Molecular Epidemiology of the Norwegian SARS-CoV-2 Delta Lineage AY.63
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sequence Data and Metadata Collection
2.2. Phylogenetic Analysis
2.3. Plotting and Visualization
2.4. Pairwise SNP Distance and Import Analysis
2.5. Protein Structure Visualization
3. Results and Discussion
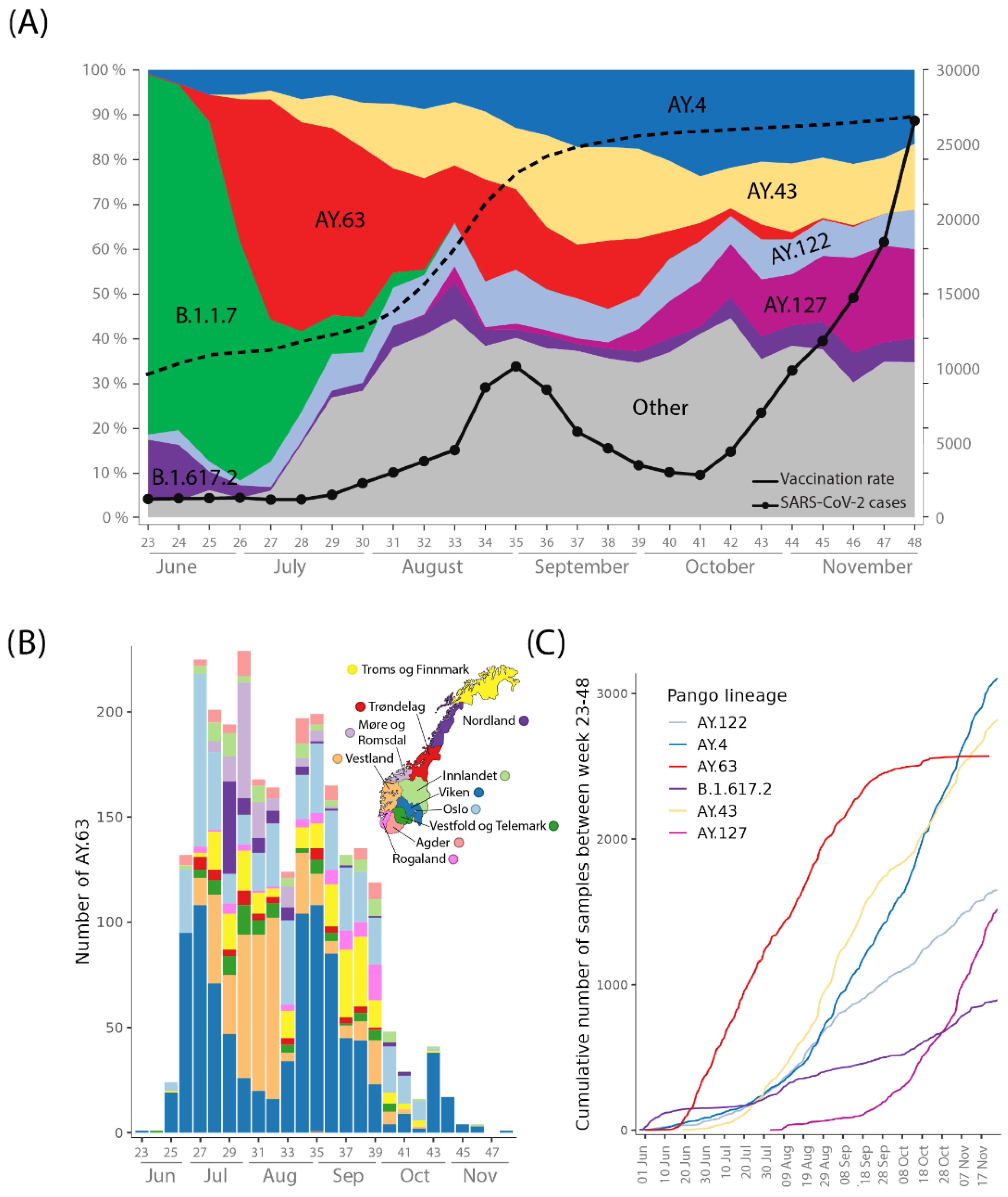
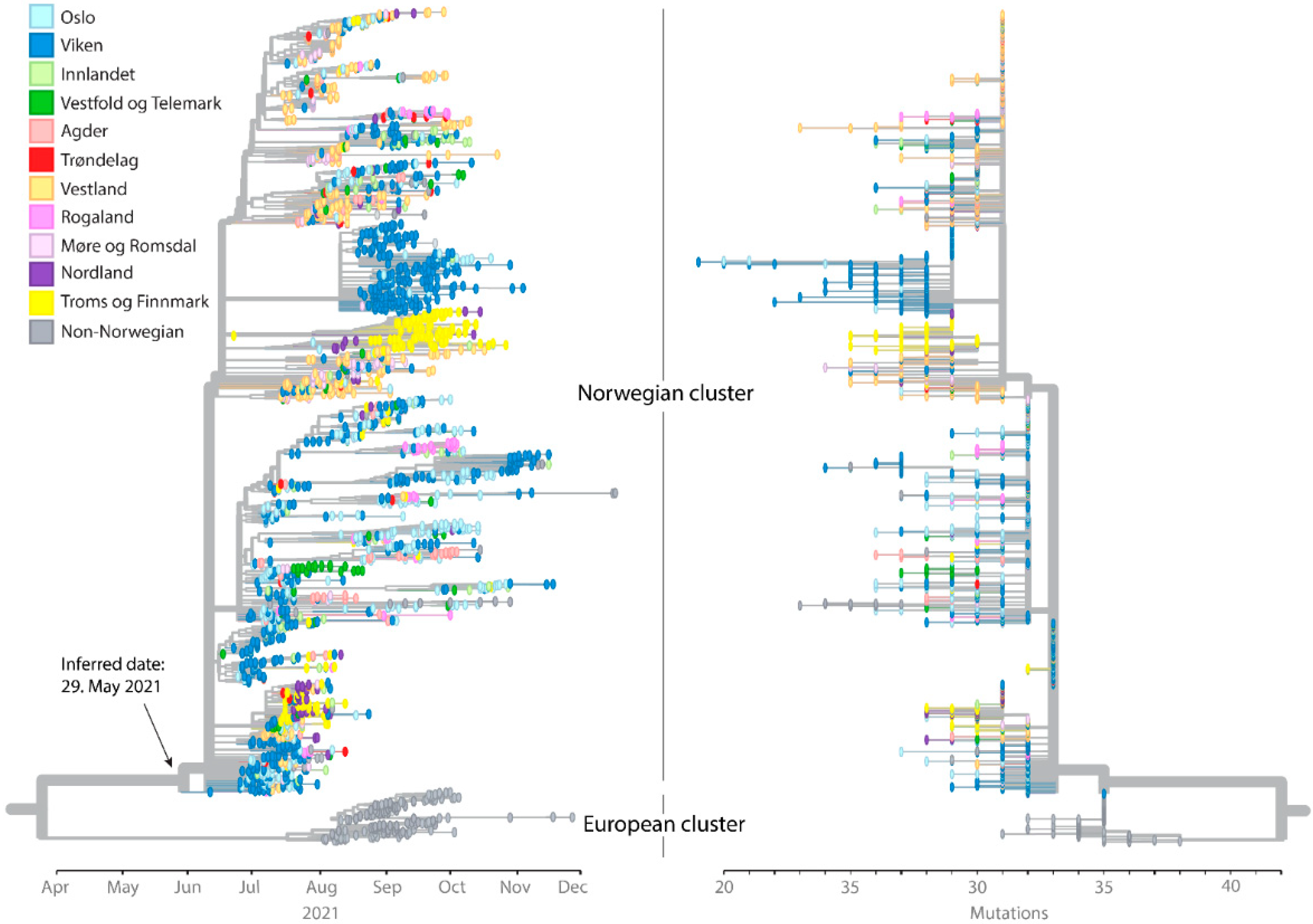
3.1. Introduction and Spread of AY.63 in Norway
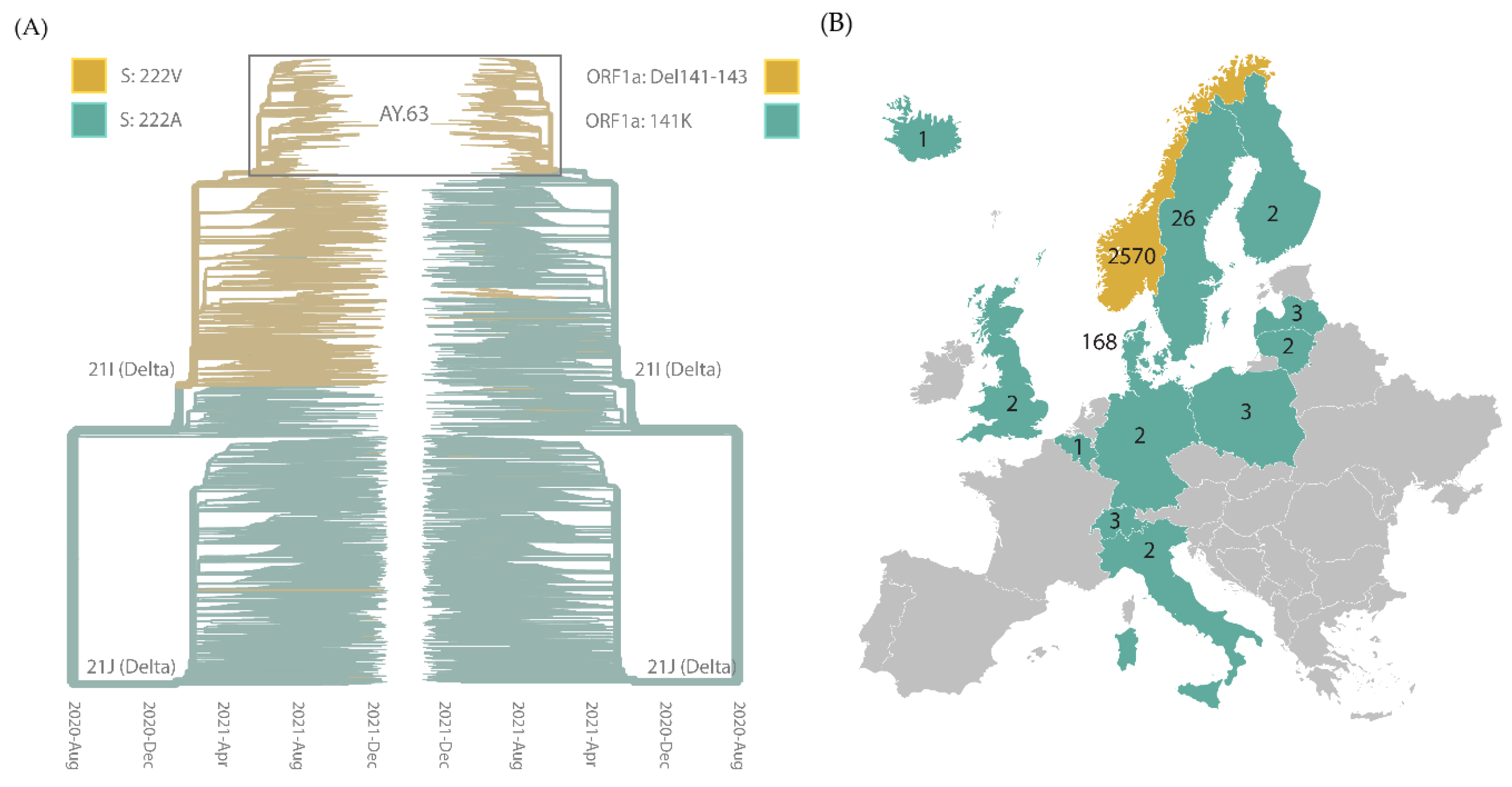
3.2. Origin, Spread, and Extinction of AY.63 in Europe
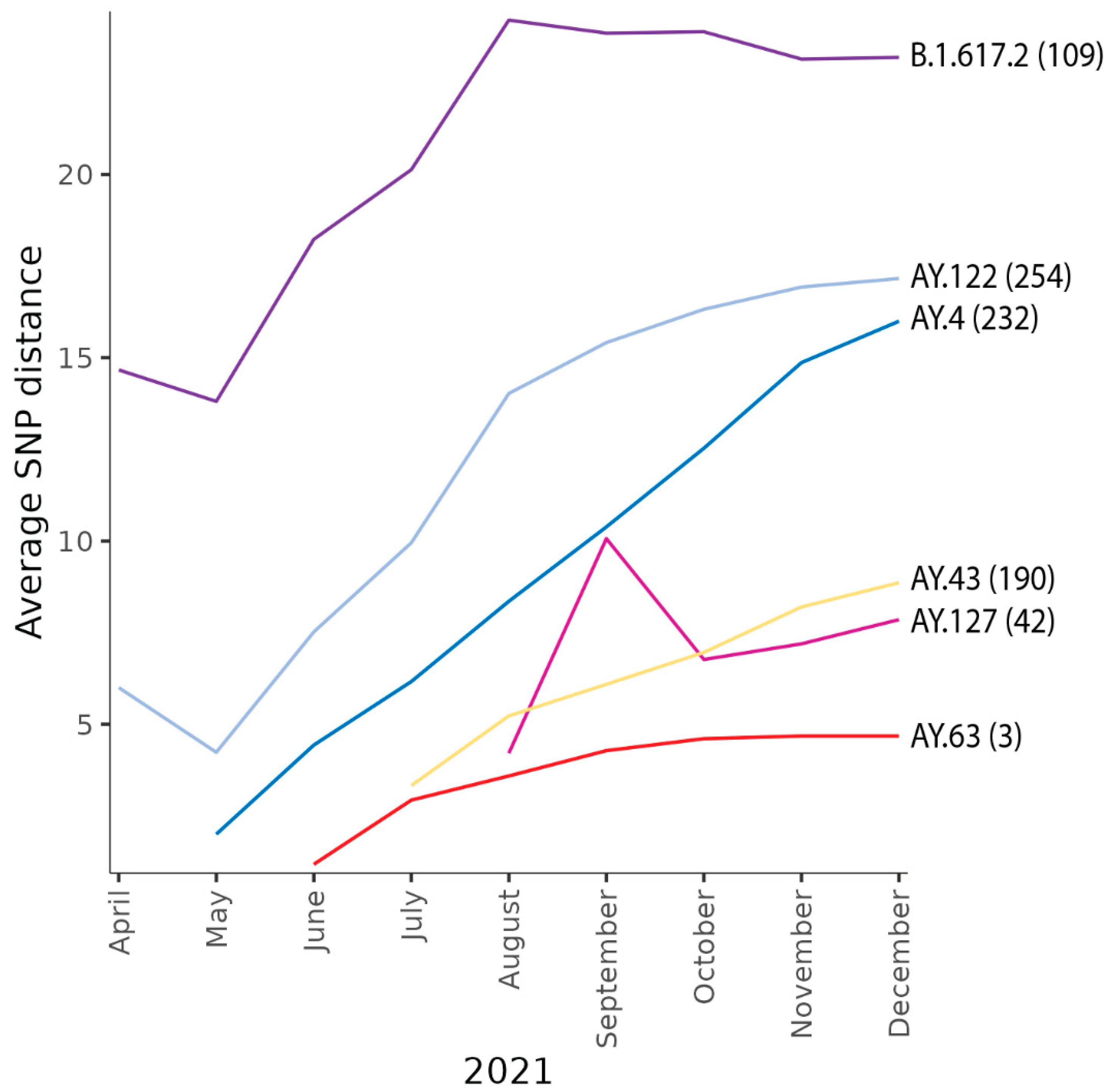
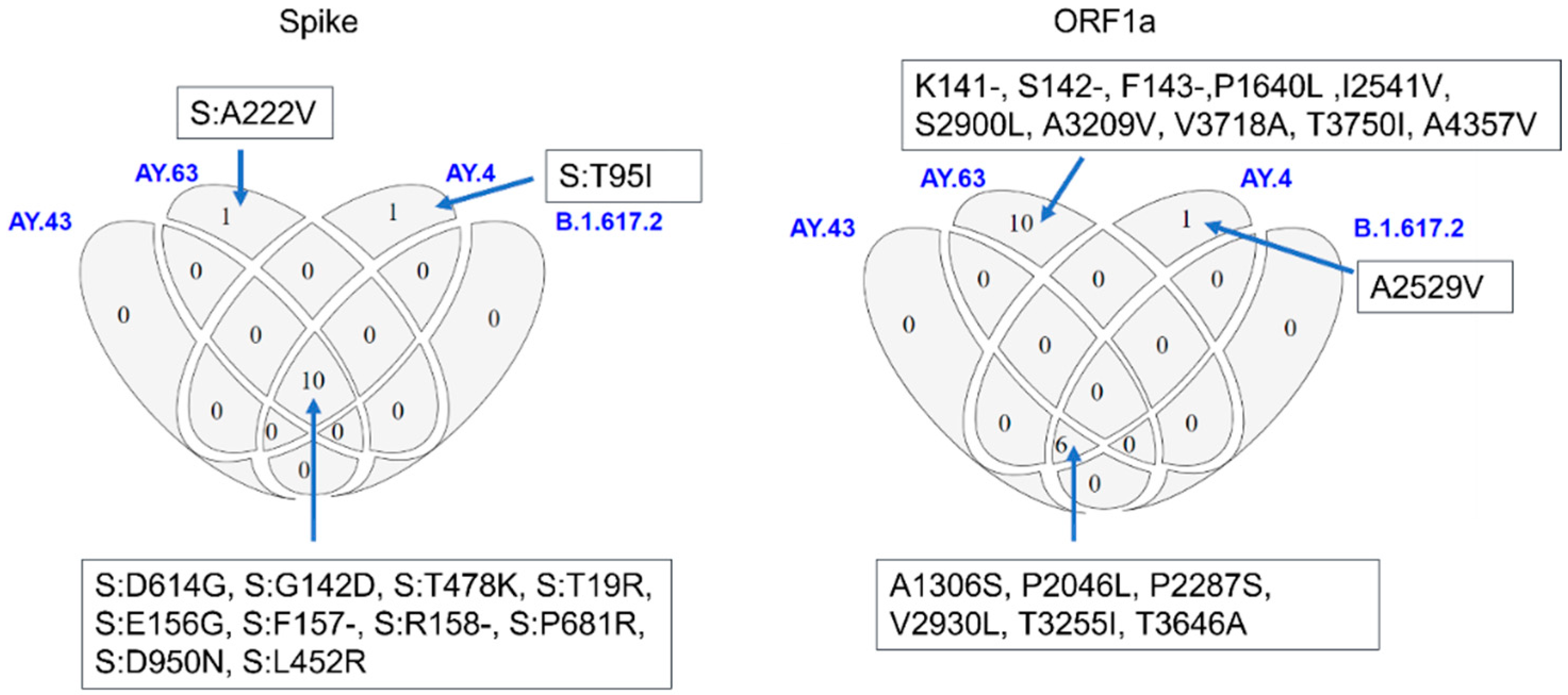
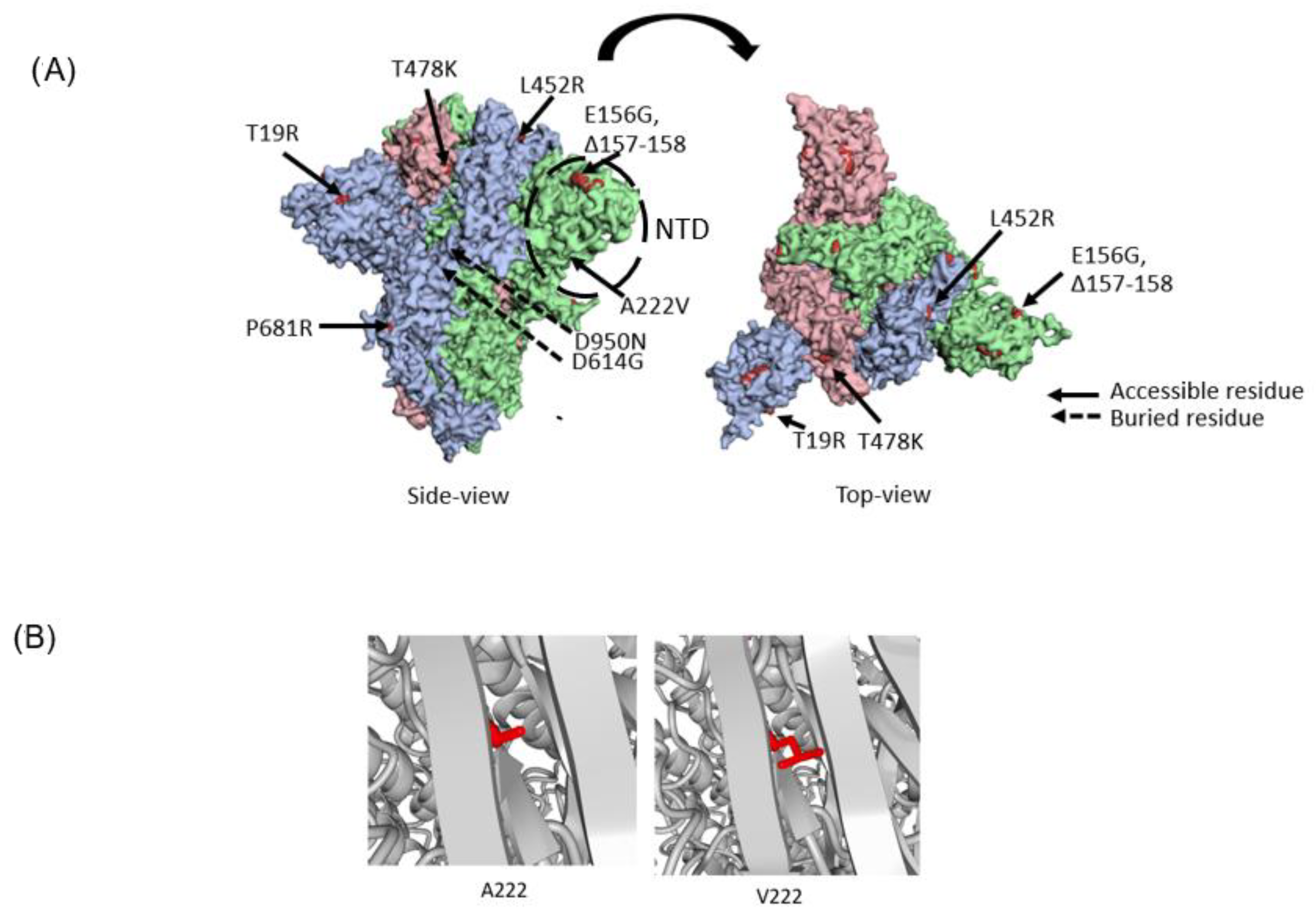
3.3. Molecular Characteristics of AY.63
3.4. Concluding Remark
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants (accessed on 10 June 2022).
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- Khare, S.; Gurry, C.; Freitas, L.; Schultz, M.B.; Bach, G.; Diallo, A.; Akite, N.; Ho, J.; Lee, R.T.; Yeo, W.; et al. GISAID’s Role in Pandemic Response. China CDC Wkly. 2021, 3, 1049–1051. [Google Scholar] [CrossRef] [PubMed]
- Tao, K.; Tzou, P.L.; Nouhin, J.; Gupta, R.K.; de Oliveira, T.; Kosakovsky Pond, S.L.; Fera, D.; Shafer, R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021, 22, 757–773. [Google Scholar] [CrossRef] [PubMed]
- Du Plessis, L.; McCrone, J.T.; Zarebski, A.E.; Hill, V.; Ruis, C.; Gutierrez, B.; Raghwani, J.; Ashworth, J.; Colquhoun, R.; Connor, T.R. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science 2021, 371, 708–712. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J.; et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Li, J.; Lai, S.; Gao, G.F.; Shi, W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature 2021, 600, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Thomas, S. Mapping the Nonstructural Transmembrane Proteins of Severe Acute Respiratory Syndrome Coronavirus 2. J. Comput. Biol. 2021, 28, 909–921. [Google Scholar] [CrossRef]
- Nagesha, S.; Ramesh, B.; Pradeep, C.; Shashidhara, K.; Ramakrishnappa, T.; Krishnaprasad, B.; Jnanashree, S.; Manohar, M.; Arunkumar, N.; Patel, D. SARS-CoV 2 spike protein S1 subunit as an ideal target for stable vaccines: A bioinformatic study. Mater. Today Proc. 2022, 49, 904–912. [Google Scholar]
- Earnest, R.; Uddin, R.; Matluk, N.; Renzette, N.; Turbett, S.E.; Siddle, K.J.; Loreth, C.; Adams, G.; Tomkins-Tinch, C.H.; Petrone, M.E.; et al. Comparative transmissibility of SARS-CoV-2 variants Delta and Alpha in New England, USA. Cell Rep. Med. 2022, 3, 100583. [Google Scholar] [CrossRef]
- WHO. COVID-19 Weekly Epidemiological Update, Edition 58. Available online: https://apps.who.int/iris/bitstream/handle/10665/345456/CoV-weekly-sitrep21Sep21-eng.pdf?sequence=1&isAllowed=y (accessed on 28 October 2022).
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.M.; Wang, W.; Song, Z.G.; Hu, Y.; Tao, Z.W.; Tian, J.H.; Pei, Y.Y.; et al. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Gangavarapu, K.; Latif, A.A.; Mullen, J.L.; Alkuzweny, M.; Hufbauer, E.; Tsueng, G.; Haag, E.; Zeller, M.; Aceves, C.M.; Zaiets, K.; et al. Outbreak.info genomic reports: Scalable and dynamic surveillance of SARS-CoV-2 variants and mutations. medRxiv 2022. [Google Scholar] [CrossRef]
- Osnes, M.N.; Alfsnes, K.; Bråte, J.; Garcia, I.; Riis, R.K.; Instefjord, K.H.; Elshaug, H.; Vollan, H.S.; Moen, L.V.; Pedersen, B.N.; et al. The impact of global lineage dynamics, border restrictions, and emergence of the B.1.1.7 lineage on the SARS-CoV-2 epidemic in Norway. Virus Evol. 2021, 7, veab086. [Google Scholar] [CrossRef] [PubMed]
- Norwegian Institute of Public Health. COVID-19 Ukerapport—Uke 29. Available online: https://www.fhi.no/contentassets/8a971e7b0a3c4a06bdbf381ab52e6157/vedlegg/alle-ukerapporter-2021/ukerapport-uke-29-19.07---25.07.21.pdf (accessed on 28 October 2022).
- Hodcroft, E.B.; Zuber, M.; Nadeau, S.; Vaughan, T.G.; Crawford, K.H.D.; Althaus, C.L.; Reichmuth, M.L.; Bowen, J.E.; Walls, A.C.; Corti, D.; et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature 2021, 595, 707–712. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [Green Version]
- Team R Core. R: A Language and Environment for Statistical Computing. 2013. Available online: https://www.r-project.org/ (accessed on 1 November 2022).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef] [Green Version]
- Villanueva, R.A.M.; Chen, Z.J. ggplot2: Elegant graphics for data analysis. Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Chen, C.; Nadeau, S.; Yared, M.; Voinov, P.; Xie, N.; Roemer, C.; Stadler, T. CoV-Spectrum: Analysis of globally shared SARS-CoV-2 data to identify and characterize new variants. Bioinformatics 2021, 38, 1735–1737. [Google Scholar] [CrossRef]
- Cao, Y.; Choi, Y.K.; Frank, M.; Woo, H.; Park, S.-J.; Yeom, M.S.; Seok, C.; Im, W. Dynamic Interactions of Fully Glycosylated SARS-CoV-2 Spike Protein with Various Antibodies. J. Chem. Theory Comput. 2021, 17, 6559–6569. [Google Scholar] [CrossRef] [PubMed]
- Krieger, E.; Vriend, G. YASARA View—Molecular graphics for all devices—From smartphones to workstations. Bioinformatics 2014, 30, 2981–2982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vriend, G. WHAT IF: A molecular modeling and drug design program. J. Mol. Graph. 1990, 8, 52–56. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell 2020, 181, 281–292.e286. [Google Scholar] [CrossRef]
- Rego, N.; Koes, D. 3Dmol.js: Molecular visualization with WebGL. Bioinformatics 2014, 31, 1322–1324. [Google Scholar] [CrossRef] [Green Version]
- Norwegian Institute of Public Health. COVID-19 Ukerapport—Uke 49. Available online: https://www.fhi.no/contentassets/8a971e7b0a3c4a06bdbf381ab52e6157/vedlegg/alle-ukerapporter-2021/ukerapport-uke-49-06.12---12.12.21.pdf (accessed on 28 October 2022).
- Baral, P.; Bhattarai, N.; Hossen, M.L.; Stebliankin, V.; Gerstman, B.S.; Narasimhan, G.; Chapagain, P.P. Mutation-induced changes in the receptor-binding interface of the SARS-CoV-2 Delta variant B.1.617.2 and implications for immune evasion. Biochem. Biophys. Res. Commun. 2021, 574, 14–19. [Google Scholar] [CrossRef]
- Aksamentov, I.; Roemer, C.; Hodcroft, E.B.; Neher, R.A. Nextclade: Clade assignment, mutation calling and quality control for viral genomes. J. Open Source Softw. 2021, 6, 3773. [Google Scholar] [CrossRef]
- Hale, T.; Angrist, N.; Kira, B.; Petherick, A.; Phillips, T.; Webster, S. Variation in Government Responses to COVID-19. Available online: https://www.bsg.ox.ac.uk/research/covid-19-government-response-tracker (accessed on 15 November 2022).
- Mathieu, E.; Ritchie, H.; Rodés-Guirao, L.; Appel, C.; Giattino, C.; Hasell, J.; Macdonald, B.; Dattani, S.; Beltekian, D.; Ortiz-Ospina, E. Coronavirus Pandemic (COVID-19). Available online: https://ourworldindata.org/covid-cases (accessed on 15 November 2022).
- Shu, Y.; McCauley, J. GISAID: Global initiative on sharing all influenza data—From vision to reality. Eurosurveillance 2017, 22, 30494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaelsen, T.Y.; Bennedbæk, M.; Christiansen, L.E.; Jørgensen, M.S.F.; Møller, C.H.; Sørensen, E.A.; Knutsson, S.; Brandt, J.; Jensen, T.B.N.; Chiche-Lapierre, C.; et al. Introduction and transmission of SARS-CoV-2 lineage B.1.1.7, Alpha variant, in Denmark. Genome Med. 2022, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Danish COVID-19 Genome Consortium. Genomic Overview of SARS-CoV-2 in Denmark. Available online: https://www.covid19genomics.dk/statistics (accessed on 28 October 2022).
- Alexiev, I.; Giovanetti, M.; Cella, E.; Ivanov, I.; Stoikov, I.; Donchev, D.; Grigorova, L.; Gancheva, A.; Dimitrova, R.; Korsun, N.; et al. Initial introduction and spread of the SARS-CoV-2 AY.4.2.1 Delta variant in Bulgaria, a genomic insight. J. Med. Virol. 2022, 94, 6060–6064. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Cohen, A.R.; Naqvi, S.H.; Chand, H.S.; Quinn, T.P.; Lorson, C.L.; Byrareddy, S.N.; Singh, K. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J. Autoimmun. 2021, 124, 102715. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Pant, A.B. Mitigating COVID-19 in the face of emerging virus variants, breakthrough infections and vaccine hesitancy. J. Autoimmun. 2022, 127, 102792. [Google Scholar] [CrossRef] [PubMed]
- Klinakis, A.; Cournia, Z.; Rampias, T. N-terminal domain mutations of the spike protein are structurally implicated in epitope recognition in emerging SARS-CoV-2 strains. Comput. Struct. Biotechnol. J. 2021, 19, 5556–5567. [Google Scholar] [CrossRef] [PubMed]
- Nell, S.; Delphine, P.; William, H.B.; Christophe, R.; Slim, F.; Julian, B.; Cyril, P.; Matthieu, P.; Isabelle, S.; Florence, G.-B.; et al. Fusogenicity and neutralization sensitivity of the SARS-CoV-2 Delta sublineage AY.4.2. EBioMedicine. 2022, 77, 103934. [Google Scholar] [CrossRef]
- Yan, W.; Zheng, Y.; Zeng, X.; He, B.; Cheng, W. Structural biology of SARS-CoV-2: Open the door for novel therapies. Signal Transduct. Target. Ther. 2022, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; Schexnaydre, E.; Rafie, K.; Kurata, T.; Terenin, I.; Hauryliuk, V.; Carlson, L.A. Clinically observed deletions in SARS-CoV-2 Nsp1 affect its stability and ability to inhibit translation. FEBS Lett. 2022, 596, 1203–1213. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moen, L.V.; Vollan, H.S.; Bråte, J.; Hungnes, O.; Bragstad, K. Molecular Epidemiology of the Norwegian SARS-CoV-2 Delta Lineage AY.63. Viruses 2022, 14, 2734. https://doi.org/10.3390/v14122734
Moen LV, Vollan HS, Bråte J, Hungnes O, Bragstad K. Molecular Epidemiology of the Norwegian SARS-CoV-2 Delta Lineage AY.63. Viruses. 2022; 14(12):2734. https://doi.org/10.3390/v14122734
Chicago/Turabian StyleMoen, Line Victoria, Hilde Synnøve Vollan, Jon Bråte, Olav Hungnes, and Karoline Bragstad. 2022. "Molecular Epidemiology of the Norwegian SARS-CoV-2 Delta Lineage AY.63" Viruses 14, no. 12: 2734. https://doi.org/10.3390/v14122734




