Soft X-ray Tomography Reveals HSV-1-Induced Remodeling of Human B Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Viral Infection
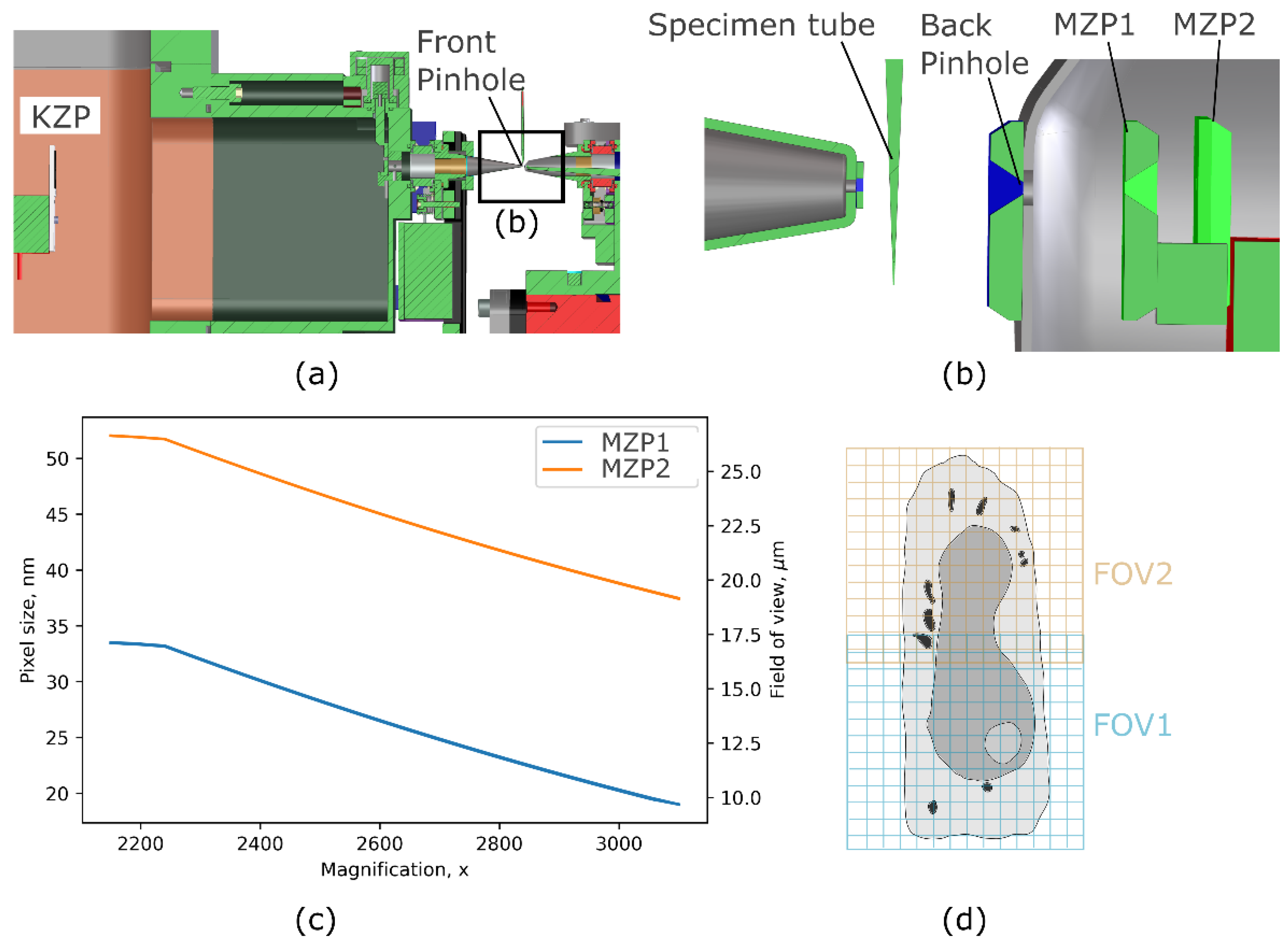
2.2. Soft X-ray Microscopy
2.3. Data Analysis
3. Results
3.1. Remodelling of Entire Cells by HSV-1
3.2. Quantitative Analysis of HSV-1 Viral Particles
3.3. Heterogeneity of Replication Compartments
3.4. Complex Three-Dimensional Membrane Structural Alterations
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eisenreich, W.; Rudel, T.; Heesemann, J.; Goebel, W. How Viral and Intracellular Bacterial Pathogens Reprogram the Metabolism of Host Cells to Allow Their Intracellular Replication. Front. Cell. Infect. Microbiol. 2019, 9, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorvel, J.-P. Pathogen–Host Cell Molecular Interactions: Knowledge and Challenge. Cell 2000, 103, 550–552. [Google Scholar] [CrossRef] [Green Version]
- Glingston, R.S.; Deb, R.; Kumar, S.; Nagotu, S. Organelle Dynamics and Viral Infections: At Cross Roads. Microbes Infect. 2019, 21, 20–32. [Google Scholar] [CrossRef]
- Robb, N.C. Virus Morphology: Insights from Super-Resolution Fluorescence Microscopy. Biochim. Biophys. Acta Mol. Basis Dis. 2022, 1868, 166347. [Google Scholar] [CrossRef]
- Richert-Pöggeler, K.R.; Franzke, K.; Hipp, K.; Kleespies, R.G. Electron Microscopy Methods for Virus Diagnosis and High Resolution Analysis of Viruses. Front. Microbiol. 2019, 9, 3255. [Google Scholar] [CrossRef] [Green Version]
- Baena, V.; Conrad, R.; Friday, P.; Fitzgerald, E.; Kim, T.; Bernbaum, J.; Berensmann, H.; Harned, A.; Nagashima, K.; Narayan, K. FIB-SEM as a Volume Electron Microscopy Approach to Study Cellular Architectures in SARS-CoV-2 and Other Viral Infections: A Practical Primer for a Virologist. Viruses 2021, 13, 611. [Google Scholar] [CrossRef]
- Peddie, C.J.; Genoud, C.; Kreshuk, A.; Meechan, K.; Micheva, K.D.; Narayan, K.; Pape, C.; Parton, R.G.; Schieber, N.L.; Schwab, Y.; et al. Volume Electron Microscopy. Nat. Rev. Methods Primer 2022, 2, 1–23. [Google Scholar] [CrossRef]
- Ronchi, P.; Mizzon, G.; Machado, P.; D’Imprima, E.; Best, B.T.; Cassella, L.; Schnorrenberg, S.; Montero, M.G.; Jechlinger, M.; Ephrussi, A.; et al. High-Precision Targeting Workflow for Volume Electron Microscopy. J. Cell Biol. 2021, 220, e202104069. [Google Scholar] [CrossRef]
- Xu, C.S.; Hayworth, K.J.; Lu, Z.; Grob, P.; Hassan, A.M.; García-Cerdán, J.G.; Niyogi, K.K.; Nogales, E.; Weinberg, R.J.; Hess, H.F. Enhanced FIB-SEM Systems for Large-Volume 3D Imaging. eLife 2017, 6, e25916. [Google Scholar] [CrossRef]
- Harkiolaki, M.; Darrow, M.C.; Spink, M.C.; Kosior, E.; Dent, K.; Duke, E. Cryo-Soft X-ray Tomography: Using Soft X-rays to Explore the Ultrastructure of Whole Cells. Emerg. Top. Life Sci. 2018, 2, 81–92. [Google Scholar] [CrossRef]
- Guo, J.; Larabell, C.A. Soft X-ray Tomography: Virtual Sculptures from Cell Cultures. Curr. Opin. Struct. Biol. 2019, 58, 324–332. [Google Scholar] [CrossRef] [PubMed]
- Schneider, G.; Niemann, B.; Guttmann, P.; Rudolph, D.; Schmahl, G. Cryo X-ray Microscopy. Synchrotron Radiat. News 1995, 8, 19–28. [Google Scholar] [CrossRef]
- Weiß, D.; Schneider, G.; Niemann, B.; Guttmann, P.; Rudolph, D.; Schmahl, G. Computed Tomography of Cryogenic Biological Specimens Based on X-ray Microscopic Images. Ultramicroscopy 2000, 84, 185–197. [Google Scholar] [CrossRef] [PubMed]
- McDermott, G.; Fox, D.M.; Epperly, L.; Wetzler, M.; Barron, A.E.; le Gros, M.A.; Larabell, C.A. Visualizing and Quantifying Cell Phenotype Using Soft X-ray Tomography. BioEssays News Rev. Mol. Cell. Dev. Biol. 2012, 34, 320–327. [Google Scholar] [CrossRef] [PubMed]
- Le Gros, M.A.; Clowney, E.J.; Magklara, A.; Yen, A.; Markenscoff-Papadimitriou, E.; Colquitt, B.; Myllys, M.; Kellis, M.; Lomvardas, S.; Larabell, C.A. Soft X-ray Tomography Reveals Gradual Chromatin Compaction and Reorganization during Neurogenesis In Vivo. Cell Rep. 2016, 17, 2125–2136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, E.A.; McDermott, G.; Do, M.; Leung, K.; Panning, B.; le Gros, M.A.; Larabell, C.A. Quantitatively Imaging Chromosomes by Correlated Cryo-Fluorescence and Soft X-ray Tomographies. Biophys. J. 2014, 107, 1988–1996. [Google Scholar] [CrossRef] [Green Version]
- Aho, V.; Mäntylä, E.; Ekman, A.; Hakanen, S.; Mattola, S.; Chen, J.-H.; Weinhardt, V.; Ruokolainen, V.; Sodeik, B.; Larabell, C.; et al. Quantitative Microscopy Reveals Stepwise Alteration of Chromatin Structure during Herpesvirus Infection. Viruses 2019, 11, 935. [Google Scholar] [CrossRef] [Green Version]
- Kounatidis, I.; Stanifer, M.L.; Phillips, M.A.; Paul-Gilloteaux, P.; Heiligenstein, X.; Wang, H.; Okolo, C.A.; Fish, T.M.; Spink, M.C.; Stuart, D.I.; et al. 3D Correlative Cryo-Structured Illumination Fluorescence and Soft X-ray Microscopy Elucidates Reovirus Intracellular Release Pathway. Cell 2020, 182, 515–530.e17. [Google Scholar] [CrossRef]
- Loconte, V.; Chen, J.-H.; Cortese, M.; Ekman, A.; le Gros, M.A.; Larabell, C.; Bartenschlager, R.; Weinhardt, V. Using Soft X-ray Tomography for Rapid Whole-Cell Quantitative Imaging of SARS-CoV-2-Infected Cells. Cell Rep. Methods 2021, 1, 100117. [Google Scholar] [CrossRef]
- Schneider, G.; Guttmann, P.; Rehbein, S.; Werner, S.; Follath, R. Cryo X-ray Microscope with Flat Sample Geometry for Correlative Fluorescence and Nanoscale Tomographic Imaging. J. Struct. Biol. 2012, 177, 212–223. [Google Scholar] [CrossRef]
- Pereiro, E. Correlative Cryo-Soft X-ray Tomography of Cells. Biophys. Rev. 2019, 11, 529–530. [Google Scholar] [CrossRef] [PubMed]
- Suzich, J.B.; Cliffe, A.R. Strength in Diversity: Understanding the Pathways to Herpes Simplex Virus Reactivation. Virology 2018, 522, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Grünewald, K.; Desai, P.; Winkler, D.C.; Heymann, J.B.; Belnap, D.M.; Baumeister, W.; Steven, A.C. Three-Dimensional Structure of Herpes Simplex Virus from Cryo-Electron Tomography. Science 2003, 302, 1396–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beilstein, F.; Cohen, G.H.; Eisenberg, R.J.; Nicolas, V.; Esclatine, A.; Pasdeloup, D. Dynamic Organization of Herpesvirus Glycoproteins on the Viral Envelope Revealed by Super-Resolution Microscopy. PLoS Pathog. 2019, 15, e1008209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laine, R.F.; Albecka, A.; van de Linde, S.; Rees, E.J.; Crump, C.M.; Kaminski, C.F. Structural Analysis of Herpes Simplex Virus by Optical Super-Resolution Imaging. Nat. Commun. 2015, 6, 5980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, A.; Kulkarni, S.; Mukherjee, A. Herpes Simplex Virus: The Hostile Guest That Takes Over Your Home. Front. Microbiol. 2020, 11, 733. [Google Scholar] [CrossRef] [PubMed]
- Myllys, M.; Ruokolainen, V.; Aho, V.; Smith, E.A.; Hakanen, S.; Peri, P.; Salvetti, A.; Timonen, J.; Hukkanen, V.; Larabell, C.A.; et al. Herpes Simplex Virus 1 Induces Egress Channels through Marginalized Host Chromatin. Sci. Rep. 2016, 6, 28844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nahas, K.L.; Connor, V.; Scherer, K.M.; Kaminski, C.F.; Harkiolaki, M.; Crump, C.M.; Graham, S.C. Near-Native State Imaging by Cryo-Soft-X-ray Tomography Reveals Remodelling of Multiple Cellular Organelles during HSV-1 Infection. PLoS Pathog. 2022, 18, e1010629. [Google Scholar] [CrossRef]
- Everett, R.D.; Sourvinos, G.; Orr, A. Recruitment of Herpes Simplex Virus Type 1 Transcriptional Regulatory Protein ICP4 into Foci Juxtaposed to ND10 in Live, Infected Cells. J. Virol. 2003, 77, 3680–3689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.-H.; Vanslembrouck, B.; Loconte, V.; Ekman, A.; Cortese, M.; Bartenschlager, R.; McDermott, G.; Larabell, C.A.; le Gros, M.A.; Weinhardt, V. A Protocol for Full-Rotation Soft X-ray Tomography of Single Cells. Star Protoc. 2022, 3, 101176. [Google Scholar] [CrossRef]
- Parkinson, D.Y.; Knoechel, C.; Yang, C.; Larabell, C.A.; le Gros, M.A. Automatic Alignment and Reconstruction of Images for Soft X-ray Tomography. J. Struct. Biol. 2012, 177, 259–266. [Google Scholar] [CrossRef] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDermott, G.; le Gros, M.A.; Knoechel, C.G.; Uchida, M.; Larabell, C.A. Soft X-ray Tomography and Cryogenic Light Microscopy: The Cool Combination in Cellular Imaging. Trends Cell Biol. 2009, 19, 587–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scherer, K.M.; Manton, J.D.; Soh, T.K.; Mascheroni, L.; Connor, V.; Crump, C.M.; Kaminski, C.F. A Fluorescent Reporter System Enables Spatiotemporal Analysis of Host Cell Modification during Herpes Simplex Virus-1 Replication. J. Biol. Chem. 2021, 296, 100236. [Google Scholar] [CrossRef] [PubMed]
- Burgess, H.M.; Mohr, I. Defining the Role of Stress Granules in Innate Immune Suppression by the Herpes Simplex Virus 1 Endoribonuclease VHS. J. Virol. 2018, 92, e00829-18. [Google Scholar] [CrossRef] [Green Version]
- Wheeler, J.R.; Matheny, T.; Jain, S.; Abrisch, R.; Parker, R. Distinct Stages in Stress Granule Assembly and Disassembly. eLife 2016, 5, e18413. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.H.; Dougherty, M.; Jakana, J.; He, J.; Rixon, F.J.; Chiu, W. Seeing the Herpesvirus Capsid at 8.5 A. Science 2000, 288, 877–880. [Google Scholar] [CrossRef]
- Netherton, C.; Moffat, K.; Brooks, E.; Wileman, T. A Guide to Viral Inclusions, Membrane Rearrangements, Factories, and Viroplasm Produced during Virus Replication. Adv. Virus Res. 2007, 70, 101–182. [Google Scholar] [CrossRef]
- Hutchinson, I.; Whiteley, A.; Browne, H.; Elliott, G. Sequential Localization of Two Herpes Simplex Virus Tegument Proteins to Punctate Nuclear Dots Adjacent to ICP0 Domains. J. Virol. 2002, 76, 10365–10373. [Google Scholar] [CrossRef] [PubMed]
- Livingston, C.M.; Ifrim, M.F.; Cowan, A.E.; Weller, S.K. Virus-Induced Chaperone-Enriched (VICE) Domains Function as Nuclear Protein Quality Control Centers during HSV-1 Infection. PLoS Pathog. 2009, 5, e1000619. [Google Scholar] [CrossRef] [Green Version]
- Baines, J.D.; Jacob, R.J.; Simmerman, L.; Roizman, B. The Herpes Simplex Virus 1 UL11 Proteins Are Associated with Cytoplasmic and Nuclear Membranes and with Nuclear Bodies of Infected Cells. J. Virol. 1995, 69, 825–833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wild, P.; Leisinger, S.; de Oliveira, A.P.; Doehner, J.; Schraner, E.M.; Fraevel, C.; Ackermann, M.; Kaech, A. Nuclear Envelope Impairment Is Facilitated by the Herpes Simplex Virus 1 Us3 Kinase. F1000Research 2019, 8, 198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollinshead, M.; Johns, H.L.; Sayers, C.L.; Gonzalez-Lopez, C.; Smith, G.L.; Elliott, G. Endocytic Tubules Regulated by Rab GTPases 5 and 11 Are Used for Envelopment of Herpes Simplex Virus. EMBO J. 2012, 31, 4204–4220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calistri, A.; Sette, P.; Salata, C.; Cancellotti, E.; Forghieri, C.; Comin, A.; Göttlinger, H.; Campadelli-Fiume, G.; Palù, G.; Parolin, C. Intracellular Trafficking and Maturation of Herpes Simplex Virus Type 1 GB and Virus Egress Require Functional Biogenesis of Multivesicular Bodies. J. Virol. 2007, 81, 11468–11478. [Google Scholar] [CrossRef] [Green Version]
- Loconte, V.; White, K.L. The Use of Soft X-ray Tomography to Explore Mitochondrial Structure and Function. Mol. Metab. 2022, 57, 101421. [Google Scholar] [CrossRef] [PubMed]
- Walters, A.D.; Amoateng, K.; Wang, R.; Chen, J.-H.; McDermott, G.; Larabell, C.A.; Gadal, O.; Cohen-Fix, O. Nuclear Envelope Expansion in Budding Yeast Is Independent of Cell Growth and Does Not Determine Nuclear Volume. Mol. Biol. Cell 2019, 30, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Uchida, M.; Sun, Y.; McDermott, G.; Knoechel, C.; le Gros, M.A.; Parkinson, D.; Drubin, D.G.; Larabell, C.A. Quantitative Analysis of Yeast Internal Architecture Using Soft X-ray Tomography. Yeast Chichester Engl. 2011, 28, 227–236. [Google Scholar] [CrossRef] [Green Version]
- Remesh, S.G.; Verma, S.C.; Chen, J.-H.; Ekman, A.A.; Larabell, C.A.; Adhya, S.; Hammel, M. Nucleoid Remodeling during Environmental Adaptation Is Regulated by HU-Dependent DNA Bundling. Nat. Commun. 2020, 11, 2905. [Google Scholar] [CrossRef] [PubMed]
- Salmon, B.; Cunningham, C.; Davison, A.J.; Harris, W.J.; Baines, J.D. The Herpes Simplex Virus Type 1 U(L)17 Gene Encodes Virion Tegument Proteins That Are Required for Cleavage and Packaging of Viral DNA. J. Virol. 1998, 72, 3779–3788. [Google Scholar] [CrossRef] [PubMed]
- Fahy, K.; Weinhardt, V.; Vihinen-Ranta, M.; Fletcher, N.; Skoko, D.; Pereiro, E.; Gastaminza, P.; Bartenschlager, R.; Scholz, D.; Ekman, A.; et al. Compact Cell Imaging Device (CoCID) to Provide Insights into the Cellular Origins of Viral Infections. J. Phys. Photonics 2021, 3, 031002. [Google Scholar] [CrossRef]
- Dehlinger, A.; Braenzel, J.; Groetzsch, D.; Feigl, T.; Jung, R.; KanngieBer, B.; Rehbein, S.; Seim, C.; Stiel, H. Towards High Performance Soft X-ray Cryo-Tomography in the Laboratory. Microsc. Microanal. 2018, 24, 248–249. [Google Scholar] [CrossRef] [Green Version]
- Hertz, H.M.; von Hofsten, O.; Bertilson, M.; Vogt, U.; Holmberg, A.; Reinspach, J.; Martz, D.; Selin, M.; Christakou, A.E.; Jerlström-Hultqvist, J.; et al. Laboratory Cryo Soft X-ray Microscopy. J. Struct. Biol. 2012, 177, 267–272. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.-H.; Vanslembrouck, B.; Ekman, A.; Aho, V.; Larabell, C.A.; Le Gros, M.A.; Vihinen-Ranta, M.; Weinhardt, V. Soft X-ray Tomography Reveals HSV-1-Induced Remodeling of Human B Cells. Viruses 2022, 14, 2651. https://doi.org/10.3390/v14122651
Chen J-H, Vanslembrouck B, Ekman A, Aho V, Larabell CA, Le Gros MA, Vihinen-Ranta M, Weinhardt V. Soft X-ray Tomography Reveals HSV-1-Induced Remodeling of Human B Cells. Viruses. 2022; 14(12):2651. https://doi.org/10.3390/v14122651
Chicago/Turabian StyleChen, Jian-Hua, Bieke Vanslembrouck, Axel Ekman, Vesa Aho, Carolyn A. Larabell, Mark A. Le Gros, Maija Vihinen-Ranta, and Venera Weinhardt. 2022. "Soft X-ray Tomography Reveals HSV-1-Induced Remodeling of Human B Cells" Viruses 14, no. 12: 2651. https://doi.org/10.3390/v14122651
APA StyleChen, J.-H., Vanslembrouck, B., Ekman, A., Aho, V., Larabell, C. A., Le Gros, M. A., Vihinen-Ranta, M., & Weinhardt, V. (2022). Soft X-ray Tomography Reveals HSV-1-Induced Remodeling of Human B Cells. Viruses, 14(12), 2651. https://doi.org/10.3390/v14122651





