021–1030. [Google Scholar] [CrossRef]
- Xu, T.; Gao, X.; Wu, Z.; Selinger, D.W.; Zhou, Z. Indomethacin has a potent antiviral activity against SARS CoV-2 in vitro and canine coronavirus in vivo. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gomeni, R.; Xu, T.; Gao, X.; Bressolle-Gomeni, F. Model based approach for estimating the dosage regimen of indomethacin a potential antiviral treatment of patients infected with SARS CoV-2. J. Pharmacokinet. Pharmacodyn. 2020, 47, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Ruggieri, A.; Di Trani, L.; Gatto, I.; Franco, M.; Vignolo, E.; Bedini, B.; Elia, G.; Buonavoglia, C. Canine coronavirus induces apoptosis in cultured cells. Vet. Microbiol. 2007, 121, 64–72. [Google Scholar] [CrossRef] [PubMed]
- McBride, R.; van Zyl, M.; Fielding, B.C. The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 2014, 6, 2991–3018. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Liu, M.; Zhao, W.; Zhang, J.; Zhang, X.; Wang, K.; Gu, C.; Wu, K.; Li, Y.; Zheng, C.; et al. Isolation of virus from a SARS patient and genome-wide analysis of genetic mutations related to pathogenesis and epidemiology from 47 SARS-CoV isolates. Virus Genes 2005, 30, 93–102. [Google Scholar] [CrossRef]
- Dutta, N.K.; Mazumdar, K.; Gordy, J.T.; Dutch, R.E. The nucleocapsid protein of SARSCoV-2: A target for vaccine development. J. Virol. 2020, 94, e00647-20. [Google Scholar] [CrossRef]
- Tseng, Y.Y.; Liao, G.R.; Lien, A.; Hsu, W.L. Current concepts in the development of therapeutics against human and animal coronavirus diseases by targeting NP. Comput. Struct. Biotechnol. J. 2021, 19, 1072–1080. [Google Scholar] [CrossRef]


| Host | Genus | CoVs | Model | Cell Type | References |
|---|---|---|---|---|---|
| Mouse | Betacoronavirus | M-CoV | In vitro | Bone-marrow-derived macrophage | [23] |
| Mouse | Betacoronavirus | M-CoV | In vitro | Bone-marrow-derived dendritic cells | [23] |
| Mouse | Betacoronavirus | M-CoV | In vitro | Liver (C57BL-6 mice) | [23] |
| Mouse | Betacoronavirus | M-CoV | In vitro | Bone-marrow-derived macrophage | [24] |
| Human | Alphacoronavirus | H-CoV-229E | In vitro | Human hepatoma (Huh7) | [22] |
| Human | Alphacoronavirus | H-CoV-229E | In vitro | Human lung adenocarcinoma (A549) | [24] |
| Human | Betacoronavirus | MERS-CoV | In vitro | Human lung adenocarcinoma (Calu-3) | [24] |
| Human | Betacoronavirus | SARS CoV-1 | In vitro | Human hepatoma (Huh7) | [22] |
| Human | Betacoronavirus | SARS-CoV-2 | In vitro | Primary human lung epithelium (NHBE) | [24] |
| Human | Betacoronavirus | SARS-CoV-2 | In vitro | Human lung adenocarcinoma (A549) | [24] |
| Human | Betacoronavirus | SARS-CoV-2 | In vitro | Human lung adenocarcinoma (Calu-3) | [24] |
| Human | Betacoronavirus | SARS-CoV-2 | Patients | Nasal swabs | [24] |
| Dog | Alphacoronavirus | CCoV-IIa | In vitro | Canine Fibrosarcoma (A72) | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cerracchio, C.; Serra, F.; Amoroso, M.G.; Fiorito, F. Canine Coronavirus Activates Aryl Hydrocarbon Receptor during In Vitro Infection. Viruses 2022, 14, 2437. https://doi.org/10.3390/v14112437
Cerracchio C, Serra F, Amoroso MG, Fiorito F. Canine Coronavirus Activates Aryl Hydrocarbon Receptor during In Vitro Infection. Viruses. 2022; 14(11):2437. https://doi.org/10.3390/v14112437
Chicago/Turabian StyleCerracchio, Claudia, Francesco Serra, Maria Grazia Amoroso, and Filomena Fiorito. 2022. "Canine Coronavirus Activates Aryl Hydrocarbon Receptor during In Vitro Infection" Viruses 14, no. 11: 2437. https://doi.org/10.3390/v14112437
APA StyleCerracchio, C., Serra, F., Amoroso, M. G., & Fiorito, F. (2022). Canine Coronavirus Activates Aryl Hydrocarbon Receptor during In Vitro Infection. Viruses, 14(11), 2437. https://doi.org/10.3390/v14112437






