Novel Aeromonas Phage Ahy-Yong1 and Its Protective Effects against Aeromonas hydrophila in Brocade Carp (Cyprinus aka Koi)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strain
2.2. Antibiotic Susceptibility Test
2.3. Phage Isolation and Purification
2.4. Phage Morphology
2.5. Host Range Test
2.6. Optimal Multiplicity of Infection (MOI), Optimum Adsorption Time, and One-Step Growth Curve
2.7. Temperature, pH, and Chloroform Stability
2.8. The Ability of A. hydrophila A18 to Form Biofilm and the Biofilm-Removal Ability of Aeromonas Phage Ahy-Yong1
2.9. Genome Sequencing and Analysis
2.10. Taxonomic Analysis
2.11. Protective Effects of Aeromonas Phage Ahy-Yong1 against A. hydrophila in Brocade Carp (Cyprinus aka Koi)
2.12. Statistical Analysis
3. Results
3.1. Antibiotic Susceptibility of A. hydrophila A18
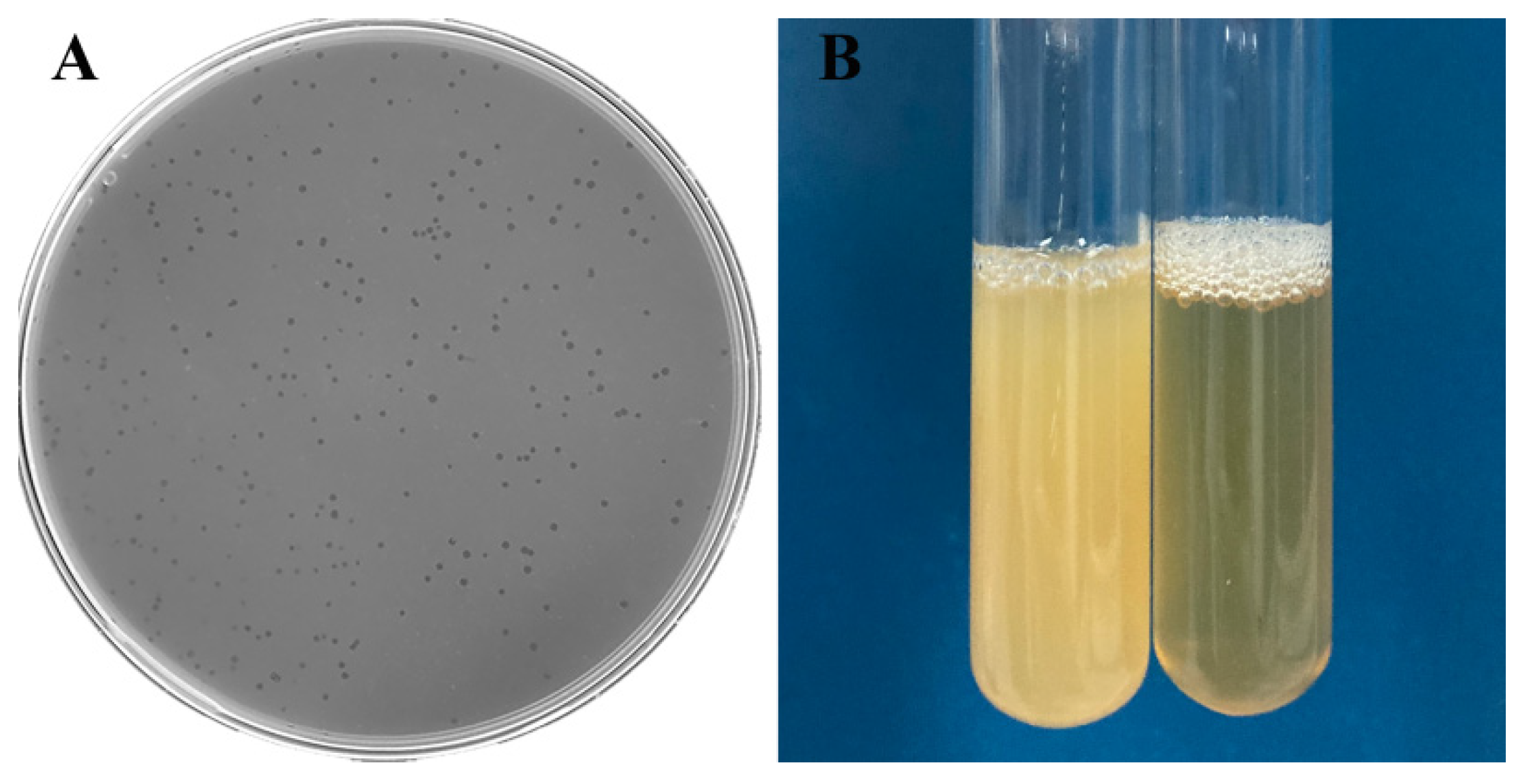
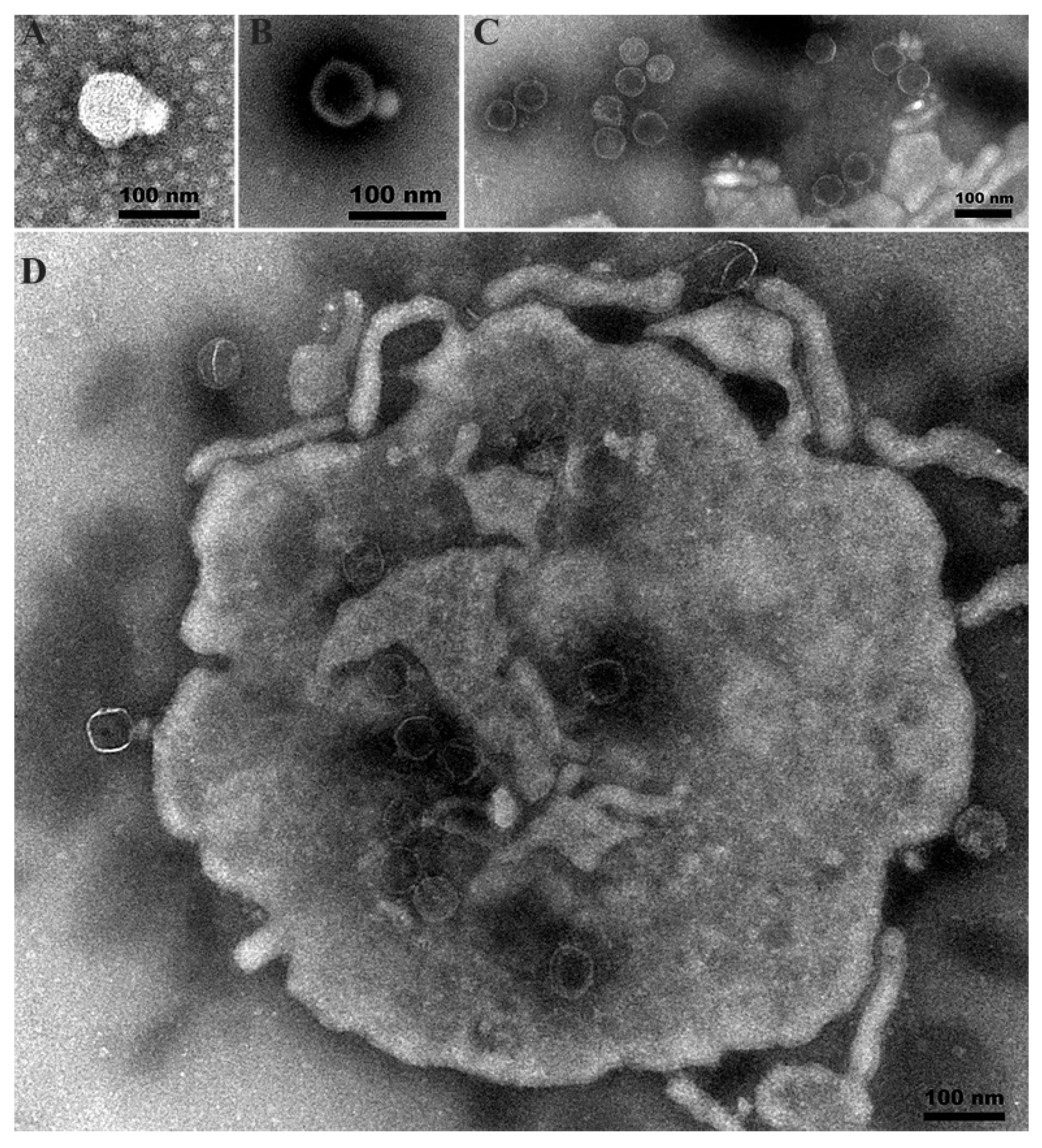
3.2. Plaque and Phage Morphology
3.3. Host Range
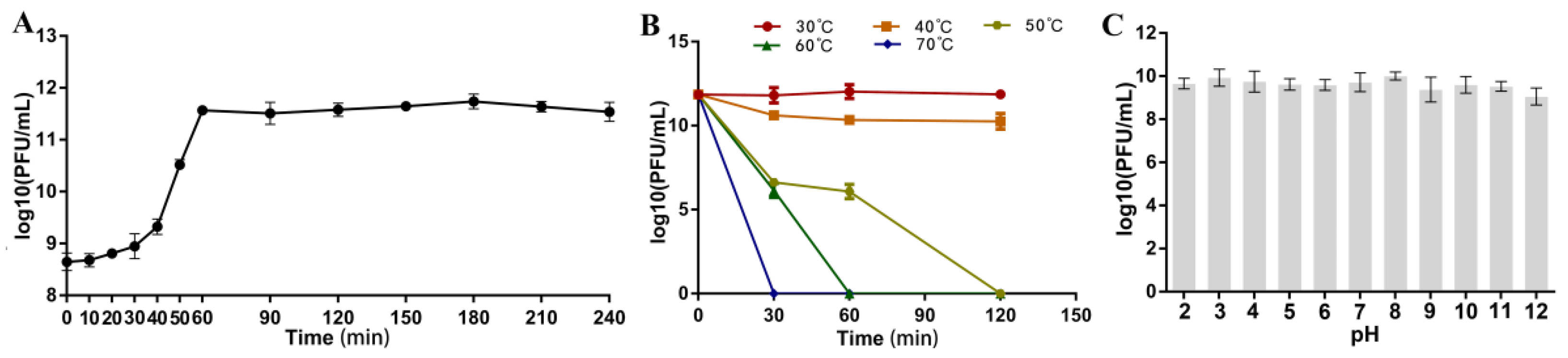
3.4. Optimal Multiplicity of Infection (MOI), Optimal Adsorption Time, and One-Step Growth Curve
3.5. Temperature, pH, and Chloroform Stability
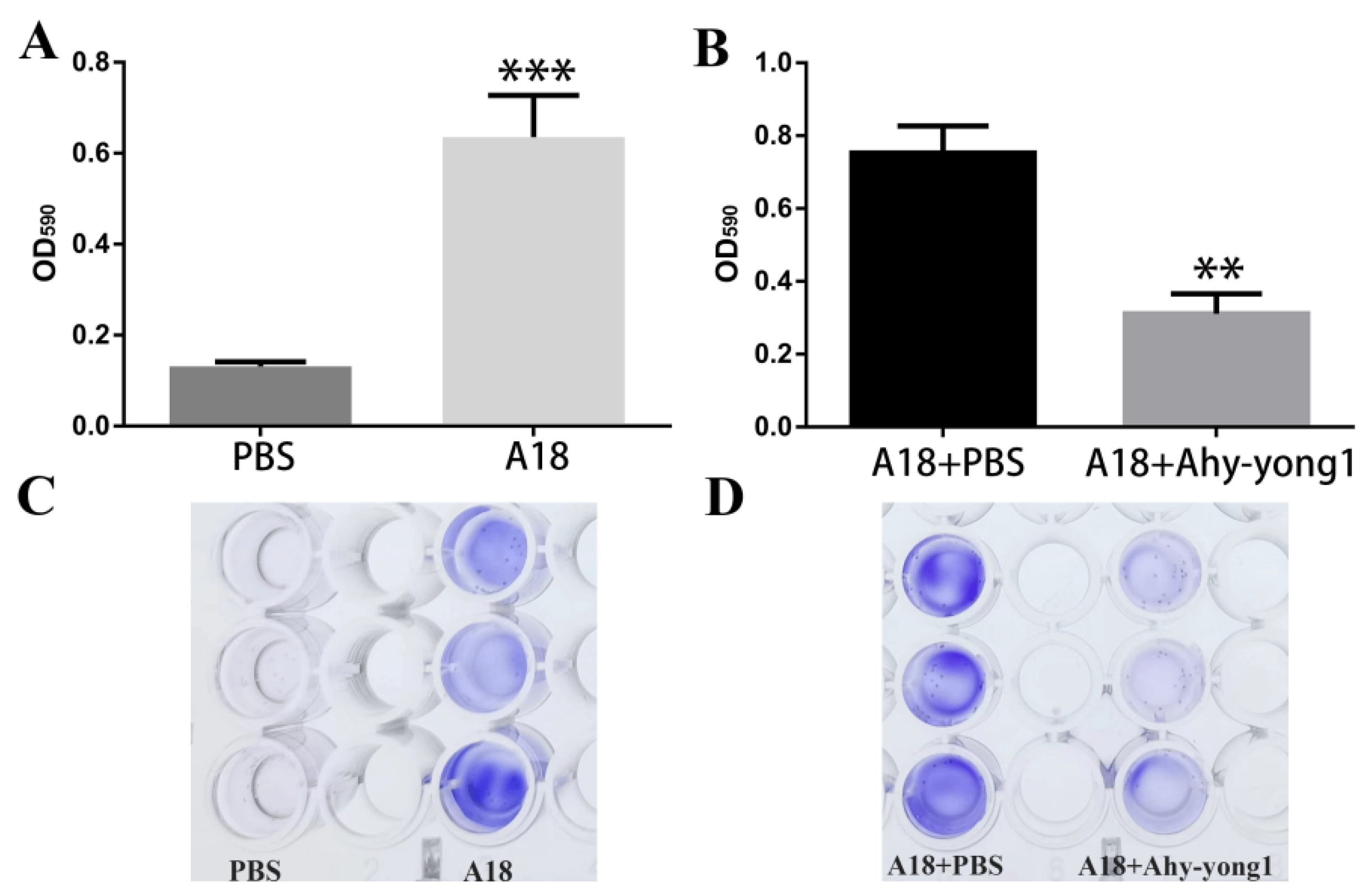
3.6. The Ability of A. hydrophila A18 to Form Biofilm and the Biofilm-Removal Ability of Aeromonas Phage Ahy-Yong1
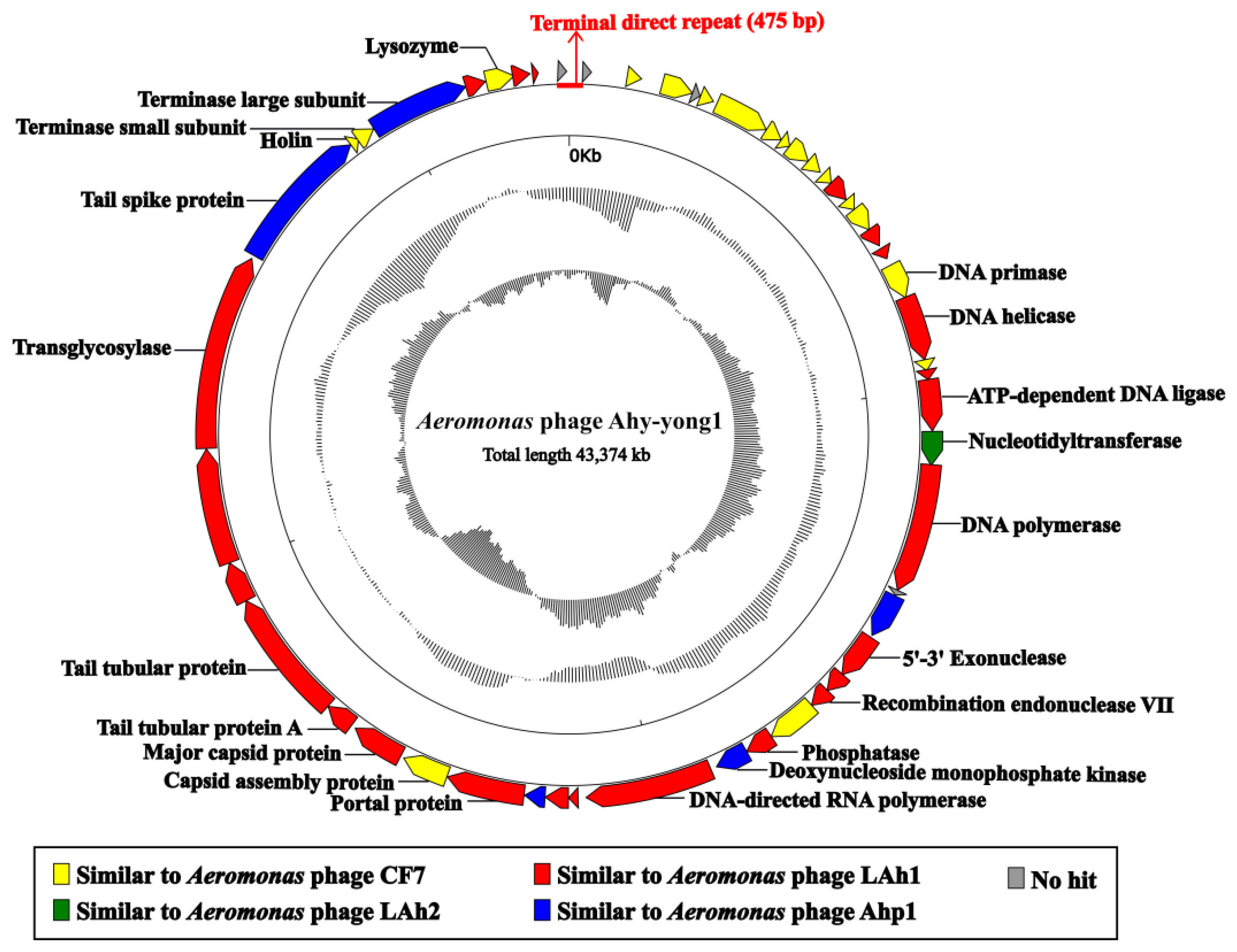
3.7. Genome Analysis
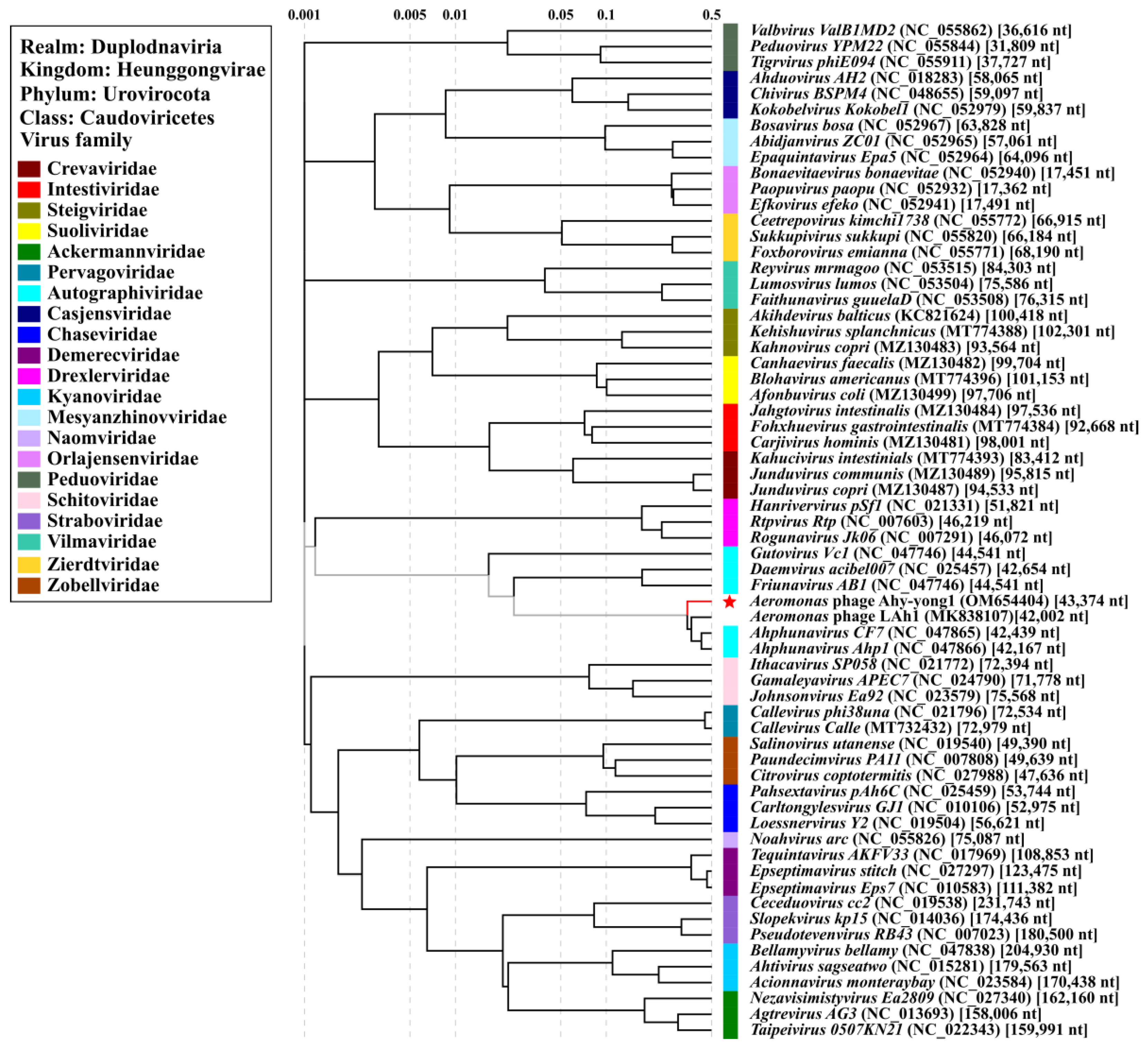
3.8. Taxonomic Analysis
3.9. Comparison of Phage Ahy-Yong1 and the Seven Most Related Aeromonas Phages
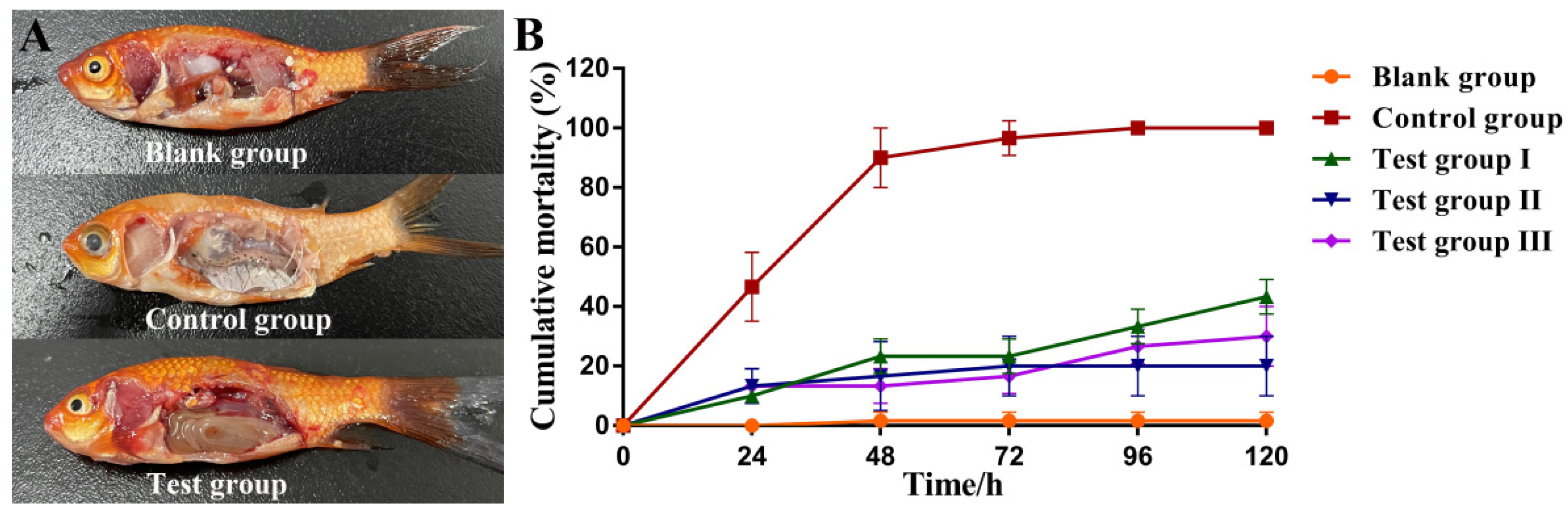
3.10. Protective Effects of Aeromonas Phage Ahy-Yong1 in Brocade Carp (Cyprinus aka Koi)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Pérez-Sánchez, T.; Mora-Sánchez, B.; Balcázar, J.L. Biological approaches for disease control in aquaculture: Advantages, limitations and challenges. Trends Microbiol. 2018, 26, 896–903. [Google Scholar] [CrossRef]
- Cabello, F.C.; Godfrey, H.P.; Buschmann, A.H.; Dölz, H.J. Aquaculture as yet another environmental gateway to the development and globalization of antimicrobial resistance. Lancet Infect. Dis. 2016, 16, e127–e133. [Google Scholar] [CrossRef]
- Janelidze, N.; Jaiani, E.; Didebulidze, E.; Kusradze, I.; Kotorashvili, A.; Chalidze, K.; Porchkhidze, K.; Khukhunashvili, T.; Tsertsvadze, G.; Jgenti, D.; et al. Phenotypic and genetic characterization of Aeromonas hydrophila phage AhMtk13a and evaluation of its therapeutic potential on simulated Aeromonas infection in Danio rerio. Viruses 2022, 14, 412. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Van Belleghem, J.D.; de Vries, C.R.; Burgener, E.; Chen, Q.; Manasherob, R.; Aronson, J.R.; Amanatullah, D.F.; Tamma, P.D.; Suh, G.A. The safety and toxicity of phage therapy: A review of animal and clinical studies. Viruses 2021, 13, 1268. [Google Scholar] [CrossRef] [PubMed]
- Donati, V.L.; Dalsgaard, I.; Sundell, K.; Castillo, D.; Er-Rafik, M.; Clark, J.; Wiklund, T.; Middelboe, M.; Madsen, L. Phage-mediated control of Flavobacterium psychrophilum in aquaculture: In vivo experiments to compare delivery methods. Front. Microbiol. 2021, 12, 628309. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Cisek, A.A.; Dąbrowska, I.; Gregorczyk, K.P.; Wyżewski, Z. Phage therapy in bacterial infections treatment: One hundred years after the discovery of bacteriophages. Curr. Microbiol. 2017, 74, 277–283. [Google Scholar] [CrossRef] [Green Version]
- Wittebole, X.; De Roock, S.; Opal, S.M. A historical overview of bacteriophage therapy as an alternative to antibiotics for the treatment of bacterial pathogens. Virulence 2014, 5, 226–235. [Google Scholar] [CrossRef]
- Fernández-Bravo, A.; Figueras, M.J. An update on the genus Aeromonas: Taxonomy, epidemiology, and pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef] [Green Version]
- Janda, J.M.; Abbott, S.L. The genus Aeromonas: Taxonomy, pathogenicity, and infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Aly, S.M. A Review of Fish Diseases in the Egyptian Aquaculture Sector: Working Report; WorldFish: Bayan Lepas, Malaysia, 2013. [Google Scholar]
- Anjur, N.; Sabran, S.F.; Daud, H.M.; Othman, N.Z. An update on the ornamental fish industry in Malaysia: Aeromonas hydrophila-associated disease and its treatment control. Vet. World 2021, 14, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lin, Y.; Zhang, T.; Wu, Y.; Fang, P.; Wang, S.; Wu, Z.; Hao, J.; Li, A. Etiological characteristics of “tail blister disease” of Australian redclaw crayfish (Cheraxquadricarinatus). J. Invertebr. Pathol. 2021, 184, 107643. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.L.; Meng, X.L.; Lu, R.H.; Wu, C.; Luo, Y.T.; Yan, X.; Li, X.J.; Kong, X.H.; Ni, G.X. Effects of Rehmanniaglutinosa on growth performance, immunological parameters and disease resistance to Aeromonas hydrophila in common carp (Cyprinus carpio L.). Aquaculture 2015, 435, 293–300. [Google Scholar] [CrossRef]
- Lee, S.W.; Wendy, W. Antibiotic and heavy metal resistance of Aeromonas hydrophila and Edwardsiella tarda isolated from red hybrid tilapia (Oreochromis spp.) coinfected with motile aeromonas septicemia and edwardsiellosis. Vet. World 2017, 10, 803–807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vivekanandhan, G.; Savithamani, K.; Hatha, A.A.; Lakshmanaperumalsamy, P. Antibiotic resistance of Aeromonas hydrophila isolated from marketed fish and prawn of South India. Int. J. Food Microbiol. 2002, 76, 165–168. [Google Scholar] [CrossRef]
- Zhang, Y.; Jia, K.X.; Zhang, X.W.; Gao, N.; Liu, Z.Q.; Liu, C.Y.; Shan, X.F.; Qian, A.D.; Zhang, X.; Xu, F.Y.; et al. Biological characteristics of phage PZL-Ah1 and its therapeutic effect on Aeromonas hydrophila infection. Chin. J. Prev. Vet. Med. 2021, 43, 7. [Google Scholar]
- Easwaran, M.; Dananjaya, S.H.S.; Park, S.C.; Lee, J.; Shin, H.J.; De Zoysa, M. Characterization of bacteriophage pAh-1 and its protective effects on experimental infection of Aeromonas hydrophila in Zebrafish (Danio rerio). J. Fish Dis. 2017, 40, 841–846. [Google Scholar] [CrossRef]
- Jun, J.W.; Kim, J.H.; Shin, S.P.; Han, J.E.; Chai, J.Y.; Park, S.C. Protective effects of the Aeromonas phages pAh1-C and pAh6-C against mass mortality of the cyprinid loach (Misgurnus anguillicaudatus) caused by Aeromonas hydrophila. Aquaculture 2013, 416–417, 289–295. [Google Scholar] [CrossRef]
- Dien, L.T.; Ky, L.B.; Huy, B.T.; Mursalim, M.F.; Kayansamruaj, P.; Senapin, S.; Rodkhum, C.; Dong, H.T. Characterization and protective effects of lytic bacteriophage pAh6.2TG against a pathogenic multidrug-resistant Aeromonas hydrophila in Nile tilapia (Oreochromis niloticus). Transbound. Emerg. Dis. 2022, 69, e435–e450. [Google Scholar] [CrossRef]
- Akmal, M.; Rahimi-Midani, A.; Hafeez-Ur-Rehman, M.; Hussain, A.; Choi, T.J. Isolation, characterization, and application of a bacteriophage infecting the fish pathogen Aeromonas hydrophila. Pathogens 2020, 9, 215. [Google Scholar] [CrossRef] [Green Version]
- Tu, V.Q.; Nguyen, T.T.; Tran, X.T.T.; Millard, A.D.; Phan, H.T.; Le, N.P.; Dang, O.T.H.; Hoang, H.A. Complete genome sequence of a novel lytic phage infecting Aeromonas hydrophila, an infectious agent in striped catfish (Pangasianodon hypophthalmus). Arch. Virol. 2020, 165, 2973–2977. [Google Scholar] [CrossRef] [PubMed]
- Le, T.S.; Nguyen, T.H.; Vo, H.P.; Doan, V.C.; Nguyen, H.L.; Tran, M.T.; Tran, T.T.; Southgate, P.C.; Kurtböke, D.I. Protective effects of bacteriophages against Aeromonas hydrophila species causing Motile Aeromonas Septicemia (MAS) in striped catfish. Antibiotics 2018, 7, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.N.; Li, J.M.; Sang, R.X.; Ding, Q.H.; Xu, H.Y. Isolation, identification and biological characteristics of a bacteriophage against Aeromonas hydrophila. China Feed 2021, 13, 18–23. [Google Scholar]
- Jia, K.X. Isolation, Identification and Genomic Analysis of Aeromonas hydrophila Phage PZL-Ah152 and Functional Exploration of Depolymerase Dep47. Master’s Thesis, Jilin Agricultural University, Jilin, China, 2021. [Google Scholar]
- Huo, S.T.; Jiao, H.Q.; Li, Q.; Gu, Z.M.; Liu, X.Q. Isolation, identification and preliminary application of Aeromonas hydrophila phage from Procambarus clarkii. Acta Hydrobiol. Sin. 2021, 45, 366–375. [Google Scholar]
- Lv, S.J.; Liu, L.; Cao, Z.; Lu, S.J.; Lin, F. Isolation and functional identification of a phage against Aeromonas hydrophila isolated from Trionyx sinensis. Jiangsu Agric. Sci. 2018, 46, 108–111. [Google Scholar]
- Gao, S.S. Isolation and Biological Characteristics Analysis of Aeromonas hydrophila Bacteriophages. Master’s Thesis, Nanjing Agricultural University, Nanjing, China, 2016. [Google Scholar]
- Shen, C.J. Biological Characteristics and Genomic Analysis of Aeromonas hydrophila Bacteriophages. Master’s Thesis, Nanjing Agricultural University, Nanjing, China, 2013. [Google Scholar]
- Pereira, C.; Duarte, J.; Costa, P.; Braz, M.; Almeida, A. Bacteriophages in the control of Aeromonas sp. in aquaculture systems: An integrative view. Antibiotics 2022, 11, 2. [Google Scholar] [CrossRef]
- Lin, W.; Li, D.F.; Gao, M.M.; Qin, W.N.; Xu, L.H.; Pan, L.T.; Liu, W.C.; Fan, H.; Mi, Z.Q.; Tong, Y.G. Isolation, characterization and biocontrol efficacy of a T4-like phage virulent to multidrug-resistant Enterobacter hormaechei. Dis. Aquat. Organ. 2021, 147, 97–109. [Google Scholar] [CrossRef]
- Ghosh, K.; Senevirathne, A.; Kang, H.S.; Hyun, W.B.; Kim, J.E.; Kim, K. Complete nucleotide sequence analysis of a novel Bacillus subtilis-infecting bacteriophage BSP10 and its effect on poly-gamma-glutamic acid degradation. Viruses 2018, 10, 240. [Google Scholar] [CrossRef] [Green Version]
- Gutiérrez, S.; Pirolles, E.; Yvon, M.; Baecker, V.; Michalakis, Y.; Blanc, S. The multiplicity of cellular infection changes depending on the route of cell infection in a plant virus. J. Virol. 2015, 89, 9665–9675. [Google Scholar] [CrossRef] [Green Version]
- Walakira, J.K.; Carrias, A.A.; Hossain, M.J.; Jones, E.; Terhune, J.S.; Liles, M.R. Identification and characterization of bacteriophages specific to the catfish pathogen, Edwardsiellaictaluri. J. Appl. Microbiol. 2008, 105, 2133–2142. [Google Scholar] [CrossRef]
- Cramton, S.E.; Gerke, C.; Götz, F. In vitro methods to study staphylococcal biofilm formation. Method Enzymol. 2001, 336, 239–255. [Google Scholar]
- Garneau, J.R.; Depardieu, F.; Fortier, L.C.; Bikard, D.; Monot, M. PhageTerm: A tool for fast and accurate determination of phage termini and packaging mechanism using next-generation sequencing data. Sci. Rep. 2017, 7, 8292. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fan, H.; An, X.; Fan, H.; Jiang, H.; Chen, Y.; Tong, Y. Scrutinizing virus genome termini by high-throughput sequencing. PLoS ONE 2014, 9, e85806. [Google Scholar] [CrossRef] [Green Version]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, 686–689. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 4, W200–W204. [Google Scholar] [CrossRef] [Green Version]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kübler, J.; Lozajic, M. A completely reimplemented MPI Bioinformatics Toolkit with a new HHpred server at its core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [Green Version]
- Figueras, M.J.; Beaz-Hidalgo, R.; Hossain, M.J.; Liles, M.R. Taxonomic affiliation of new genomes should be verified using average nucleotide identity and multilocus phylogenetic analysis. Genome Announc. 2014, 2, e00927-14. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.; Chetvernin, V.; Tatusova, T. Improvements to pairwise sequence comparison (PASC): A genome-based web tool for virus classification. Arch. Virol. 2014, 159, 3293–3304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC-A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, Y.; Yoshida, T.; Kuronishi, M.; Uehara, H.; Ogata, H.; Goto, S. ViPTree: The viral proteomic tree server. Bioinformatics 2017, 33, 2379–2380. [Google Scholar] [CrossRef] [PubMed]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.Y.; Zheng, X.Y.; Cao, F.F.; Li, Y.Y.; Zheng, T.L. Antimicrobial resistance of Aeromonas hydrophila isolated from soft-shelled turtle Pelodiscus sinensis. Acta Agric. Zhejiangensis 2017, 29, 1445–1450. [Google Scholar]
- Kuhn, C.D.; Wilusz, J.E.; Zheng, Y.; Beal, P.A.; Joshua-Tor, L. On-enzyme refolding permits small RNA and tRNA surveillance by the CCA-adding enzyme. Cell 2015, 160, 644–658. [Google Scholar] [CrossRef]
- Lavysh, D.; Sokolova, M.; Minakhin, L.; Yakunina, M.; Artamonova, T.; Kozyavkin, S.; Makarova, K.S.; Koonin, E.V.; Severinov, K. The genome of AR9, a giant transducing Bacillus phage encoding two multisubunit RNA polymerases. Virology 2016, 495, 185–196. [Google Scholar] [CrossRef]
- Cuervo, A.; Fàbrega-Ferrer, M.; Machón, C.; Conesa, J.J.; Fernández, F.J.; Pérez-Luque, R.; Pérez-Ruiz, M.; Pous, J.; Vega, M.C.; Carrascosa, J.L.; et al. Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism. Nat. Commun. 2019, 10, 3746. [Google Scholar] [CrossRef] [Green Version]
- Thunnissen, A.M.; Rozeboom, H.J.; Kalk, K.H.; Dijkstra, B.W. Structure of the 70-kDa soluble lytic transglycosylase complexed with bulgecin A. Implications for the enzymatic mechanism. Biochemistry 1995, 34, 12729–12737. [Google Scholar] [CrossRef] [Green Version]
- Park, T.; Struck, D.K.; Deaton, J.F.; Young, R. Topological dynamics of holins in programmed bacterial lysis. Proc. Natl. Acad. Sci. USA 2006, 103, 19713–19718. [Google Scholar] [CrossRef] [Green Version]
- Kunstmann, S.; Scheidt, T.; Buchwald, S.; Helm, A.; Mulard, L.A.; Fruth, A.; Barbirz, S. Bacteriophage Sf6 tailspike protein for detection of Shigella flexneri pathogens. Viruses 2018, 10, 431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adriaenssens, E.; Brister, J.R. How to name and classify your phage: An informal guide. Viruses 2017, 9, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adriaenssens, E.M.; Sullivan, M.B.; Knezevic, P.; van Zyl, L.J.; Sarkar, B.L.; Dutilh, B.E. Taxonomy of prokaryotic viruses: 2018–2019 update from the ICTV Bacterial and Archaeal Viruses Subcommittee. Arch. Virol. 2020, 165, 1253–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.B.; Lin, N.T.; Tseng, Y.H.; Weng, S.F. Genomic characterization of the novel Aeromonas hydrophila phage Ahp1 suggests the derivation of a new subgroup from phiKMV-Like family. PLoS ONE 2016, 11, e0162060. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabwe, M.; Brown, T.; Speirs, L.; Ku, H.; Leach, M.; Chan, H.T.; Petrovski, S.; Lock, P.; Tucci, J. Novel bacteriophages capable of disrupting biofilms from clinical strains of Aeromonas hydrophila. Front. Microbiol. 2020, 11, 194. [Google Scholar] [CrossRef]
- Turner, D.; Kropinski, A.M.; Alfernas-Zerbini, P.; Buttimer, C.; Lavigne, R.; Bister, J.R.; Tolstoy, I.; Morozova, V.V.; Babkin, I.V.; Kozlova, Y.N.; et al. Create One New Family (Autographiviridae) including Nine Subfamilies and 132 Genera in the Order Caudovirales. Available online: https://ictv.global/ICTV/proposals/2019.103B.zip (accessed on 17 May 2020).
- Lavigne, R.; Seto, D.; Mahadevan, P.; Ackermann, H.W.; Kropinski, A.M. Unifying classical and molecular taxonomic classification: Analysis of the Podoviridae using BLASTP-based tools. Res. Microbiol. 2008, 159, 406–414. [Google Scholar] [CrossRef]
- De Jonge, P.A.; Nobrega, F.L.; Brouns, S.J.J.; Dutilh, B.E. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 2018, 27, 51–63. [Google Scholar] [CrossRef]
- Jacquemot, L.; Bettarel, Y.; Monjol, J.; Corre, E.; Halary, S.; Desnues, C.; Bouvier, T.; Ferrier-Pagès, C.; Baudoux, A.C. Therapeutic potential of a new jumbo phage that infects Vibrio coralliilyticus, a widespread coral pathogen. Front. Microbiol. 2018, 9, 2501. [Google Scholar] [CrossRef] [Green Version]
- Abedon, S.T. Selection for bacteriophage latent period length by bacterial density: A theoretical examination. Microb. Ecol. 1989, 182, 79–88. [Google Scholar] [CrossRef]


| Strain | Infectivity |
|---|---|
| Aeromonas hydrophila A18 | + |
| Aeromonas hydrophila Ah2 | - |
| Aeromonas hydrophila ATCC49140 | - |
| Aeromonas hydrophila AS1.1801 | - |
| Aeromonas sobria ATCC43979 | - |
| Aeromonas salmonicida CGMCC1.16015 | - |
| Aeromonas veronii CGMCC 1.927 | - |
| Aeromonas veronii AV4 | - |
| Aeromonas Veyron | - |
| Hafnia paralvei LY-23 | - |
| Hafnia psychrotolerans CGMCC 1.12806 | - |
| Hafnia alvei CGMCC 1.2026 | - |
| Shigella dysenteriae | - |
| Salmonella paratyphi | - |
| Escherichia coli DH5α | - |
| Pseudomonas aeruginosa | - |
| Lactococcus garvieae | - |
| Shewanellaputrefaciens | - |
| Citrobacter freundii | - |
| Enterobacter cloacae | - |
| Enterobacter hormaechei 1322 | - |
| Vibrio harveyis | - |
| Vibrio harveyi 1–5 | - |
| Vibrio alginolyticuss | - |
| Vibrio alginolyticus WY | - |
| Vibrio alginolyticus LDF | - |
| Vibrio mediterranei 117-T6 | - |
| Vibrio Parahemolyticus MCCC 1A11655 | - |
| Vibrio anguillarum | - |
| Vibrio campbellii MCCC 1A08161 | - |
| Edwardsiellatarda ET | - |
| Edwardsiellaictaluri ATCC 33202 | - |
| Edwardsiellahoshinae ATCC 33379 | - |
| Edwardsiella piscicida MCCC 1K03230 | - |
| Edwardsiellatarda MCCC235 | - |
| Groups | Treatment | Mortality |
|---|---|---|
| Blank group | Successive injection (n = 2) with 100 µL of 0.01 M PBS (Injection time interval: 2 h) | 1.7 ± 2.4% |
| Control group | Successive injection with 100 µL of A. hydrophila A18 (108 CFU/mL) and 0.01 M PBS(Injection time interval: 2) | 100.0% |
| Test group I | Successive injection with 100 µL of A. hydrophila A18 (108 CFU/mL) and phage Ahy-yong1 (107 PFU/mL) (Injection time interval: 2 h) | 43.3 ± 4.7% |
| Test group II | Successive injection with 100 µL ofphage Ahy-yong1 (107 PFU/mL) and A. hydrophila A18 (108 CFU/mL) (Injection time interval: 2 h) | 20.0 ± 8.2% |
| Test group III | Injected with 100 µL of phage Ahy-yong1 (107 PFU/mL) immediately after the injection of 100 µL of A. hydrophila A18 (108 CFU/mL) | 30.0 ± 8.2% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, L.; Li, D.; Lin, W.; Liu, W.; Qu, C.; Qian, M.; Cai, R.; Zhou, Q.; Wang, F.; Tong, Y. Novel Aeromonas Phage Ahy-Yong1 and Its Protective Effects against Aeromonas hydrophila in Brocade Carp (Cyprinus aka Koi). Viruses 2022, 14, 2498. https://doi.org/10.3390/v14112498
Pan L, Li D, Lin W, Liu W, Qu C, Qian M, Cai R, Zhou Q, Wang F, Tong Y. Novel Aeromonas Phage Ahy-Yong1 and Its Protective Effects against Aeromonas hydrophila in Brocade Carp (Cyprinus aka Koi). Viruses. 2022; 14(11):2498. https://doi.org/10.3390/v14112498
Chicago/Turabian StylePan, Lingting, Dengfeng Li, Wei Lin, Wencai Liu, Chenxin Qu, Minhua Qian, Ruqian Cai, Qin Zhou, Fei Wang, and Yigang Tong. 2022. "Novel Aeromonas Phage Ahy-Yong1 and Its Protective Effects against Aeromonas hydrophila in Brocade Carp (Cyprinus aka Koi)" Viruses 14, no. 11: 2498. https://doi.org/10.3390/v14112498






