High APOBEC3B mRNA Expression Is Associated with Human Papillomavirus Type 18 Infection in Cervical Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Population
2.2. Total RNA Isolation and cDNA Synthesis
2.3. Real-Time PCR for APOBEC3B mRNA
2.4. Statistical Analyses
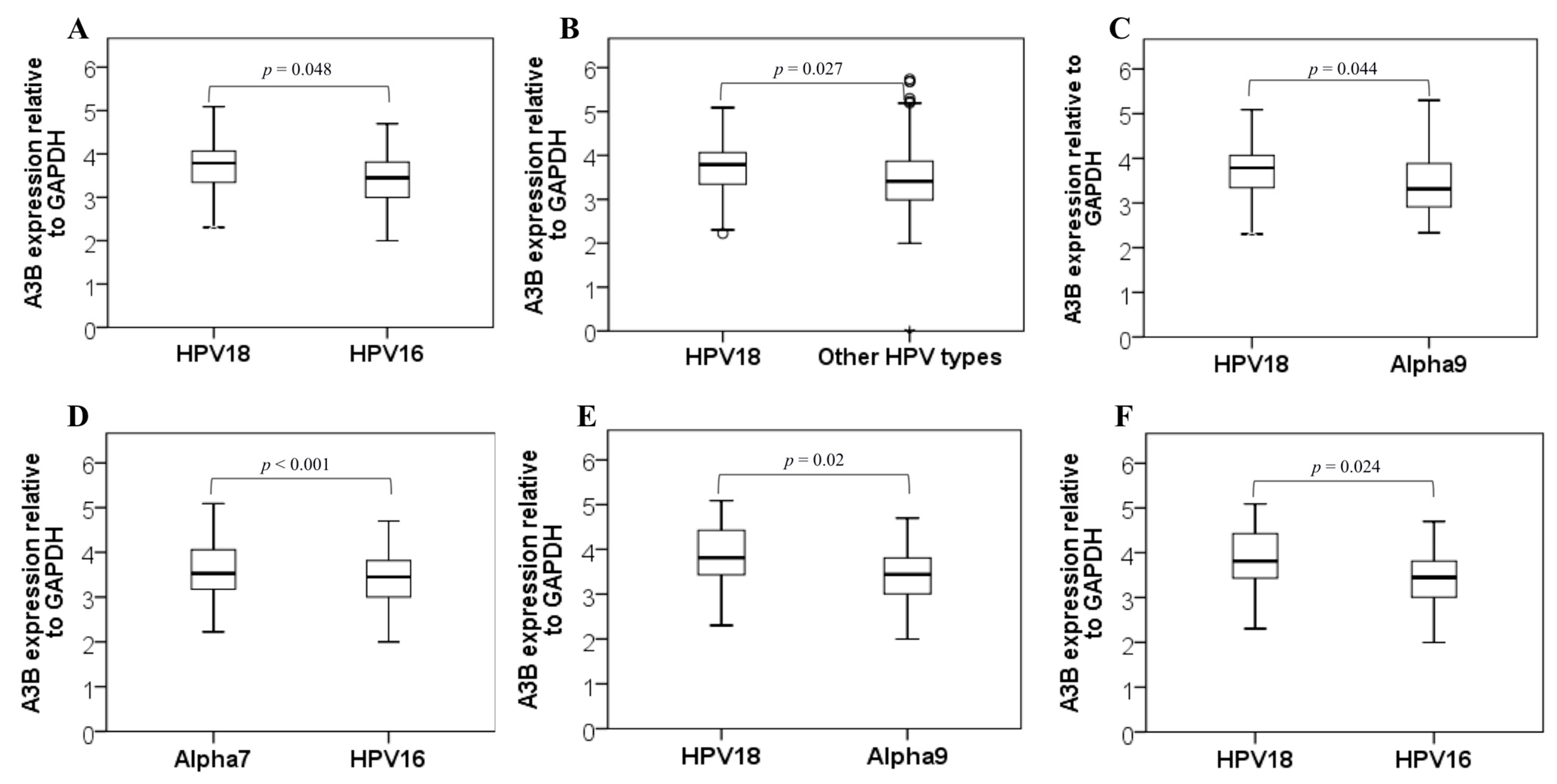
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Doorslaer, K.; Chen, Z.; Bernard, H.-U.; Chan, P.K.S.; DeSalle, R.; Dillner, J.; Forslund, O.; Haga, T.; McBride, A.A.; Villa, L.L.; et al. ICTV Virus Taxonomy Profile: Papillomaviridae. J. Gen. Virol. 2018, 99, 989–990. [Google Scholar] [CrossRef]
- zur Hausen, H. Condylomata acuminata and human genital cancer. Cancer Res. 1976, 36, 794. [Google Scholar]
- Arroyo-Mühr, L.S.; Lagheden, C.; Hultin, E.; Eklund, C.; Adami, H.-O.; Dillner, J.; Sundström, K. Human papillomavirus type 16 genomic variation in women with subsequent in situ or invasive cervical cancer: Prospective population-based study. Br. J. Cancer 2018, 119, 1163–1168. [Google Scholar] [CrossRef] [Green Version]
- Harari, A.; Chen, Z.; Burk, R.D. Human Papillomavirus Genomics: Past, Present and Future. Curr. Probl. Dermatol. 2014, 45, 1–18. [Google Scholar]
- Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 2006, 110, 525–541. [Google Scholar] [CrossRef] [Green Version]
- De Villiers, E.-M.; Fauquet, C.; Broker, T.R.; Bernard, H.-U.; Zur Hausen, H. Classification of papillomaviruses. Virology 2004, 324, 17–27. [Google Scholar] [CrossRef] [Green Version]
- Clifford, G.M.; Smith, J.S.; Plummer, M.; Muñoz, N.; Franceschi, S. Human papillomavirus types in invasive cervical cancer worldwide: A meta-analysis. Br. J. Cancer 2003, 88, 63–73. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Roberts, S.A.; Lawrence, M.S.; Klimczak, L.J.; Grimm, S.A.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.V.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 970–976. [Google Scholar] [CrossRef]
- Warren, C.J.; Van Doorslaer, K.; Pandey, A.; Espinosa, J.M.; Pyeon, D. Role of the host restriction factor APOBEC3 on papillomavirus evolution. Virus Evol. 2015, 1, vev015. [Google Scholar] [CrossRef]
- Harris, R.S.; Hultquist, J.F.; Evans, D.T. The Restriction Factors of Human Immunodeficiency Virus. J. Biol. Chem. 2012, 287, 40875–40883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Refsland, E.W.; Harris, R.S. The APOBEC3 Family of Retroelement Restriction Factors. Curr. Top. Microbiol. Immunol. 2013, 371, 1–27. [Google Scholar] [PubMed]
- Tsuboi, M.; Yamane, A.; Horiguchi, J.; Yokobori, T.; Kawabata-Iwakawa, R.; Yoshiyama, S.; Rokudai, S.; Odawara, H.; Tokiniwa, H.; Oyama, T.; et al. APOBEC3B high expression status is associated with aggressive phenotype in Japanese breast cancers. Breast Cancer 2016, 23, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.B.; Temiz, N.A.; Harris, R.S. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat. Genet. 2013, 45, 977–983. [Google Scholar] [CrossRef] [Green Version]
- Nik-Zainal, S.; Wedge, D.C.; Alexandrov, L.B.; Petljak, M.; Butler, A.P.; Bolli, N.; Davies, H.R.; Knappskog, S.; Martin, S.; Papaemmanuil, E.; et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat. Genet. 2014, 46, 487–491. [Google Scholar] [CrossRef] [Green Version]
- Harris, R.S.; Petersen-Mahrt, S.K.; Neuberger, M.S. RNA Editing Enzyme APOBEC1 and Some of Its Homologs Can Act as DNA Mutators. Mol. Cell 2002, 10, 1247–1253. [Google Scholar] [CrossRef]
- Cescon, D.W.; Haibe-Kains, B.; Mak, T.W. APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc. Natl. Acad. Sci. USA 2015, 112, 2841–2846. [Google Scholar] [CrossRef] [Green Version]
- Refsland, E.W.; Stenglein, M.D.; Shindo, K.; Albin, J.S.; Brown, W.L.; Harris, R.S. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: Implications for HIV-1 restriction. Nucleic Acids Res. 2010, 38, 4274–4284. [Google Scholar] [CrossRef] [Green Version]
- Vieira, V.C.; Leonard, B.; White, E.A.; Starrett, G.J.; Temiz, N.A.; Lorenz, L.D.; Lee, D.; Soares, M.A.; Lambert, P.F.; Howley, P.M.; et al. Human Papillomavirus E6 Triggers Upregulation of the Antiviral and Cancer Genomic DNA Deaminase APOBEC3B. mBio 2014, 5, e02234-14. [Google Scholar] [CrossRef] [Green Version]
- de Almeida, L.M.; Martins, L.F.L.; Pontes, V.B.; Corrêa, F.M.; Montenegro, R.C.; Pinto, L.C.; Soares, B.M.; Vidal, J.P.C.B.; Félix, S.P.; Bertoni, N.; et al. Human Papillomavirus Genotype Distribution among Cervical Cancer Patients prior to Brazilian National HPV Immunization Program. J. Environ. Public Health 2017, 2017, 1645074. [Google Scholar] [CrossRef] [Green Version]
- Vidal, J.P.C.B.; Felix, S.P.; Chaves, C.B.P.; Patury, P.; Franco, V.F.; de Morais, E.A.; de Carvalho, N.A.; Carvalho, A.C.L.; Almeida Neto, O.F.; Vieira, L.M.T.M.; et al. Genetic diversity of HPV16 and HPV18 in Brazilian patients with invasive cervical cancer. J. Med. Virol. 2016, 88, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Burns, M.B.; Lackey, L.; Carpenter, M.A.; Rathore, A.; Land, A.M.; Leonard, B.; Refsland, E.W.; Kotandeniya, D.; Tretyakova, N.; Nikas, J.B.; et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 2013, 494, 366–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonard, B.; Hart, S.N.; Burns, M.B.; Carpenter, M.A.; Temiz, N.A.; Rathore, A.; Vogel, R.I.; Nikas, J.B.; Law, E.K.; Brown, W.L.; et al. APOBEC3B Upregulation and Genomic Mutation Patterns in Serous Ovarian Carcinoma. Cancer Res. 2013, 73, 7222–7231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, S.V. The human papillomavirus replication cycle, and its links to cancer progression: A comprehensive review. Clin. Sci. 2017, 131, 2201–2221. [Google Scholar] [CrossRef] [Green Version]
- Groves, I.J.; Coleman, N. Pathogenesis of human papillomavirus-associated mucosal disease. J. Pathol. 2015, 235, 527–538. [Google Scholar] [CrossRef] [Green Version]
- Albertini, S.; Lo Cigno, I.; Calati, F.; De Andrea, M.; Borgogna, C.; Dell’Oste, V.; Landolfo, S.; Gariglio, M. HPV18 Persistence Impairs Basal and DNA Ligand–Mediated IFN-β and IFN-λ1 Production through Transcriptional Repression of Multiple Downstream Effectors of Pattern Recognition Receptor Signaling. J. Immunol. 2018, 200, 2076–2089. [Google Scholar] [CrossRef] [Green Version]
- Ho, G.Y.F.; Burk, R.D.; Klein, S.; Kadish, A.S.; Chang, C.J.; Palan, P.; Basu, J.; Tachezy, R.; Lewis, R.; Romney, S. Persistent Genital Human Papillomavirus Infection as a Risk Factor for Persistent Cervical Dysplasia. JNCI J. Natl. Cancer Inst. 1995, 87, 1365–1371. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.J.; Xu, T.; Guo, K.; Griffin, L.M.; Westrich, J.A.; Lee, D.; Lambert, P.F.; Santiago, M.L.; Pyeon, D. APOBEC3A Functions as a Restriction Factor of Human Papillomavirus. J. Virol. 2015, 89, 688–702. [Google Scholar] [CrossRef] [Green Version]
- Warren, C.; Westrich, J.; Doorslaer, K.; Pyeon, D. Roles of APOBEC3A and APOBEC3B in Human Papillomavirus Infection and Disease Progression. Viruses 2017, 9, 233. [Google Scholar] [CrossRef] [Green Version]
- Mori, S.; Takeuchi, T.; Ishii, Y.; Yugawa, T.; Kiyono, T.; Nishina, H.; Kukimoto, I. Human Papillomavirus 16 E6 Upregulates APOBEC3B via the TEAD Transcription Factor. J. Virol. 2017, 91, e02413-16. [Google Scholar] [CrossRef] [Green Version]
- Ohba, K.; Ichiyama, K.; Yajima, M.; Gemma, N.; Nikaido, M.; Wu, Q.; Chong, P.; Mori, S.; Yamamoto, R.; Wong, J.E.L.; et al. In Vivo and In Vitro Studies Suggest a Possible Involvement of HPV Infection in the Early Stage of Breast Carcinogenesis via APOBEC3B Induction. PLoS ONE 2014, 9, e97787. [Google Scholar] [CrossRef]
- Jia, Q.; Yan, C.; Zheng, X.; Pan, X.; Cao, X.; Cao, L. Upregulation of MTA1 expression by human papillomavirus infection promotes CDDP resistance in cervical cancer cells via modulation of NF-κB/APOBEC3B cascade. Cancer Chemother. Pharmacol. 2019, 83, 625–637. [Google Scholar] [CrossRef]
- Periyasamy, M.; Singh, A.K.; Gemma, C.; Kranjec, C.; Farzan, R.; Leach, D.A.; Navaratnam, N.; Pálinkás, H.L.; Vértessy, B.G.; Fenton, T.R.; et al. p53 controls expression of the DNA deaminase APOBEC3B to limit its potential mutagenic activity in cancer cells. Nucleic Acids Res. 2017, 45, 11056–11069. [Google Scholar] [CrossRef] [Green Version]
- Fischer, M.; Uxa, S.; Stanko, C.; Magin, T.M.; Engeland, K. Human papilloma virus E7 oncoprotein abrogates the p53-p21-DREAM pathway. Sci. Rep. 2017, 7, 2603. [Google Scholar] [CrossRef] [Green Version]
- Muto, V.; Stellacci, E.; Lamberti, A.G.; Perrotti, E.; Carrabba, A.; Matera, G.; Sgarbanti, M.; Battistini, A.; Liberto, M.C.; Focà, A. Human Papillomavirus Type 16 E5 Protein Induces Expression of Beta Interferon through Interferon Regulatory Factor 1 in Human Keratinocytes. J. Virol. 2011, 85, 5070–5080. [Google Scholar] [CrossRef] [Green Version]
- Sadler, A.J.; Williams, B.R.G. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 2008, 8, 559–568. [Google Scholar] [CrossRef]
- Li, J.; Hakata, Y.; Takeda, E.; Liu, Q.; Iwatani, Y.; Kozak, C.A.; Miyazawa, M. Two Genetic Determinants Acquired Late in Mus Evolution Regulate the Inclusion of Exon 5, which Alters Mouse APOBEC3 Translation Efficiency. PLoS Pathog. 2012, 8, e1002478. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Lei, K.J.; Jin, W.; Greenwell-Wild, T.; Wahl, S.M. Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti–HIV-1 activity. J. Exp. Med. 2006, 203, 41–46. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Wakae, K.; Kitamura, K.; Aoyama, S.; Liu, G.; Koura, M.; Monjurul, A.M.; Kukimoto, I.; Muramatsu, M. APOBEC3 Deaminases Induce Hypermutation in Human Papillomavirus 16 DNA upon Beta Interferon Stimulation. J. Virol. 2014, 88, 1308–1317. [Google Scholar] [CrossRef] [Green Version]
- Bonvin, M.; Achermann, F.; Greeve, I.; Stroka, D.; Keogh, A.; Inderbitzin, D.; Candinas, D.; Sommer, P.; Wain-Hobson, S.; Vartanian, J.-P.; et al. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 2006, 43, 1364–1374. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Xiao, Y.; Yeager, M.; Clifford, G.; Wentzensen, N.; Cullen, M.; Boland, J.F.; Bass, S.; Steinberg, M.K.; Raine-Bennett, T.; et al. Mutations in the HPV16 genome induced by APOBEC3 are associated with viral clearance. Nat. Commun. 2020, 11, 886. [Google Scholar] [CrossRef] [PubMed]
- Faden, D.L.; Kuhs, K.A.L.; Lin, M.; Langenbucher, A.; Pinheiro, M.; Yeager, M.; Cullen, M.; Boland, J.F.; Steinberg, M.; Bass, S.; et al. APOBEC Mutagenesis Is Concordant between Tumor and Viral Genomes in HPV-Positive Head and Neck Squamous Cell Carcinoma. Viruses 2021, 13, 1666. [Google Scholar] [CrossRef] [PubMed]
- Lagström, S.; Løvestad, A.H.; Umu, S.U.; Ambur, O.H.; Nygård, M.; Rounge, T.B.; Christiansen, I.K. HPV16 and HPV18 type-specific APOBEC3 and integration profiles in different diagnostic categories of cervical samples. Tumour Virus Res. 2021, 12, 200221. [Google Scholar] [CrossRef]
| HPV16 (n = 139) | HPV18 (n = 30) | Alpha-7 * (HPV18 Excluded) (n = 24) | Alpha-9 ** (HPV16 Excluded) (n = 18) | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Tumor Histological Type | ADC | 10.8 | 5.6–16 | 43.3 | 24.8–61.8 | 12.5 | 0–26.5 | 5.6 | 0–16.9 | 15.2 | 10.3–20 |
| SCC | 89.2 | 84–94.4 | 56.7 | 38.2–75.2 | 87.5 | 73.5–100 | 94.4 | 83.1–100 | 84.8 | 80–89.7 | |
| Staging | I | 18.7 | 12.2–25.2 | 16.7 | 2.8–30.6 | 12.5 | 0–26.5 | 16.7 | 0–35.2 | 17.5 | 12.4–22.7 |
| II | 41.7 | 33.5–50 | 36.7 | 18.7–54.7 | 50 | 28.9–71.1 | 38.9 | 14.6–63.1 | 41.7 | 35–48.4 | |
| III | 33.1 | 25.2–41 | 43.3 | 24.8–61. 8 | 33.3 | 13.4–53.2 | 44.4 | 19.7–69.2 | 35.5 | 29–42 | |
| IV | 6.5 | 2.3–10.6 | 3.3 | 0–100 | 4.2 | 0–100 | 0 | - | 5.2 | 0–11.7 | |
| Variables | Categories | Adjusted p-Value | OR | IC95% |
|---|---|---|---|---|
| HPV Type | HPV16 | 1 | ||
| HPV18 | 0.011 * | 1.26 | 1.05–1.50 | |
| HPV31 | 0.001 | 0.46 | 0.29–0.73 | |
| HPV33 | 0.096 | 1.28 | 0.95–1.71 | |
| HPV35 | 0.233 | 1.27 | 0.85–1.90 | |
| HPV39 | 0.408 | 1.21 | 0.76–1.90 | |
| HPV45 | 0.893 | 0.98 | 0.77–1.25 | |
| HPV52 | 0.821 | 0.93 | 0.50–1.71 | |
| HPV56 | <0.001 | 0.65 | 0.52–0.81 | |
| HPV58 | 0.714 | 0.87 | 0.41–1.83 | |
| HPV59 | 0.664 | 0.89 | 0.52–1.50 | |
| HPV73 | 0.088 | 0,91 | 0.82–1.01 | |
| Tumor Type | SCC | 1 | ||
| ADC | 0.564 | 0.94 | 0.76–1.15 | |
| Stage | I | 0.344 | 0.84 | 0.57–1.15 |
| II | 0.652 | 0.92 | 0.67–1.28 | |
| III | 0.412 | 0.87 | 0.63–1.20 | |
| IV | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Oliveira, G.R.; Carvalho, P.S.; Vieira, V.C.; Curty, G.; Basto, D.L.; Moreira, M.Â.M.; Soares, M.A. High APOBEC3B mRNA Expression Is Associated with Human Papillomavirus Type 18 Infection in Cervical Cancer. Viruses 2022, 14, 2653. https://doi.org/10.3390/v14122653
de Oliveira GR, Carvalho PS, Vieira VC, Curty G, Basto DL, Moreira MÂM, Soares MA. High APOBEC3B mRNA Expression Is Associated with Human Papillomavirus Type 18 Infection in Cervical Cancer. Viruses. 2022; 14(12):2653. https://doi.org/10.3390/v14122653
Chicago/Turabian Stylede Oliveira, Gisele R., Pedro S. Carvalho, Valdimara C. Vieira, Gislaine Curty, Diogo L. Basto, Miguel Ângelo M. Moreira, and Marcelo A. Soares. 2022. "High APOBEC3B mRNA Expression Is Associated with Human Papillomavirus Type 18 Infection in Cervical Cancer" Viruses 14, no. 12: 2653. https://doi.org/10.3390/v14122653






