A National Catalogue of Viruses Associated with Indigenous Species Reveals High-Throughput Sequencing as a Driver of Indigenous Virus Discovery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Catalogued Viruses
2.2. Virus Detection Methods
2.3. Designation of Virus Biostatus
2.4. Virus Taxonomic and Physical Metadata
3. Results
3.1. Discovery of Viruses Associated with Aotearoa New Zealand Indigenous Species Is Influenced by Discovery Method
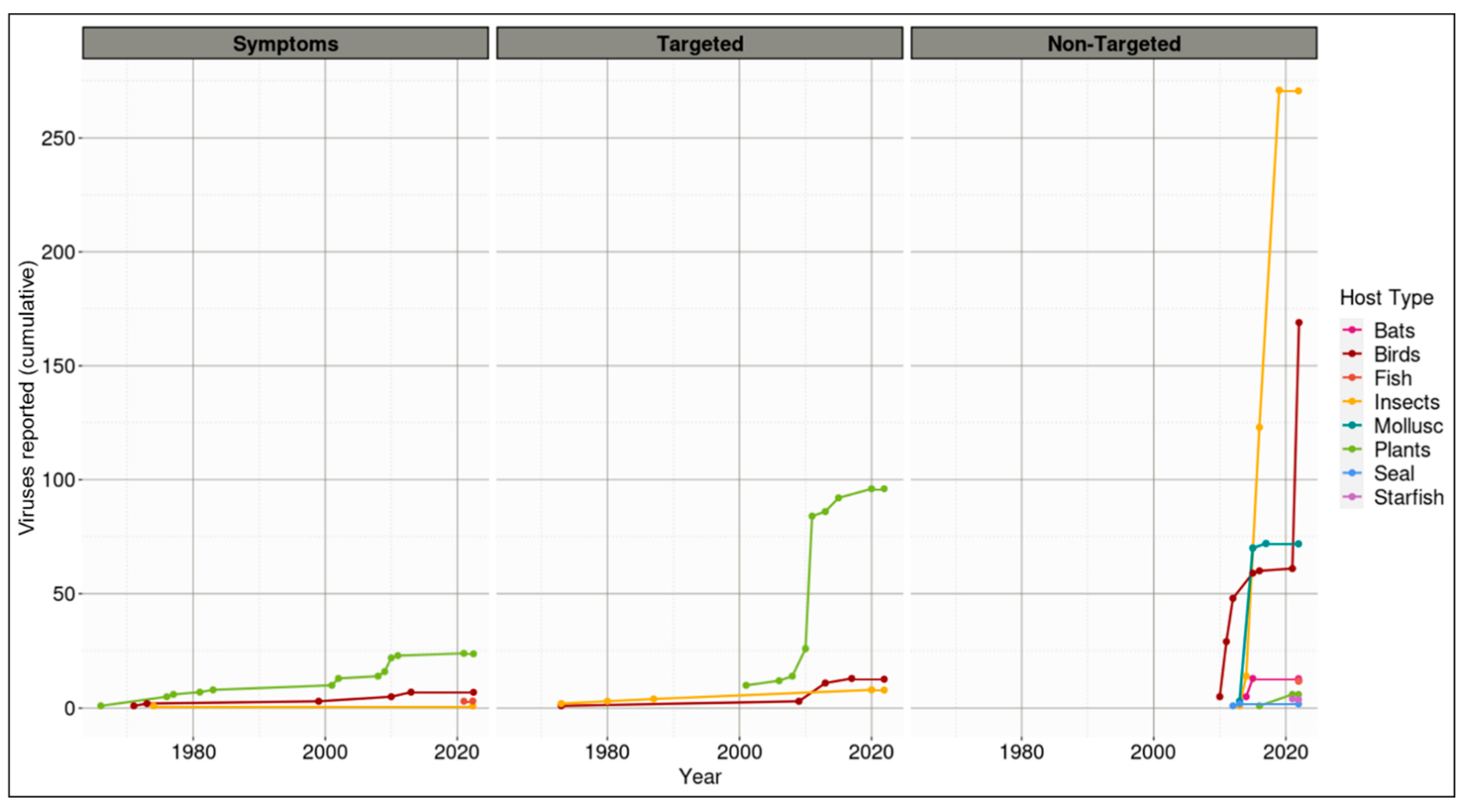
3.2. To Date, Vast Viral Diversity Has Been Described Associated with Aotearoa New Zealand Indigenous Macro- But Not Micro-Flora and Fauna
3.2.1. Plants
3.2.2. Insects
3.2.3. Birds
3.2.4. Bats
3.2.5. Marine Life
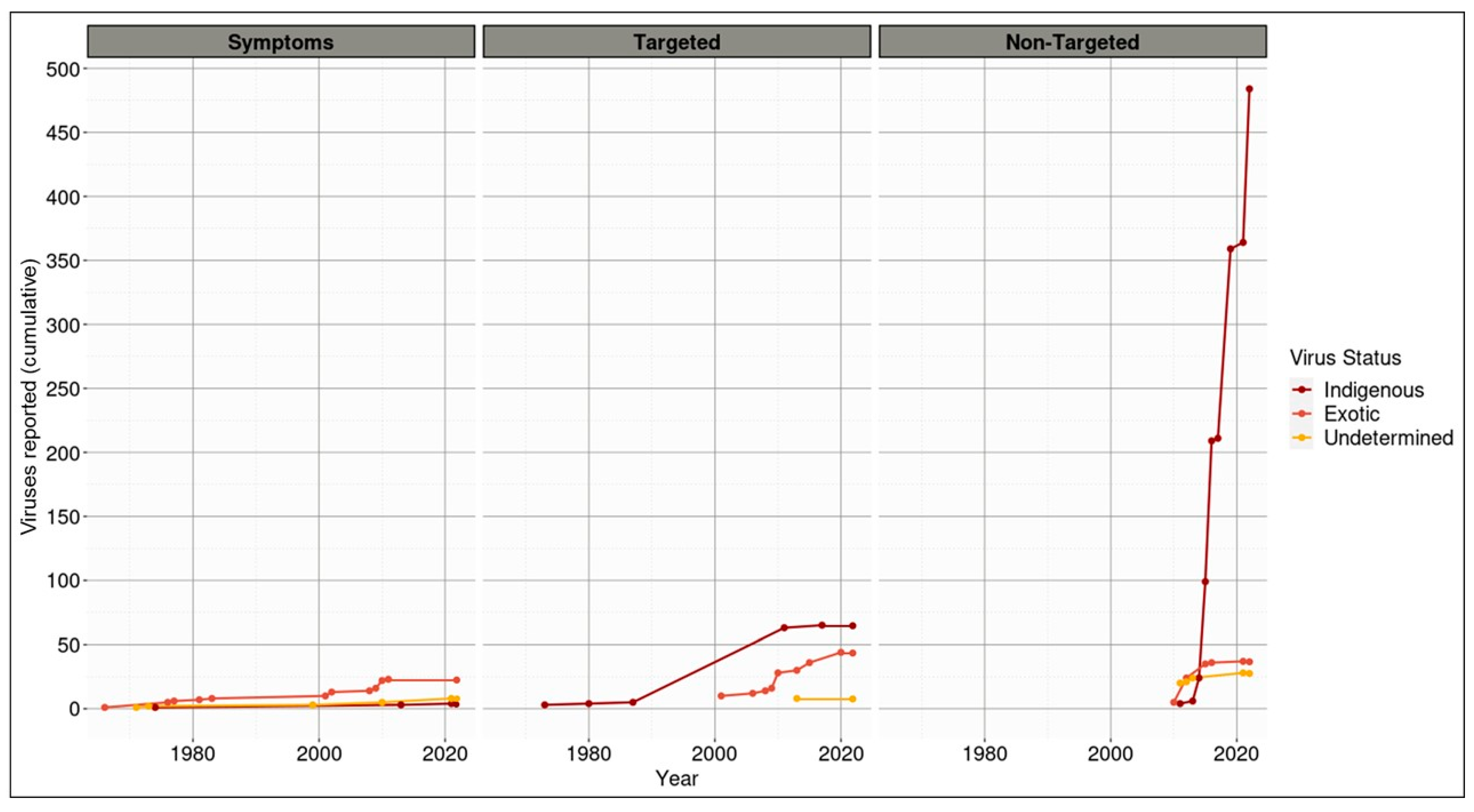
3.3. The Majority of Viruses Associated with Indigenous Species in Aotearoa New Zealand Are Themselves Classified as Indigenous
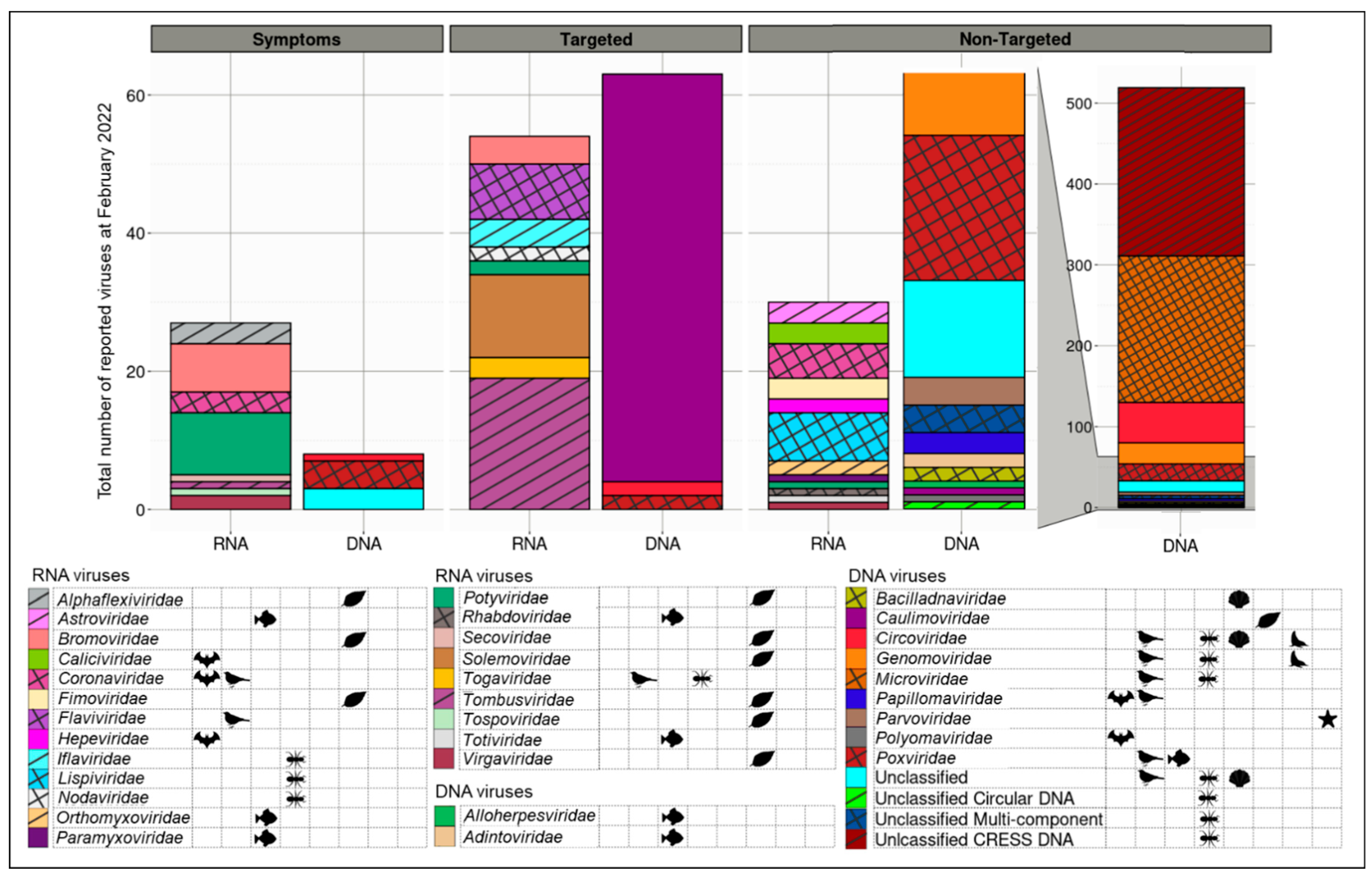
3.4. Detection Ease and Sum of Viruses Described Associated with Aotearoa New Zealand Indigenous Hosts Varies between Virus and Host Types
4. Discussion
4.1. Indigenous and Endemic Viruses of Aotearoa New Zealand
4.2. Implications of New Virus Discovery in Aotearoa New Zealand
4.3. Virus Abundances within Aotearoa New Zealand
4.4. Impacts on Virus Management
4.5. Current and Future Methods of Detection
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pearson, M.N.; Clover GR, G.; Guy, P.L.; Fletcher, J.D.; Beever, R.E. A review of the plant virus, viroid and mollicute records for New Zealand. Australas. Plant Pathol. 2006, 35, 217–252. [Google Scholar] [CrossRef]
- Pennycook, S.R. Plant Diseases Recorded in New Zealand; Plant Diseases Division, DSIR: Auckland, New Zealand, 1989. [Google Scholar]
- Veerakone, S.; Tang, J.Z.; Ward, L.I.; Liefting, L.W.; Perez-Egusquiza, Z.; Lebas, B.S.M.; Delmiglio, C.; Fletcher, J.D.; Guy, P.L. A review of the plant virus, viroid, liberibacter and phytoplasma records for New Zealand. Australas. Plant Pathol. 2015, 44, 463–514. [Google Scholar] [CrossRef]
- Kreuze, J.F.; Perez, A.; Untiveros, M.; Quispe, D.; Fuentes, S.; Barker, I.; Simon, R. Complete viral genome sequence and discovery of novel viruses by deep sequencing of small RNAs: A generic method for diagnosis, discovery and sequencing of viruses. Virology 2009, 388, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, W.; Li, S.; Massart, S. Is There a “Biological Desert” With the Discovery of New Plant Viruses? A Retrospective Analysis for New Fruit Tree Viruses. Front. Microbiol. 2020, 11, 592816. [Google Scholar] [CrossRef] [PubMed]
- Maclot, F.; Candresse, T.; Filloux, D.; Malmstrom, C.M.; Roumagnac, P.; van der Vlugt, R.; Massart, S. Illuminating an Ecological Blackbox: Using High Throughput Sequencing to Characterize the Plant Virome Across Scales. Front. Microbiol. 2020, 11, 578064. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Losada, M.; Arenas, M.; Galán, J.C.; Bracho, M.A.; Hillung, J.; García-González, N.; González-Candelas, F. High-throughput sequencing (HTS) for the analysis of viral populations. Infect. Genet. Evol. 2020, 80, 104208. [Google Scholar] [CrossRef]
- Rumbou, A.; Vainio, E.J.; Büttner, C. Towards the Forest Virome: High-Throughput Sequencing Drastically Expands Our Understanding on Virosphere in Temperate Forest Ecosystems. Microorganisms 2021, 9, 1730. [Google Scholar] [CrossRef]
- Smith, S.E.; Huang, W.; Tiamani, K.; Unterer, M.; Khan Mirzaei, M.; Deng, L. Emerging technologies in the study of the virome. Curr. Opin. Virol. 2022, 54, 101231. [Google Scholar] [CrossRef]
- Márquez, L.M.; Redman, R.S.; Rodriguez, R.J.; Roossinck, M.J. A Virus in a Fungus in a Plant: Three-Way Symbiosis Required for Thermal Tolerance. Science 2007, 315, 513–515. [Google Scholar] [CrossRef] [Green Version]
- Ewers, R.M.; Kliskey, A.D.; Walker, S.; Rutledge, D.; Harding, J.S.; Didham, R.K. Past and future trajectories of forest loss in New Zealand. Biol. Conserv. 2006, 133, 312–325. [Google Scholar] [CrossRef]
- Seersholm, F.V.; Cole, T.L.; Grealy, A.; Rawlence, N.J.; Greig, K.; Knapp, M.; Stat, M.; Hansen, A.J.; Easton, L.J.; Shepherd, L.; et al. Subsistence practices, past biodiversity, and anthropogenic impacts revealed by New Zealand-wide ancient DNA survey. Proc. Natl. Acad. Sci. 2018, 115, 7771–7776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilmshurst, J.M.; Hunt, T.L.; Lipo, C.P.; Anderson, A.J. High-precision radiocarbon dating shows recent and rapid initial human colonization of East Polynesia. Proc. Natl. Acad. Sci. USA 2011, 108, 1815–1820. [Google Scholar] [CrossRef] [Green Version]
- UNESCO. The New Zealand Sub-Antarctic Islands. 2022. Available online: https://whc.unesco.org/en/list/877/ (accessed on 18 October 2022).
- Goldberg, J.; Trewick, S.A.; Paterson, A.M. Evolution of New Zealand’s terrestrial fauna: A review of molecular evidence. Philos. Trans. R. Soc. B: Biol. Sci. 2008, 363, 3319–3334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christensen, N.H.; Worthington, B. Import Risk Analysis: Budgerigars (Melopsittacus undulatus) from the United Kingdom; MAF Biosecurity: Wellington, New Zealand, 2008. [Google Scholar]
- Gummow, B.; Tan, R.; Joice, R.; Burgess, G.; Picard, J. Seroprevalence and associated risk factors of mosquito-borne alphaviruses in horses in northern Queensland. Aust. Vet. J. 2018, 96, 243–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tompkins, D.; Paterson, R.; Massey, B.; Gleeson, D. Whataroa virus four decades on: Emerging, persisting, or fading out? J. R. Soc. N. Z. 2010, 40, 1–9. [Google Scholar] [CrossRef]
- O’Donnell, C.F.J. The ecology and conservation of New Zealand bats. In Island Bats. Evolution, Ecology and Conservation; Fleming, T.H., Racey, P.A., Eds.; Chicago University Press: Chicago, IL, USA, 2009; pp. 460–495. [Google Scholar]
- Fritsche, L.G.; Igl, W.; Bailey JN, C.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016, 48, 134–143. [Google Scholar] [CrossRef] [Green Version]
- Sikorski, A.; Dayaram, A.; Varsani, A. Identification of a Novel Circular DNA Virus in New Zealand Fur Seal (Arctocephalus forsteri) Fecal Matter. Genome Announc. 2013, 1, e00558-13. [Google Scholar] [CrossRef] [Green Version]
- Tribunal, W. Ko Aotearoa Tēnei: Report on the Wai 262 Claim. 2011. Available online: https://www.waitangitribunal.govt.nz/news/ko-aotearoa-tenei-report-on-the-wai-262-claim-released (accessed on 3 November 2022).
- Ngaaho. The Mataatua Declaration on Cultural and Intellectual Property Rights of Indigenous Peoples. 1993. Available online: https://ngaaho.maori.nz/cms/resources/mataatua.pdf (accessed on 3 November 2022).
- Carroll, E.L.; McGowen, M.R.; McCarthy, M.L.; Marx, F.G.; Aguilar, N.; Dalebout, M.L.; Dreyer, S.; Gaggiotti, O.E.; Hansen, S.S.; van Helden, A.; et al. Speciation in the deep: Genomics and morphology reveal a new species of beaked whale Mesoplodon eueu. Proc. R. Soc. B Biol. Sci. 2021, 288, 20211213. [Google Scholar] [CrossRef]
- McAllister, T.; Beggs, J.; Ogilvie, S.; Kirikiri, R.; Black, A.; Wehi, P. Kua takoto te mānuka: Mātauranga Māori in New Zealand ecology. N. Z. J. Ecol. 2020, 43, 1–7. [Google Scholar] [CrossRef]
- Rabbidge, L.O.; Blouin, A.G.; Chooi, K.M.; Higgins, C.M.; MacDiarmid, R.M. Characterisation and Distribution of Karaka Ōkahu Purepure Virus—A Novel Emaravirus Likely to Be Endemic to New Zealand. Viruses 2021, 13, 1611. [Google Scholar] [CrossRef]
- Veale, A.; de Lange, P.; Buckley, T.; Cracknell, M.; Hohaia, H.; Parry, K.; Raharaha-Nehemia, K.; Reihana, K.; Seldon, D.; Tawiri, K.; et al. Using te reo Māori and ta re Moriori in taxonomy. N. Z. J. Ecol. 2019, 43, 1–11. [Google Scholar] [CrossRef]
- Fuentes, S.; Gibbs, A.J.; Adams, I.P.; Hajizadeh, M.; Kreuze, J.; Fox, A.; Blouin, A.G.; Jones, R.A.C. Phylogenetics and Evolution of Potato Virus V: Another Potyvirus that Originated in the Andes. Plant Dis. 2022, 106, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, S.; Jones RA, C.; Matsuoka, H.; Ohshima, K.; Kreuze, J.; Gibbs, A.J. Potato virus Y; the Andean connection. Virus Evol. 2019, 5, vez037. [Google Scholar] [CrossRef] [Green Version]
- Zarghani, S.N.; Hily, J.M.; Glasa, M.; Marais, A.; Wetzel, T.; Faure, C.; Vigne, E.; Velt, A.; Lemaire, O.; Boursiquot, J.M.; et al. Grapevine virus T diversity as revealed by full-length genome sequences assembled from high-throughput sequence data. PLoS ONE 2018, 13, e0206010. [Google Scholar] [CrossRef]
- Pereyra, P.J. Rethinking the native range concept. Conserv. Biol. 2020, 34, 373–377. [Google Scholar] [CrossRef]
- Austin, F.J.; Bull, P.C.; Chaudry, M.A. A poxvirus isolated from silvereyes (Zosterops lateralis) from Lower Hutte, New Zealand. J. Wildl. Dis. 1973, 9, 111–114. [Google Scholar] [CrossRef] [Green Version]
- Department of Conservation. Silvereye or Wax-Eye/Tauhou. Available online: https://www.doc.govt.nz/nature/native-animals/birds/birds-a-z/silvereye-or-wax-eye/ (accessed on 18 October 2022).
- Estoup, A.; Clegg, S.M. Bayesian inferences on the recent island colonization history by the bird Zosterops lateralis lateralis. Mol. Ecol. 2003, 12, 657–674. [Google Scholar] [CrossRef]
- Mortimer, N.; Campbell, H.J.; Tulloch, A.J.; King, P.R.; Stagpoole, V.M.; Wood, R.A.; Rattenbury, M.S.; Sutherland, R.; Adams, C.J.; Collot, J.; et al. Zealandia: Earth’s Hidden Continent. GSA Today 2017, 27, 27–35. [Google Scholar] [CrossRef]
- Wallis, G.P.; Trewick, S.A. New Zealand phylogeography: Evolution on a small continent. Mol. Ecol. 2009, 18, 3548–3580. [Google Scholar] [CrossRef]
- Smyth, J.A.; Todd, D.; Scott, A.; Beckett, A.; Twentyman, C.M.; Bröjer, C.; Uhlhorn, H.; Gavier-Widen, D. Identification of circovirus infection in three species of gull. Vet. Rec. 2006, 159, 212–214. [Google Scholar] [CrossRef]
- Twentyman, C.M.; Alley, M.R.; Meers, J.; Cooke, M.M.; Duignan, P.J. Circovirus-like infection in a southern black-backed gull (Larus dominicanus). Avian Pathol. 1999, 28, 513–516. [Google Scholar] [CrossRef] [PubMed]
- MacDiarmid, R.; Rodoni, B.; Melcher, U.; Ochoa-Corona, F.; Roossinck, M. Biosecurity Implications of New Technology and Discovery in Plant Virus Research. PLoS Pathog. 2013, 9, e1003337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- New Zealand Government. Hazardous Substances and New Organisms Act 1996. 2022. Available online: https://www.legislation.govt.nz/act/public/1996/0030/latest/DLM381222.html (accessed on 18 October 2022).
- New Zealand Government. Hazardous Substances and New Organisms Act 1996 No 30 (as at 07 August 2020), Public Act 2A Meaning of Term New Organism—New Zealand Legislation. 2022. Available online: https://www.legislation.govt.nz/act/public/1996/0030/111.0/DLM382982.html (accessed on 18 October 2022).
- Roossinck, M.J. Plants, viruses and the environment: Ecology and mutualism. Virology 2015, 479–480, 271–277. [Google Scholar] [CrossRef] [Green Version]
- Safari, M.; Roossinck, M.J. Coevolution of a Persistent Plant Virus and Its Pepper Hosts. Mol. Plant-Microbe Interact. MPMI 2018, 31, 766–776. [Google Scholar] [CrossRef] [Green Version]
- Massart, S.; Candresse, T.; Gil, J.; Lacomme, C.; Predajna, L.; Ravnikar, M.; Reynard, J.S.; Rumbou, A.; Saldarelli, P.; Škorić, D.; et al. A framework for the evaluation of biosecurity, commercial, regulatory, and scientific impacts of plant viruses and viroids identified by NGS technologies. Front. Microbiol. 2017, 8, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coutts, B.A.; Kehoe, M.A.; Webster, C.G.; Wylie, S.J.; Jones, R.A.C. Indigenous and introduced potyviruses of legumes and Passiflora spp. from Australia: Biological properties and comparison of coat protein nucleotide sequences. Arch. Virol. 2011, 156, 1757–1774. [Google Scholar] [CrossRef]
- Vincent, S.J.; Coutts, B.A.; Jones, R.A. Effects of introduced and indigenous viruses on native plants: Exploring their disease causing potential at the agro-ecological interface. PLoS ONE 2014, 9, e91224. [Google Scholar] [CrossRef] [PubMed]
- Guy, P.L.; Delmiglio, C.; Pearson, M.N. Virus invasions of the New Zealand flora. Biol. Invasions 2022, 24, 1599–1609. [Google Scholar] [CrossRef]
- Debat, H.; Zavallo, D.; Brisbane, R.S.; Vončina, D.; Almeida RP, P.; Blouin, A.G.; Al Rwahnih, M.; Gomez-Talquenca, S.; Asurmendi, S. Grapevine virus L.: A novel vitivirus in grapevine. Eur. J. Plant Pathol. 2019, 155, 319–328. [Google Scholar] [CrossRef]
- Li, X.; Cheng, Z.; Dang, C.; Zhang, M.; Zheng, Y.; Yu, X. Metagenomic and viromic data mining reveals viral threats in biologically treated domestic wastewater. Environ. Sci. Ecotechnol. 2021, 7, 100105. [Google Scholar] [CrossRef]
- Edgar, R.C.; Taylor, J.; Lin, V.; Altman, T.; Barbera, P.; Meleshko, D.; Lohr, D.; Novakovsky, G.; Buchfink, B.; Al-Shayeb, B.; et al. Petabase-scale sequence alignment catalyses viral discovery. Nature 2022, 602, 7895. [Google Scholar] [CrossRef] [PubMed]
- Samarth Lee, R.; Kelly, D.; Turnbull, M.H.; Macknight, R.C.; Poole, A.M.; Jameson, P.E. Molecular control of the floral transition in the mast seeding plant Celmisia lyallii (Asteraceae). Mol. Ecol. 2021, 30, 1846–1863. [Google Scholar] [CrossRef] [PubMed]
- Correa-Garhwal, S.M.; Clarke, T.H.; Janssen, M.; Crevecoeur, L.; McQuillan, B.N.; Simpson, A.H.; Vink, C.J.; Hayashi, C.Y. Spidroins and Silk Fibers of Aquatic Spiders. Sci. Rep. 2019, 9, 1. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Jordan, M.D.; Newcomb, R.D.; Gemmell, N.J.; Bank, S.; Meusemann, K.; Dearden, P.K.; Duncan, E.J.; Grosser, S.; Rutherford, K.; et al. Analysis of the genome of the New Zealand giant collembolan (Holacanthella duospinosa) sheds light on hexapod evolution. BMC Genom. 2017, 18, 795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, R.J.; Wang, J.; Peacey, M.; Moore, N.E.; McInnes, K.; Tompkins, D.M. New Alphacoronavirus in Mystacina tuberculata Bats, New Zealand. Emerg. Infect. Dis. 2014, 20, 697–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Moore, N.E.; Murray, Z.L.; McInnes, K.; White, D.J.; Tompkins, D.M.; Hall, R.J. Discovery of novel virus sequences in an isolated and threatened bat species, the New Zealand lesser short-tailed bat (Mystacina tuberculata). J. Gen. Virol. 2015, 96 Pt 8, 2442–2452. [Google Scholar] [CrossRef] [Green Version]
- Sikorski, A.; Kearvell, J.; Elkington, S.; Dayaram, A.; Argüello-Astorga, G.R.; Varsani, A. Novel ssDNA viruses discovered in yellow-crowned parakeet (Cyanoramphus auriceps) nesting material. Arch. Virol. 2013, 158, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Sikorski, A.; Massaro, M.; Kraberger, S.; Young, L.M.; Smalley, D.; Martin, D.P.; Varsani, A. Novel myco-like DNA viruses discovered in the faecal matter of various animals. Virus Res. 2013, 177, 209–216. [Google Scholar] [CrossRef]
- Hewson, I.; Sewell, M.A. Surveillance of densoviruses and mesomycetozoans inhabiting grossly normal tissues of three Aotearoa New Zealand asteroid species. PLoS ONE 2021, 16, e0241026. [Google Scholar] [CrossRef]
- Dayaram, A.; Goldstien, S.; Zawar-Reza, P.; Gomez, C.; Harding, J.S.; Varsani, A. Novel ssDNA virus recovered from estuarine Mollusc (Amphibola crenata) whose replication associated protein (Rep) shares similarities with Rep-like sequences of bacterial origin. J. Gen. Virol. 2013, 94, 1104–1110. [Google Scholar] [CrossRef]
- Kazlauskas, D.; Dayaram, A.; Kraberger, S.; Goldstien, S.; Varsani, A.; Krupovic, M. Evolutionary history of ssDNA bacilladnaviruses features horizontal acquisition of the capsid gene from ssRNA nodaviruses. Virology 2017, 504, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Dayaram, A.; Goldstien, S.; Argüello-Astorga, G.R.; Zawar-Reza, P.; Gomez, C.; Harding, J.S.; Varsani, A. Diverse small circular DNA viruses circulating amongst estuarine molluscs. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2015, 31, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Catedral, L.; Kurenbach, B.; Massaro, M.; McInnes, K.; Brunton, D.H.; Hauber, M.E.; Martin, D.P.; Varsani, A. A new isolate of beak and feather disease virus from endemic wild red-fronted parakeets (Cyanoramphus novaezelandiae) in New Zealand. Arch. Virol. 2010, 155, 613–620. [Google Scholar] [CrossRef]
- Munday, J.S.; Hardcastle, M.R.; Hunter, S.; Harvey, C.J. Papillomas and probable in situ carcinoma in association with a novel papillomavirus in a red-billed gull (Chroicocephalus novaehollandiae scopulinus). Arch. Virol. 2021, 166, 1157–1161. [Google Scholar] [CrossRef]
- Massaro, M.; Ortiz-Catedral, L.; Julian, L.; Galbraith, J.A.; Kurenbach, B.; Kearvell, J.; Kemp, J.; van Hal, J.; Elkington, S.; Taylor, G.; et al. Molecular characterisation of beak and feather disease virus (BFDV) in New Zealand and its implications for managing an infectious disease. Arch. Virol. 2012, 157, 1651–1663. [Google Scholar] [CrossRef] [PubMed]
- Jackson, B.; Varsani, A.; Holyoake, C.; Jakob-Hoff, R.; Robertson, I.; McInnes, K.; Empson, R.; Gray, R.; Nakagawa, K.; Warren, K. Emerging infectious disease or evidence of endemicity? A multi-season study of beak and feather disease virus in wild red-crowned parakeets (Cyanoramphus novaezelandiae). Arch. Virol. 2015, 160, 2283–2292. [Google Scholar] [CrossRef]
- Alley, M.; Hale, K.; Cash, W.; Ha, H.; Howe, L. Concurrent avian malaria and avipox virus infection in translocated South Island saddlebacks (Philesturnus carunculatus carunculatus). N. Z. Vet. J. 2010, 58, 218–223. [Google Scholar] [CrossRef]
- Alley, M.R.; Suepaul, R.B.; McKinlay, B.; Young, M.J.; Wang, J.; Morgan, K.J.; Hunter, S.A.; Gartrell, B.D. Diphtheritic stomatitis in yellow-eyed penguins (Megadyptes antipodes) in New Zealand. J. Wildl. Dis. 2017, 53, 102–110. [Google Scholar] [CrossRef]
- White, D.J.; Hall, R.J.; Wang, J.; Moore, N.E.; Park, D.; McInnes, K.; Gartrell, B.D.; Tompkins, D.M. Discovery and complete genome sequence of a novel circovirus-like virus in the endangered rowi kiwi, Apteryx rowi. Virus Genes 2016, 52, 727–731. [Google Scholar] [CrossRef]
- Ortiz-Catedral, L.; McInnes, K.; Hauber, M.E.; Brunton, D.H.; Ortiz-Catedral, L.; McInnes, K.; Hauber, M.E.; Brunton, D.H. First report of beak and feather disease virus (BFDV) in wild Red-fronted Parakeets (Cyanoramphus novaezelandiae) in New Zealand. Emu 2009, 109, 244–247. [Google Scholar] [CrossRef]
- Tompkins, D.; Johansen, C.; Jakob-Hoff, R.; Pulford, D.; Castro, I.; Mackereth, G. Surveillance for arboviral zoonoses in New Zealand birds. West. Pac. Surveill. Response J. WPSAR 2013, 4, 16–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, H.J.; Howe, L.; Alley, M.; Gartrell, B. The phylogenetic analysis of avipoxvirus in New Zealand. Vet. Microbiol. 2011, 150, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Ha, H.J. The Biology of Avipoxvirus in New Zealand Avifauna. Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 2013. [Google Scholar]
- Quinn, P.J. Suspected case of bird pox in a small population of New Zealand Pipits. Notornis 1971, 18, 217. [Google Scholar]
- Ha, H.; Alley, M.; Cahill, J.; Howe, L.; Gartrell, B. The prevalence of psittacine beak and feather disease virus infection in native parrots in New Zealand. N. Z. Vet. J. 2009, 57, 50–52. [Google Scholar] [CrossRef]
- Custer, J.M.; White, R.; Taylor, H.; Schmidlin, K.; Fontenele, R.S.; Stainton, D.; Kraberger, S.; Briskie, J.V.; Varsani, A. Diverse single-stranded DNA viruses identified in New Zealand (Aotearoa) South Island robin (Petroica australis) fecal samples. Virology 2022, 565, 38–51. [Google Scholar] [CrossRef]
- Moore, S.G.; Alma, P.J. Polyhedral Viruses Infecting Two Forest Insect Pests, Selidosema suavis and Heliothis armigera; New Zealand Forest Service: Wellington, New Zealand, 1974.
- Dearing, S.C.; Scotti, P.D.; Wigley, P.J.; Dhana, S.D. A small RNA virus isolated from the grass grub, Costelytra zealandica (Coleoptera: Scarabaeidae). N. Z. J. Zool. 1980, 7, 267–269. [Google Scholar] [CrossRef] [Green Version]
- Scotti, P.D.; Dearing, S.; Mossop, D.W. Flock house virus: A Nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeida). Arch. Virol. 1983, 75, 181–189. [Google Scholar] [CrossRef]
- Scotti, P.D.; Fredericksen, S. Manawatu virus: A nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae). Arch. Virol. 1987, 97, 85–92. [Google Scholar] [CrossRef]
- Miles, J.A.R. The ecology of Whataroa virus, an alphavirus, in South Westland, New Zealand. J. Hyg. 1973, 71, 701–713. [Google Scholar] [CrossRef] [Green Version]
- Dobelmann, J.; Felden, A.; Lester, P.J. Genetic Strain Diversity of Multi-Host RNA Viruses that Infect a Wide Range of Pollinators and Associates is Shaped by Geographic Origins. Viruses 2020, 12, 3. [Google Scholar] [CrossRef] [Green Version]
- Dayaram, A.; Potter, K.A.; Moline, A.B.; Rosenstein, D.D.; Marinov, M.; Thomas, J.E.; Breitbart, M.; Rosario, K.; Argüello-Astorga, G.R.; Varsani, A. High global diversity of cycloviruses amongst dragonflies. J. Gen. Virol. 2013, 94, 1827–1840. [Google Scholar] [CrossRef] [PubMed]
- Käfer, S.; Paraskevopoulou, S.; Zirkel, F.; Wieseke, N.; Donath, A.; Petersen, M.; Jones, T.C.; Liu, S.; Zhou, X.; Middendorf, M.; et al. Re-assessing the diversity of negative strand RNA viruses in insects. PLoS Pathog. 2019, 15, e1008224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dayaram, A.; Galatowitsch, M.; Harding, J.S.; Argüello-Astorga, G.R.; Varsani, A. Novel circular DNA viruses identified in Procordulia grayi and Xanthocnemis zealandica larvae using metagenomic approaches. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2014, 22, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Dayaram, A.; Galatowitsch, M.L.; Argüello-Astorga, G.R.; van Bysterveldt, K.; Kraberger, S.; Stainton, D.; Harding, J.S.; Roumagnac, P.; Martin, D.P.; Lefeuvre, P.; et al. Diverse circular replication-associated protein encoding viruses circulating in invertebrates within a lake ecosystem. Infect. Genet. Evol. 2016, 39, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Kraberger, S.; Schmidlin, K.; Fontenele, R.S.; Walters, M.; Varsani, A. Unravelling the Single-Stranded DNA Virome of the New Zealand Blackfly. Viruses 2019, 11, 532. [Google Scholar] [CrossRef] [Green Version]
- Blouin, A.G.; Ross, H.A.; Hobson-Peters, J.; O’Brien, C.A.; Warren, B.; MacDiarmid, R. A new virus discovered by immunocapture of double-stranded RNA, a rapid method for virus enrichment in metagenomic studies. Mol. Ecol. Resour. 2016, 16, 1255–1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Vianen JC CM, V.; Houliston, G.J.; Fletcher, J.D.; Heenan, P.B.; Chapman, H.M. New threats to endangered Cook’s scurvy grass (Lepidium oleraceum; Brassicaceae): Introduced crop viruses and the extent of their spread. Aust. J. Bot. 2013, 61, 161–166. [Google Scholar] [CrossRef]
- Yasaka, R.; Ohba, K.; Schwinghamer, M.W.; Fletcher, J.; Ochoa-Corona, F.M.; Thomas, J.E.; Ho SY, W.; Gibbs, A.J.; Ohshima, K. Phylodynamic evidence of the migration of turnip mosaic potyvirus from Europe to Australia and New Zealand. J. Gen. Virol. 2015, 96, 701–713. [Google Scholar] [CrossRef] [Green Version]
- Lyttle, D.J.; Orlovich, D.A.; Guy, P.L. Detection and analysis of endogenous badnaviruses in the New Zealand flora. AoB Plants 2011, 2011, plr008. [Google Scholar] [CrossRef] [Green Version]
- Guy, P.L. Detection of Cucumber mosaic virus on Clematis paniculata in lowland forest in New Zealand. Australasian Plant Disease Notes 2011, 6, 20–21. [Google Scholar] [CrossRef] [Green Version]
- Delmiglio, C.; Pearson, M.N. Effects and incidence of Cucumber mosaic virus, Watermelon mosaic virus and Zucchini yellow mosaic virus in New Zealand’s only native cucurbit, Sicyo s australis. Australas. Plant Pathol. 2006, 35, 29–35. [Google Scholar] [CrossRef]
- Fletcher, J.D.; Bulman, S.; Fletcher, P.J.; Houliston, G.J. First record of Turnip mosaic virus in Cook’s scurvy grass (Lepidium oleraceum agg.)—An endangered native plant in New Zealand. Australas. Plant Dis. Notes 2009, 4, 9–11. [Google Scholar] [CrossRef]
- Fletcher, J.D.; Lister, R.A.; Bulman, S.R.; Heenan, P.B. First record of Turnip mosaic virus in Pachycladon spp. (Brassicaceae): An endangered native plant species in New Zealand. Australas. Plant Dis. Notes 2010, 5, 9–10. [Google Scholar] [CrossRef] [Green Version]
- Podolyan, A.; Blouin, A.G.; Dhami, M.K.; Veerakone, S.; MacDiarmid, R. First report of Ageratum latent virus in Veronica species and in New Zealand. Australas. Plant Dis. Notes 2020, 15, 39. [Google Scholar] [CrossRef]
- Ashby, J.W. Infection of karaka (Corynocarpus laevigatus J. R. & G. Forst.) by cucumber mosaic virus. N. Z. J. Agric. Res. 1977, 20, 533–534. [Google Scholar] [CrossRef]
- Thomson, A.D. Virus diseases of Solanum laciniatum Ait. In New Zealand. N. Z. J. Agric. Res. 1976, 19, 521–527. [Google Scholar] [CrossRef]
- Delmiglio, C.; Pearson, M.N.; Lister, R.A.; Guy, P.L. Incidence of cereal and pasture viruses in New Zealand’s native grasses. Ann. Appl. Biol. 2010, 157, 25–36. [Google Scholar] [CrossRef]
- Davis, L.T.; Guy, P.L. Introduced Plant Viruses and the Invasion of a Native Grass Flora. Biol. Invasions 2001, 3, 89–95. [Google Scholar] [CrossRef]
- Fletcher, J.D. New plant disease records in New Zealand: Additional hosts of alfalfa mosaic virus. N. Z. J. Agric. Res. 1983, 26, 403–404. [Google Scholar] [CrossRef]
- Fletcher, J.D. New strain of potato virus. Grower 2001, 56, 35–36. [Google Scholar]
- Thomson, A.D. New plant disease record in New Zealand: Cucumber mosaic virus in Myosotidium hortensia (Decne) Baill. N. Z. J. Agric. Res. 1981, 24, 401–402. [Google Scholar] [CrossRef] [Green Version]
- Baylis, G.T.S. Solanum aviculare and S. laciniatum. Herba Hung. 1966, 5, 283–287. [Google Scholar]
- Fletcher, J.D. New hosts of Alfalfa mosaic virus, Cucumber mosaic virus, Potato virus Y, Soybean dwarf virus, and Tomato spotted wilt virus in New Zealand. N. Z. J. Crop Hortic. Sci. 2001, 29, 213–217. [Google Scholar] [CrossRef]
- Delmiglio, C. The Effects of Virus Diseases on the Growth and Distribution of Sicyos australis, and Molecular and Morphological Characterisation of the Species in New Zealand. Master’s Thesis, University of Auckland, Auckland, New Zealand, 2003. [Google Scholar]
- Delmiglio, C.; Pearson, M.N. Are common crop viruses a threat to New Zealands only native cucurbit Sicyos australis. N. Z. Plant Prot. 2002, 55, 435. [Google Scholar] [CrossRef] [Green Version]
- Veerakone, S.; Lebas, B.S.M.; Tang, J.; Clover, G.R.G. First report of Tomato mosaic virus in Griselinia lucida, an epiphytic shrub native to New Zealand. Australas. Plant Dis. Notes 2010, 5, 107–109. [Google Scholar] [CrossRef] [Green Version]
- Tang, J.; Elliott, D.R.; Quinn, B.D.; Clover, G.R.G.; Alexander, B.J.R. Occurrence of Hibiscus chlorotic ringspot virus in Hibiscus spp. In New Zealand. Plant Dis. 2008, 92, 1367. [Google Scholar] [CrossRef]
- Delmiglio, C. The Incidence and Phylogenetic Analysis of Viruses Infecting New Zealand’s Native Grasses. Ph.D. Thesis, The University of Auckland, Auckland, New Zealand, 2008. [Google Scholar]
- Ward, L.I.; Delmiglio, C.; Hill, C.F.; Clover, G.R.G. First report of Tobacco ringspot virus on Sophora microphylla, a native tree of New Zealand. Plant Pathol. 2009, 58, 784. [Google Scholar] [CrossRef]
- Veerakone, S.; Kanchiraopa, D.; Zheng, A.; Tang, J.; Liefting, L.W.; Thompson, J.R. Discovery of a new-to-science virus that infects the New Zealand native species tutu (Coriaria arborea). In Proceedings of the Australasian Plant Pathology Society Conference. Online and Hobart, Tasmania, Australia, 23–26 November 2021. [Google Scholar]
- Veerakone, S.; Tang, J.Z.; Perez-Egusquiza, Z.; Liefting, L.W.; Khan, S.; Kanchiraopally, D.; Kelly, M.; Waite, D.; Delmiglio, C.; Thompson, J.R. New records for viruses, viroids and liberibacters from New Zealand: Update 2016-2021. In Proceedings of the Australasian Plant Pathology Society Conference, Online and Hobart, Tasmania, Australia, 23–26 November 2021. [Google Scholar]
- Miller, A.K.; Mifsud, J.C.O.; Costa, V.A.; Grimwood, R.M.; Kitson, J.; Baker, C.; Brosnahan, C.L.; Pande, A.; Holmes, E.C.; Gemmell, N.J.; et al. Slippery when wet: Cross-species transmission of divergent coronaviruses in bony and jawless fish and the evolutionary history of the Coronaviridae. Virus Evol. 2021, 7, veab050. [Google Scholar] [CrossRef]
- Perry, B.J.; Darestani, M.M.; Ara, M.G.; Hoste, A.; Jandt, J.M.; Dutoit, L.; Holmes, E.C.; Ingram, T.; Geoghegan, J.L. Viromes of Freshwater Fish with Lacustrine and Diadromous Life Histories Differ in Composition. Viruses 2022, 14, 257. [Google Scholar] [CrossRef]
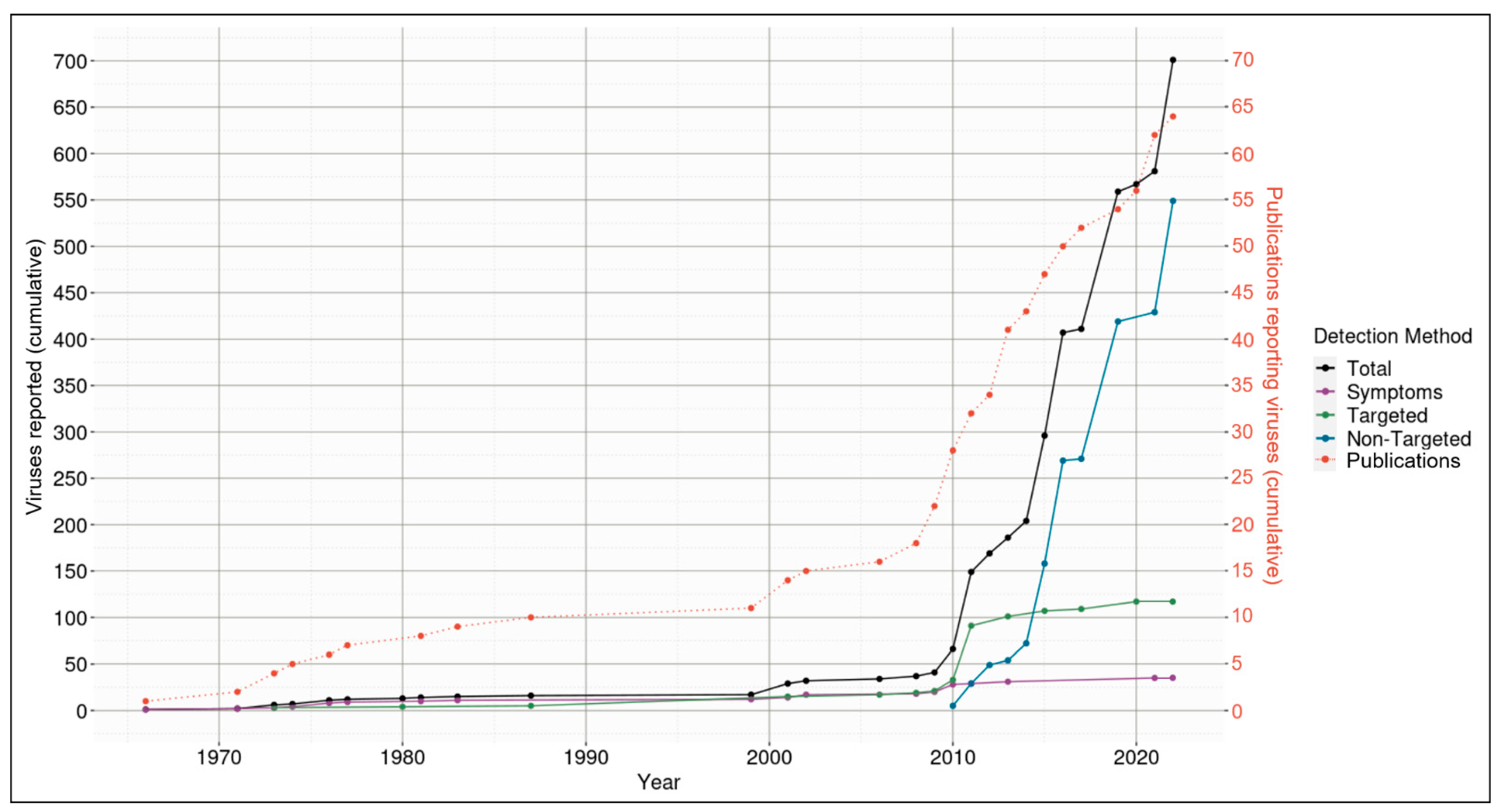
 , birds
, birds
 , fish
, fish
 , insects
, insects
 , mollusks
, mollusks
 , plants
, plants
 , seals
, seals
 , and starfish
, and starfish
 .
.
 , birds
, birds
 , fish
, fish
 , insects
, insects
 , mollusks
, mollusks
 , plants
, plants
 , seals
, seals
 , and starfish
, and starfish
 .
.
| Virus Biostatus Designation | Associated Host | Described Outside Country | Potential Spill-Over Events and Other Comments | ||
|---|---|---|---|---|---|
| Endemic | Indigenous | Exotic | |||
| Endemic | Y | N | N | N | |
| Endemic or indigenous | Y | Y | N | N | |
| Endemic or indigenous | Y | Y | Y | N | Spilt to exotic hosts |
| Indigenous | N | Y | N | N | |
| Indigenous | N | Y | N | Y | |
| Indigenous | N | Y | Y | N | Spilt to exotic hosts |
| Exotic | N | N | Y | Y | Not catalogued in this study |
| Exotic | Y | N | Y | Y | Spilt to endemic hosts |
| Exotic | N | Y | Y | Y | Spilt to indigenous hosts |
| Exotic | Y | Y | Y | Y | Spilt to endemic and indigenous hosts |
| Undetermined | N | Y | N | Y | Possibly indigenous, but found outside of NZ. May also spill to endemic and/or exotic hosts. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robson, M.; Chooi, K.M.; Blouin, A.G.; Knight, S.; MacDiarmid, R.M. A National Catalogue of Viruses Associated with Indigenous Species Reveals High-Throughput Sequencing as a Driver of Indigenous Virus Discovery. Viruses 2022, 14, 2477. https://doi.org/10.3390/v14112477
Robson M, Chooi KM, Blouin AG, Knight S, MacDiarmid RM. A National Catalogue of Viruses Associated with Indigenous Species Reveals High-Throughput Sequencing as a Driver of Indigenous Virus Discovery. Viruses. 2022; 14(11):2477. https://doi.org/10.3390/v14112477
Chicago/Turabian StyleRobson, Merlyn, Kar Mun Chooi, Arnaud Gérard Blouin, Sarah Knight, and Robin Marion MacDiarmid. 2022. "A National Catalogue of Viruses Associated with Indigenous Species Reveals High-Throughput Sequencing as a Driver of Indigenous Virus Discovery" Viruses 14, no. 11: 2477. https://doi.org/10.3390/v14112477
APA StyleRobson, M., Chooi, K. M., Blouin, A. G., Knight, S., & MacDiarmid, R. M. (2022). A National Catalogue of Viruses Associated with Indigenous Species Reveals High-Throughput Sequencing as a Driver of Indigenous Virus Discovery. Viruses, 14(11), 2477. https://doi.org/10.3390/v14112477








