Assessing Multi-Attribute Characterization of Enveloped and Non-Enveloped Viral Particles by Capillary Electrophoresis
Abstract
1. Introduction
2. Materials and Methods
2.1. Purified Samples
2.2. In-Process Samples
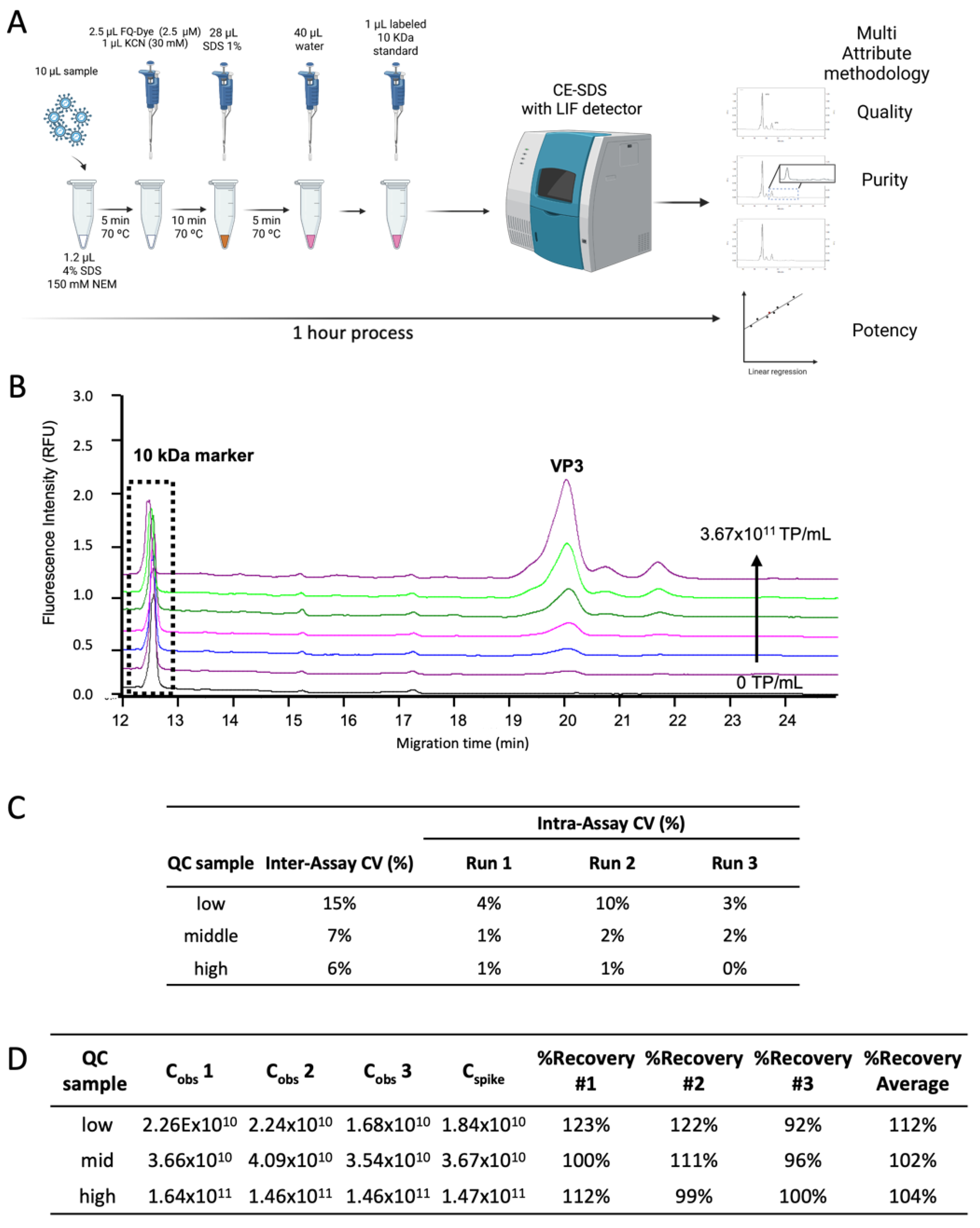
2.3. Sample Preparation for CE-SDS LIF
2.4. CE-SDS LIF Instrument Setup
2.5. CE-SDS LIF Qualification as a Quantification Method for AAV
2.6. CE-SDS UV
2.7. SDS-PAGE with Sypro Ruby Pro Staining
2.8. Quantification of Adenovirus 5 by qPCR
2.9. Quantification of Adeno-Associated Virus by ELISA
2.10. Lentivirus Infectivity Assay
2.11. Lentivirus Total Particles Quantification
3. Results
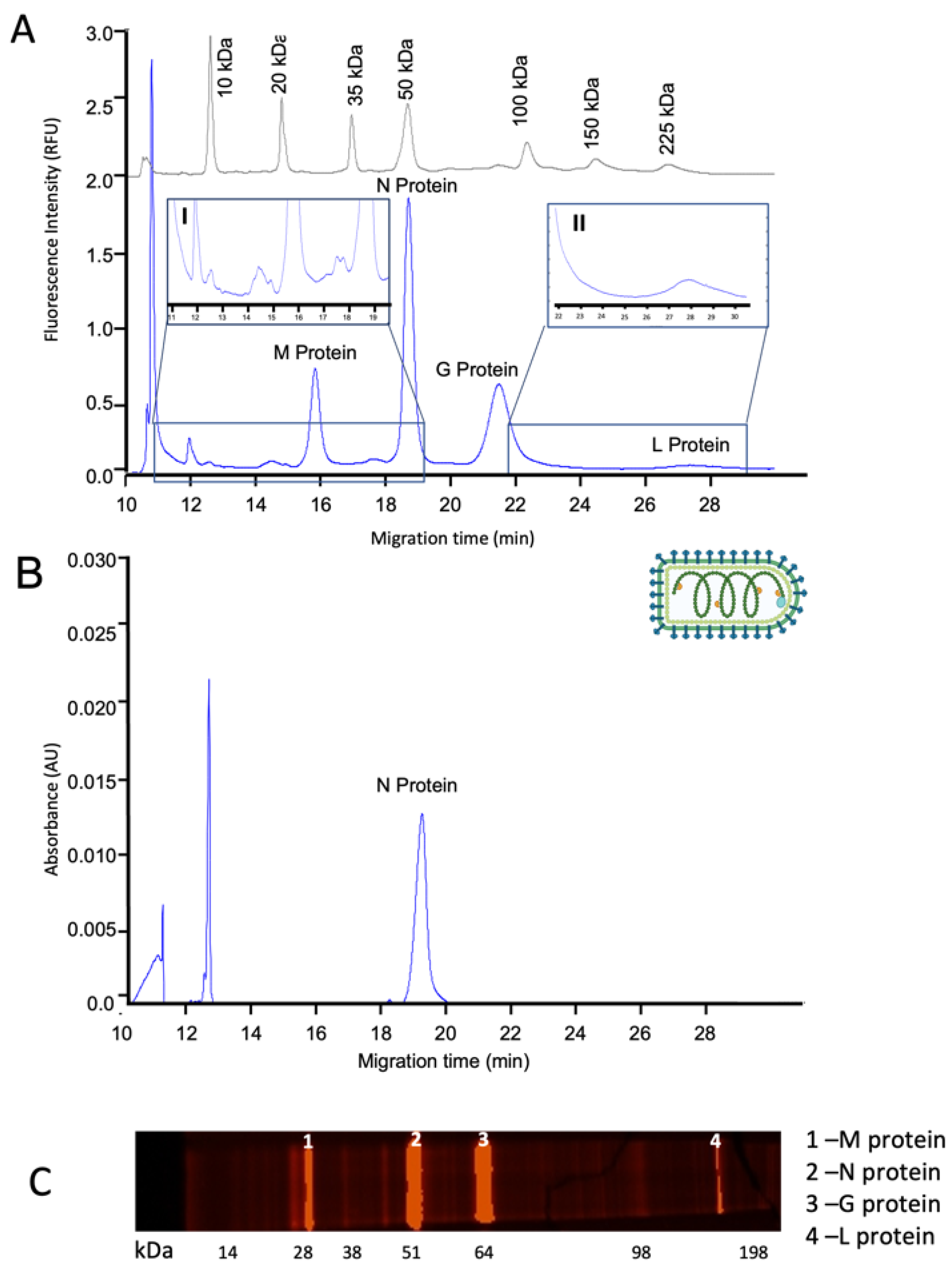
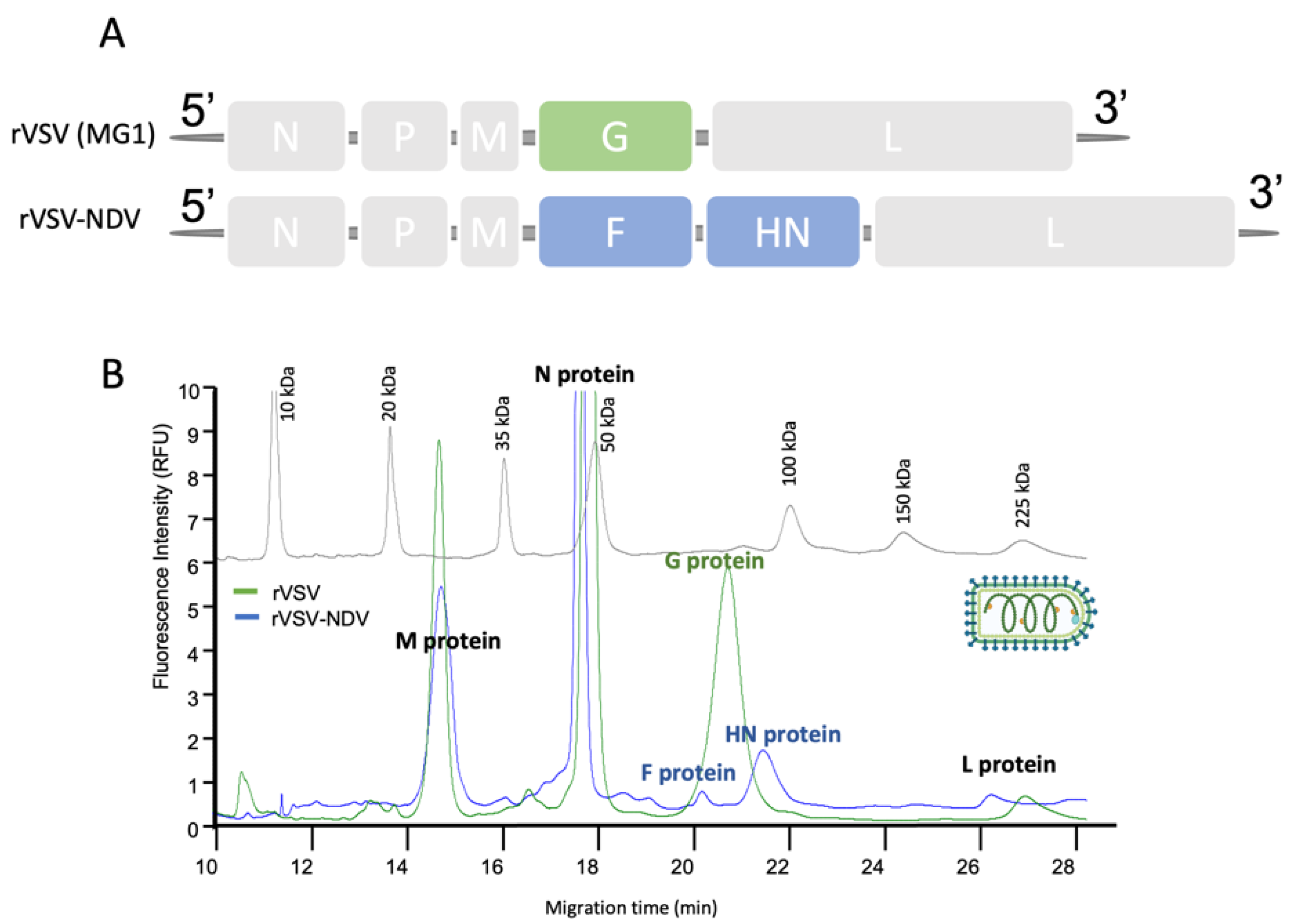
3.1. Capillary Gel Electrophoresis Analysis of Virus-Based Products
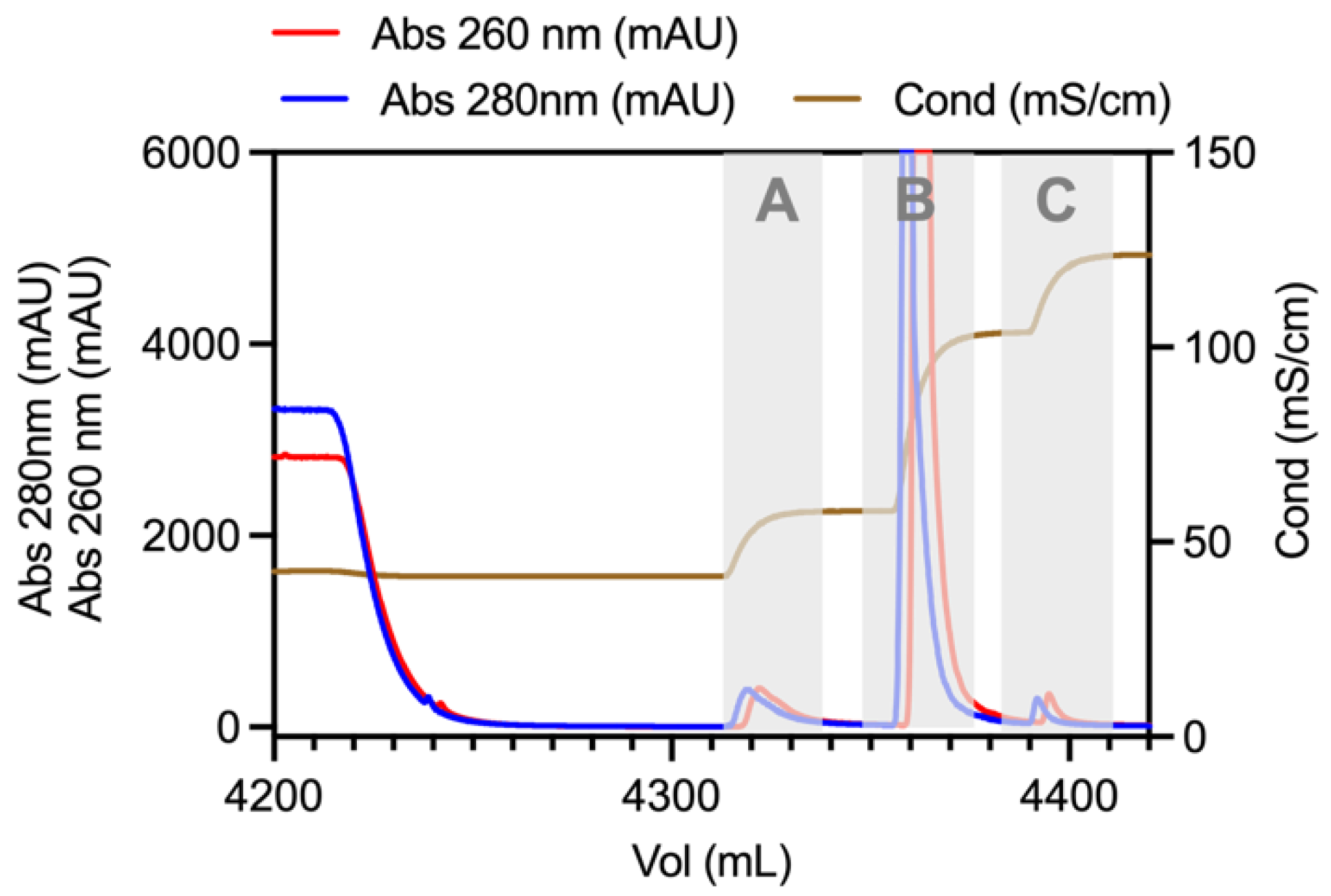
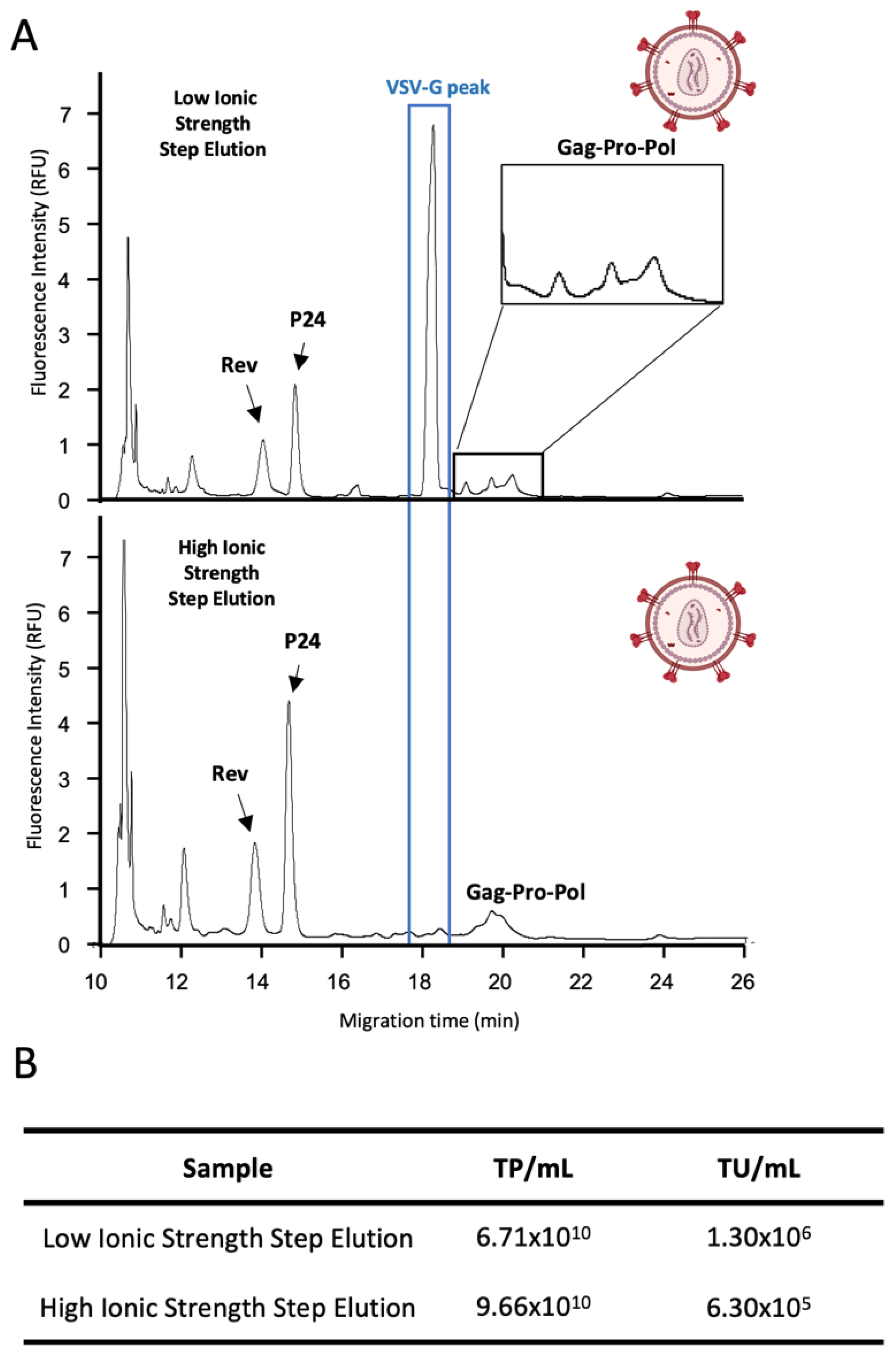
3.2. Qualitative Analysis of Virus In-Process Purification Samples
3.3. Method Qualification for Virus-Based Targets Quantification
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
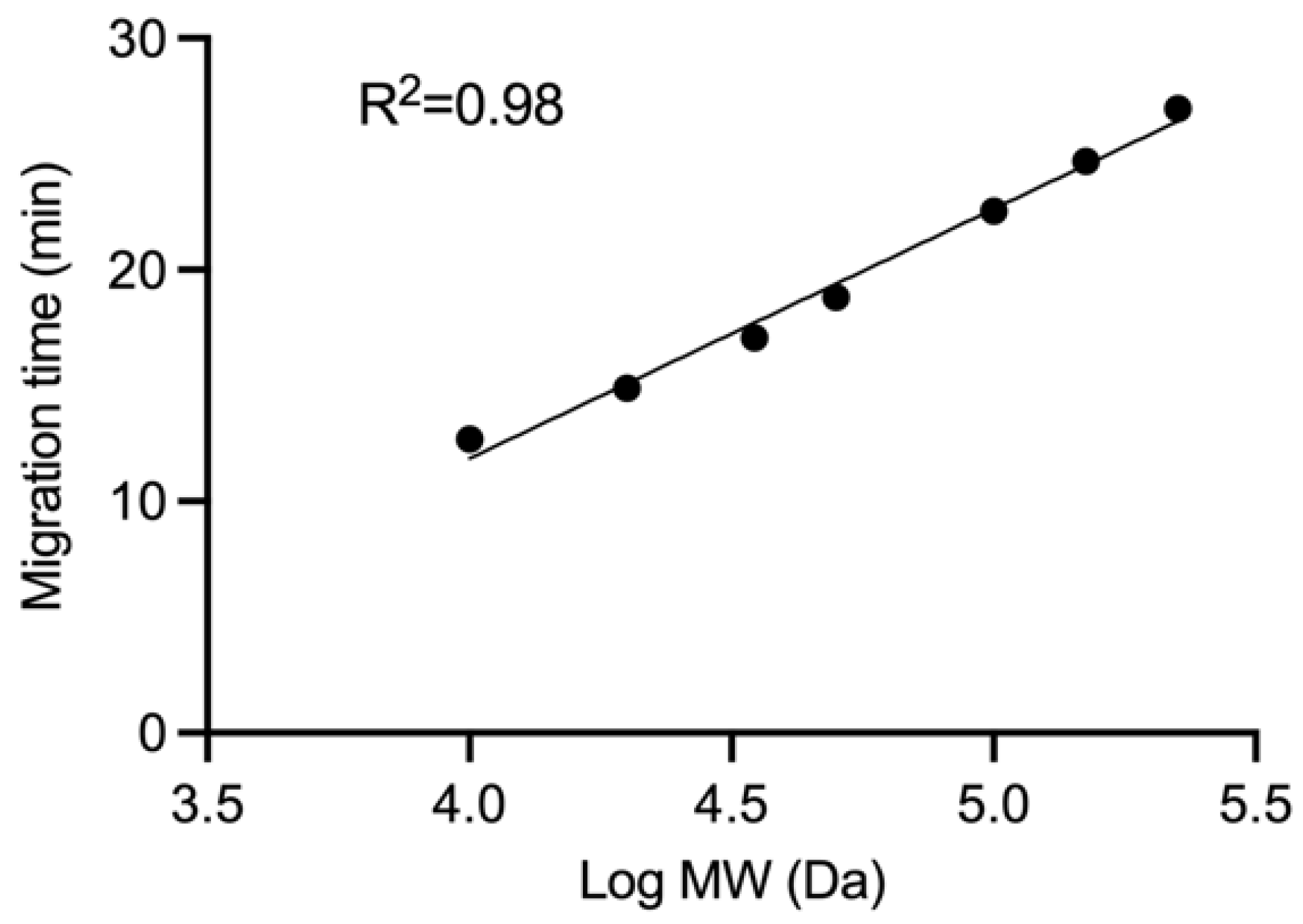
| Protein | MT (min) | log MW (Da) | MW (kDa) | |
|---|---|---|---|---|
| MG1 | M | 15.76 | 4.36 | 23 |
| N | 18.61 | 4.63 | 42 | |
| G | 21.38 | 4.88 | 77 | |
| L | 27.02 | 5.41 | 256 | |
| rVSV-NDV | M | 15.66 | 4.35 | 23 |
| N | 18.55 | 4.62 | 42 | |
| F | 20.65 | 4.82 | 66 | |
| HN | 21.70 | 4.91 | 82 | |
| L | 27.02 | 5.41 | 256 |
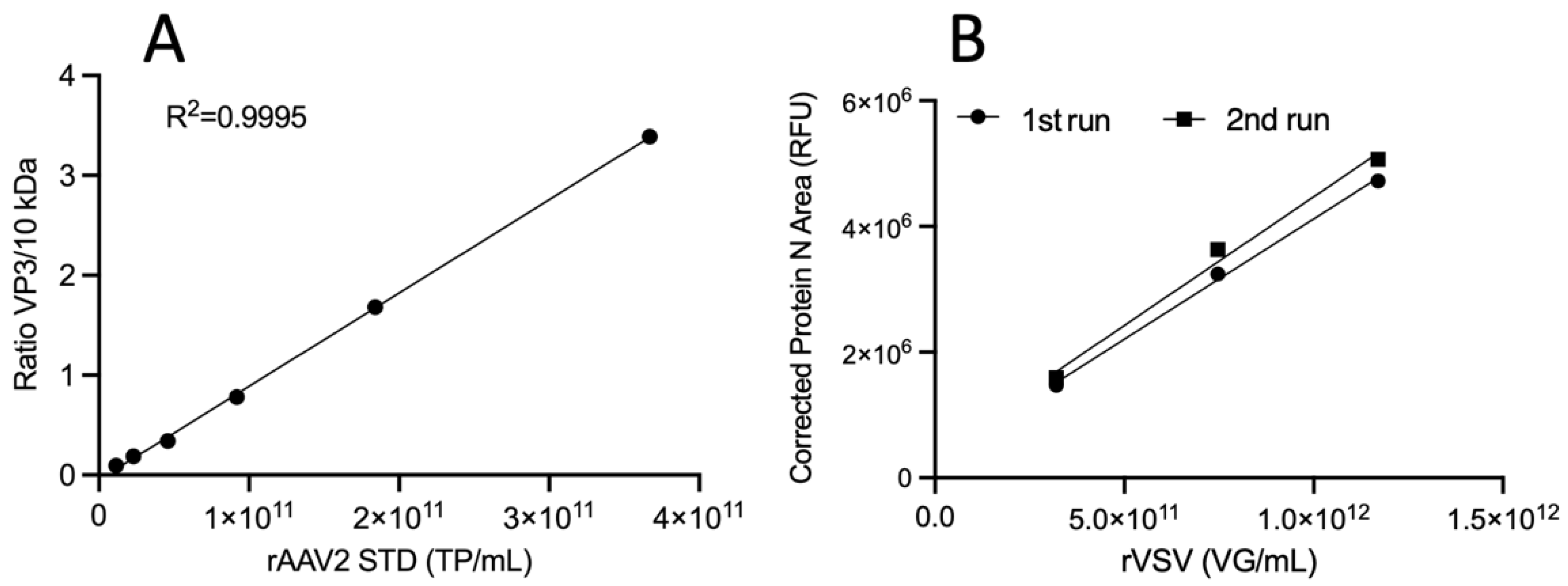
| Replicates | |||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | Average | ||
| Analyst 1 Run1 | LOD (TP/mL) | 2.86 × 109 | 3.10 × 109 | 2.31 × 109 | 2.76 × 109 |
| LOQ (TP/mL) | 8.66 × 109 | 9.40 × 109 | 6.99 × 109 | 8.35 × 109 | |
| Analyst 1 Run2 | LOD (TP/mL) | 3.41 × 109 | 6.08 × 109 | 4.49 × 109 | 4.66 × 109 |
| LOQ (TP/mL) | 1.03 × 1010 | 1.84 × 1010 | 1.36 × 1010 | 1.41 × 1010 | |
| Analyst 2 Run3 | LOD (TP/mL) | 1.98 × 109 | 2.80 × 109 | 1.71 × 109 | 2.17 × 109 |
| LOQ (TP/mL) | 6.01 × 109 | 8.50 × 109 | 5.18 × 109 | 6.56 × 109 | |
References
- Moleirinho, M.G.; Silva, R.J.S.; Alves, P.M.; Manuel, J.T.; Peixoto, C.; Moleirinho, M.G.; Silva, R.J.S.; Alves, P.M.; Manuel, J.T. Current challenges in biotherapeutic particles manufacturing. Expert Opin. Biol. Ther. 2020, 20, 451–465. [Google Scholar] [CrossRef] [PubMed]
- Merten, O.-W.; Schweizer, M.; Chahal, P.; Kamen, A. Manufacturing of viral vectors: Part II. Downstream processing and safety aspects. Pharm. Bioprocess. 2014, 2, 237–251. [Google Scholar] [CrossRef]
- Moleirinho, M.G.; Feast, S.; Moreira, A.S.; Silva, R.J.S.; Alves, P.M.; Carrondo, M.J.T.; Huber, T.; Fee, C.; Peixoto, C. 3D-printed ordered bed structures for chromatographic purification of enveloped and non-enveloped viral particles. Sep. Purif. Technol. 2021, 254, 117681. [Google Scholar] [CrossRef]
- Carvalho, S.B.; Moleirinho, M.G.; Wheatley, D.; Welsh, J.; Gantier, R.; Alves, P.M.; Peixoto, C.; Carrondo, M.J.T. Universal label-free in-process quantification of influenza virus-like particles. Biotechnol. J. 2017, 12, 1700031. [Google Scholar] [CrossRef] [PubMed]
- Lothert, K.; Eilts, F.; Wolff, M.W. Quantification methods for viruses and virus-like particles applied in biopharmaceutical production processes. Expert Rev. Vaccines 2022, 21, 1029–1044. [Google Scholar] [CrossRef] [PubMed]
- Lock, S.J.; Hendriks, K.; Berliet, J. The use of capillary electrophoresis in Gene Therapy. Cytotherapy 2020, 22, S152. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, P.; Tao, C.; Zhang, D.; Li, Z.; Yamaguchi, Y. Capillary electrophoresis of DNA with high resolution based on copoly(pentaerythritoltetra succinimidylcarboxypentyl/aminopropyl polyoxyethylene) hydrogel. Anal. Chim. Acta 2021, 1178, 338811. [Google Scholar] [CrossRef]
- Mellado, M.C.M.; Franco, C.; Coelho, A.; Alves, P.M.; Simplício, A.L. Sodium dodecyl sulfate-capillary gel electrophoresis analysis of rotavirus-like particles. J. Chromatogr. A 2008, 1192, 166–172. [Google Scholar] [CrossRef]
- Bunz, S.C.; Rapp, E.; Neusüss, C. Capillary electrophoresis/mass spectrometry of APTS-labeled glycans for the identification of unknown glycan species in capillary electrophoresis/laser-induced fluorescence systems. Anal. Chem. 2013, 85, 10218–10224. [Google Scholar] [CrossRef]
- Yu, F.; Zhang, D.; Yuan, Z.; Zhao, Q.; Wang, H. Application of affinity capillary electrophoresis in the study of protein-DNA interactions. Se pu = Chin. J. Chromatogr. 2020, 38, 1133–1142. [Google Scholar] [CrossRef]
- Hernández-Borges, J.; Neusüß, C.; Cifuentes, A.; Pelzing, M. On-line capillary electrophoresis-mass spectrometry for the analysis of biomolecules. Electrophoresis 2004, 25, 2257–2281. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wang, S.; Cong, H.; Yu, B. Analysis of metabolomics and proteomics based on capillary electrophoresis-mass spectrometry. Se pu = Chin. J. Chromatogr. 2020, 38, 1013–1021. [Google Scholar] [CrossRef]
- Kumar, R.; Guttman, A.; Rathore, A.S. Applications of capillary electrophoresis for biopharmaceutical product characterization. Electrophoresis 2022, 43, 143–166. [Google Scholar] [CrossRef]
- Escandell, J.M.; Pais, D.A.; Carvalho, S.B.; Vincent, K.; Gomes-Alves, P.; Alves, P.M. Leveraging rAAV bioprocess understanding and next generation bioanalytics development. Curr. Opin. Biotechnol. 2022, 74, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Beckman, J.; Zhang, Y.; Li, Z.J.; Szigeti, M.; Guttman, A. Capillary electrophoresis and the biopharmaceutical industry: Therapeutic protein analysis and characterization. TrAC-Trends Anal. Chem. 2021, 144, 116407. [Google Scholar] [CrossRef]
- Salas-Solano, O.; Tomlinson, B.; Du, S.; Parker, M.; Strahan, A.; Ma, S. Optimization and validation of a quantitative capillary electrophoresis sodium dodecyl sulfate method for quality control and stability monitoring of monoclonal antibodies. Anal. Chem. 2006, 78, 6583–6594. [Google Scholar] [CrossRef]
- Michels, D.A.; Parker, M.; Salas-Solano, O. Quantitative impurity analysis of monoclonal antibody size heterogeneity by CE-LIF: Example of development and validation through a quality-by-design framework. Electrophoresis 2012, 33, 815–826. [Google Scholar] [CrossRef]
- Zhang, Z.; Park, J.; Barrett, H.; Dooley, S.; Davies, C.; Verhagen, M.F. Capillary Electrophoresis-Sodium Dodecyl Sulfate with Laser-Induced Fluorescence Detection as a Highly Sensitive and Quality Control-Friendly Method for Monitoring Adeno-Associated Virus Capsid Protein Purity. Hum. Gene Ther. 2021, 32, 628–637. [Google Scholar] [CrossRef] [PubMed]
- EMA. ICH Topic Q 2 (R1) Validation of Analytical Procedures: Text and Methodology, 3rd ed.; ICH Secretariat: Geneva, Switzerland, 1995.
- Martin, J.; Frederick, A.; Luo, Y.; Jackson, R.; Joubert, M.; Sol, B.; Poulin, F.; Pastor, E.; Armentano, D.; Wadsworth, S.; et al. Generation and characterization of adeno-associated virus producer cell lines for research and preclinical vector production. Hum. Gene Ther. Methods 2013, 24, 253–269. [Google Scholar] [CrossRef] [PubMed]
- Chahal, P.S.; Schulze, E.; Tran, R.; Montes, J.; Kamen, A.A. Production of adeno-associated virus (AAV) serotypes by transient transfection of HEK293 cell suspension cultures for gene delivery. J. Virol. Methods 2014, 196, 163–173. [Google Scholar] [CrossRef]
- Tomás, H.A.; Rodrigues, A.F.; Carrondo, M.J.T.; Coroadinha, A.S. LentiPro26: Novel stable cell lines for constitutive lentiviral vector production. Sci. Rep. 2018, 8, 5271. [Google Scholar] [CrossRef] [PubMed]
- Moreira, A.S.; Faria, T.Q.; Oliveira, J.G.; Kavara, A.; Schofield, M.; Sanderson, T.; Collins, M.; Gantier, R.; Alves, P.M.; Carrondo, M.J.T.; et al. Enhancing the purification of Lentiviral vectors for clinical applications. Sep. Purif. Technol. 2021, 274, 118598. [Google Scholar] [CrossRef]
- Nass, S.A.; Mattingly, M.A.; Woodcock, D.A.; Burnham, B.L.; Ardinger, J.A.; Osmond, S.E.; Frederick, A.M.; Scaria, A.; Seng, H.; Riordan, C.R.O. Universal Method for the Purification of Recombinant AAV Vectors of Differing Serotypes. Mol. Ther. Methods Clin. Dev. 2018, 9, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Santos, M.; Guttman, A. Ultrahigh Sensitivity Analysis of Adeno-associated Virus (AAV) Capsid Proteins by Sodium Dodecyl Sulphate Capillary Gel Electrophoresis. Chromatography Today. 2020, pp. 2–4. Available online: https://www.chromatographytoday.com/article/bioanalytical/40/sciex/ultrahigh-sensitivity-analysis-of-adeno-associated-virus-aavbr-capsid-proteins-by-sodium-dodecyl-sulphate-capillary-gel-electrophoresis/2852 (accessed on 19 October 2022).
- Li, T.; Malik, M.; Yowanto, H.; Mollah, S. Purity Analysis of Adeno-Associated Virus (AAV) Capsid Proteins using CE-SDS Method. Application Note. 2019, pp. 8–13. Available online: https://lcms.cz/labrulez-bucket-strapi-h3hsga3/AAV_purity_73f90a1968/AAV-purity.pdf (accessed on 19 October 2022).
- Santos, J.M.; Heiniö, C.; Cervera-Carrascon, V.; Quixabeira, D.C.A.; Siurala, M.; Havunen, R.; Butzow, R.; Zafar, S.; de Gruijl, T.; Lassus, H.; et al. Original research: Oncolytic adenovirus shapes the ovarian tumor microenvironment for potent tumor-infiltrating lymphocyte tumor reactivity. J. Immunother. Cancer 2020, 8, 188. [Google Scholar] [CrossRef]
- Wörner, T.P.; Bennett, A.; Habka, S.; Snijder, J.; Friese, O.; Powers, T.; Agbandje-McKenna, M.; Heck, A.J.R. Adeno-associated virus capsid assembly is divergent and stochastic. Nat. Commun. 2021, 12, 1642. [Google Scholar] [CrossRef]
- Zhang, C.X.; Meagher, M.M. Sample Stacking Provides Three Orders of Magnitude Sensitivity Enhancement in SDS Capillary Gel Electrophoresis of Adeno-Associated Virus Capsid Proteins. Anal. Chem. 2017, 89, 3285–3292. [Google Scholar] [CrossRef]
- Oyama, H.; Ishii, K.; Maruno, T.; Torisu, T.; Uchiyama, S. Characterization of Adeno-Associated Virus Capsid Proteins with Two Types of VP3-Related Components by Capillary Gel Electrophoresis and Mass Spectrometry. Hum. Gene Ther. 2021, 32, 1403–1416. [Google Scholar] [CrossRef]
- Bosma, B.; du Plessis, F.; Ehlert, E.; Nijmeijer, B.; de Haan, M.; Petry, H.; Lubelski, J. Optimization of viral protein ratios for production of rAAV serotype 5 in the baculovirus system. Gene Ther. 2018, 25, 415–424. [Google Scholar] [CrossRef]
- Srivastava, A.C.; Mallela, K.M.G.; Deorkar, N.; Brophy, G. Manufacturing challenges and rational formulation development for AAV viral vectors. J. Pharm. Sci. 2021, 110, 2609–2624. [Google Scholar] [CrossRef] [PubMed]
- Saadatpour, F.; Mohammadipanah, F. Physicochemical susceptibility of SARS-CoV-2 to disinfection and physical approach of prophylaxis. Health Sci. Rep. 2020, 3, e213. [Google Scholar] [CrossRef] [PubMed]
- Penaud-Budloo, M.; Broucque, F.; Harrouet, K.; Bouzelha, M.; Saleun, S.; Douthe, S.; D’Costa, S.; Ogram, S.; Adjali, O.; Blouin, V.; et al. Stability of the adeno-associated virus 8 reference standard material. Gene Ther. 2019, 26, 211–215. [Google Scholar] [CrossRef]
- Tomás, H.A.; Mestre, D.A.; Rodrigues, A.F.; Guerreiro, M.R.; Carrondo, M.J.T.; Coroadinha, A.S. Improved GaLV-TR Glycoproteins to Pseudotype Lentiviral Vectors: Impact of Viral Protease Activity in the Production of LV Pseudotypes. Mol. Ther.-Methods Clin. Dev. 2019, 15, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Xue, C.; Kong, Q.; Zhang, C.; Bi, Y.; Cao, Y. Proteomic analysis of purified Newcastle disease virus particles. Proteome Sci. 2012, 10, 32. [Google Scholar] [CrossRef] [PubMed]
- Tözsér, J. Comparative studies on retroviral proteases: Substrate specificity. Viruses 2010, 2, 147–165. [Google Scholar] [CrossRef]
- Do Minh, A.; Star, A.T.; Stupak, J.; Fulton, K.M.; Haqqani, A.S.; Gélinas, J.F.; Li, J.; Twine, S.M.; Kamen, A.A. Characterization of extracellular vesicles secreted in lentiviral producing hek293sf cell cultures. Viruses 2021, 13, 797. [Google Scholar] [CrossRef]
- Labisch, J.J.; Philip Wiese, G.; Barnes, K.; Bollmann, F.; Pflanz, K. Infectious titer determination of lentiviral vectors using a temporal immunological real-time imaging approach. PLoS ONE 2021, 16, e0254739. [Google Scholar] [CrossRef]
- Gimpel, A.L.; Katsikis, G.; Sha, S.; Maloney, A.J.; Hong, M.S.; Nguyen, T.N.T.; Wolfrum, J.; Springs, S.L.; Sinskey, A.J.; Manalis, S.R.; et al. Analytical methods for process and product characterization of recombinant adeno-associated virus-based gene therapies. Mol. Ther.-Methods Clin. Dev. 2021, 20, 740–754. [Google Scholar] [CrossRef]
- Harron, D.W.G. Technical Requirements for Registration of Pharmaceuticals for Human Use: The ICH Process. Textb. Pharm. Med. 2013, 1994, 447–460. [Google Scholar] [CrossRef]
- Booth, B.P.; Simon, W.C. Analytical method validation. In New Drug Development; CRC Press: Boca Raton, FL, USA, 2004; pp. 138–159. [Google Scholar] [CrossRef]


| Solution | Aim | Operation Conditions (Conditioning) | Operation Conditions (Shutdown) |
|---|---|---|---|
| 0.1 M NaOH | cleaning | 10 min, 20 psi, forward | 10 min, 70 psi, forward |
| 0.1 M HCl | neutralization | 5 min, 20 psi, forward | 5 min, 50 psi, forward |
| LC-MS grade water | removal of acid residues | 2 min, 20 psi, forward | 2 min, 50 psi, forward |
| SDS-MW gel buffer | filling | 10 min, 70 psi, forward | 10 min, 70 psi, forward |
| SDS-MW gel buffer | equilibration | 10 min, 15 kV, forward | 10 min, 15 kV, reverse polarity |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, R.P.; Escandell, J.M.; Guerreiro, A.C.L.; Moura, F.; Faria, T.Q.; Carvalho, S.B.; Silva, R.J.S.; Gomes-Alves, P.; Peixoto, C. Assessing Multi-Attribute Characterization of Enveloped and Non-Enveloped Viral Particles by Capillary Electrophoresis. Viruses 2022, 14, 2539. https://doi.org/10.3390/v14112539
Fernandes RP, Escandell JM, Guerreiro ACL, Moura F, Faria TQ, Carvalho SB, Silva RJS, Gomes-Alves P, Peixoto C. Assessing Multi-Attribute Characterization of Enveloped and Non-Enveloped Viral Particles by Capillary Electrophoresis. Viruses. 2022; 14(11):2539. https://doi.org/10.3390/v14112539
Chicago/Turabian StyleFernandes, Rita P., José M. Escandell, Ana C. L. Guerreiro, Filipa Moura, Tiago Q. Faria, Sofia B. Carvalho, Ricardo J. S. Silva, Patrícia Gomes-Alves, and Cristina Peixoto. 2022. "Assessing Multi-Attribute Characterization of Enveloped and Non-Enveloped Viral Particles by Capillary Electrophoresis" Viruses 14, no. 11: 2539. https://doi.org/10.3390/v14112539
APA StyleFernandes, R. P., Escandell, J. M., Guerreiro, A. C. L., Moura, F., Faria, T. Q., Carvalho, S. B., Silva, R. J. S., Gomes-Alves, P., & Peixoto, C. (2022). Assessing Multi-Attribute Characterization of Enveloped and Non-Enveloped Viral Particles by Capillary Electrophoresis. Viruses, 14(11), 2539. https://doi.org/10.3390/v14112539





