SARS-CoV-2 Omicron (B.1.1.529) Infection of Wild White-Tailed Deer in New York City
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Surrogate Virus Neutralization Test (sVNT)
2.3. Generation of SARS-CoV-2 S Pseudotyped Viruses
2.4. Neutralization Assay of Deer Sera against Pseudotyped Virus Expressing the Spike Protein of SARS-CoV-2 Wild-Type or Variants
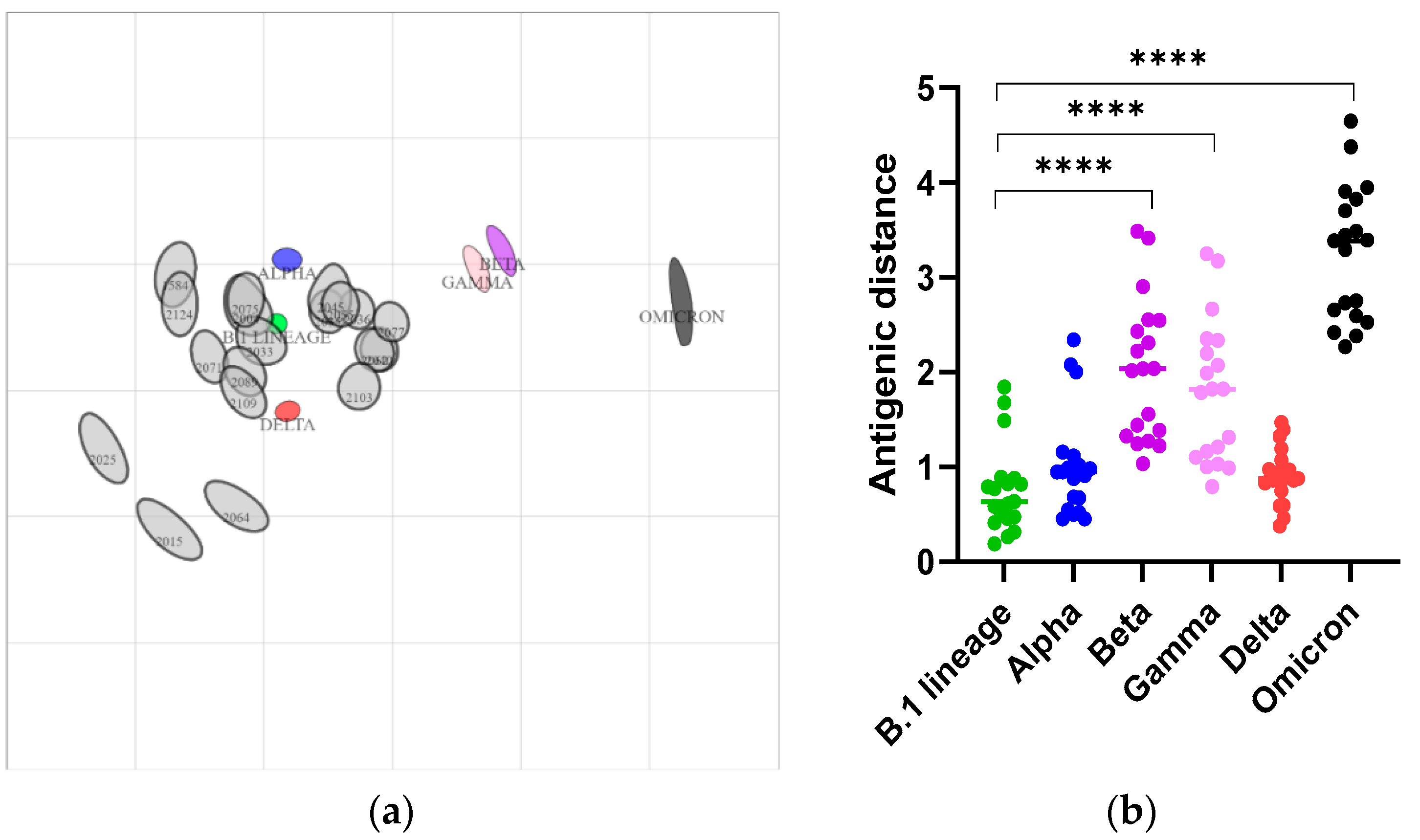
2.5. Antigen Cartography
2.6. RNA Extraction and RT-PCR for SARS-CoV-2 Detection
2.7. SARS-CoV-2 Genome Sequencing
2.8. SARS-CoV-2 Genome Sequence Analysis and Identification of Variants
2.9. Data Analysis and Visualization
3. Results
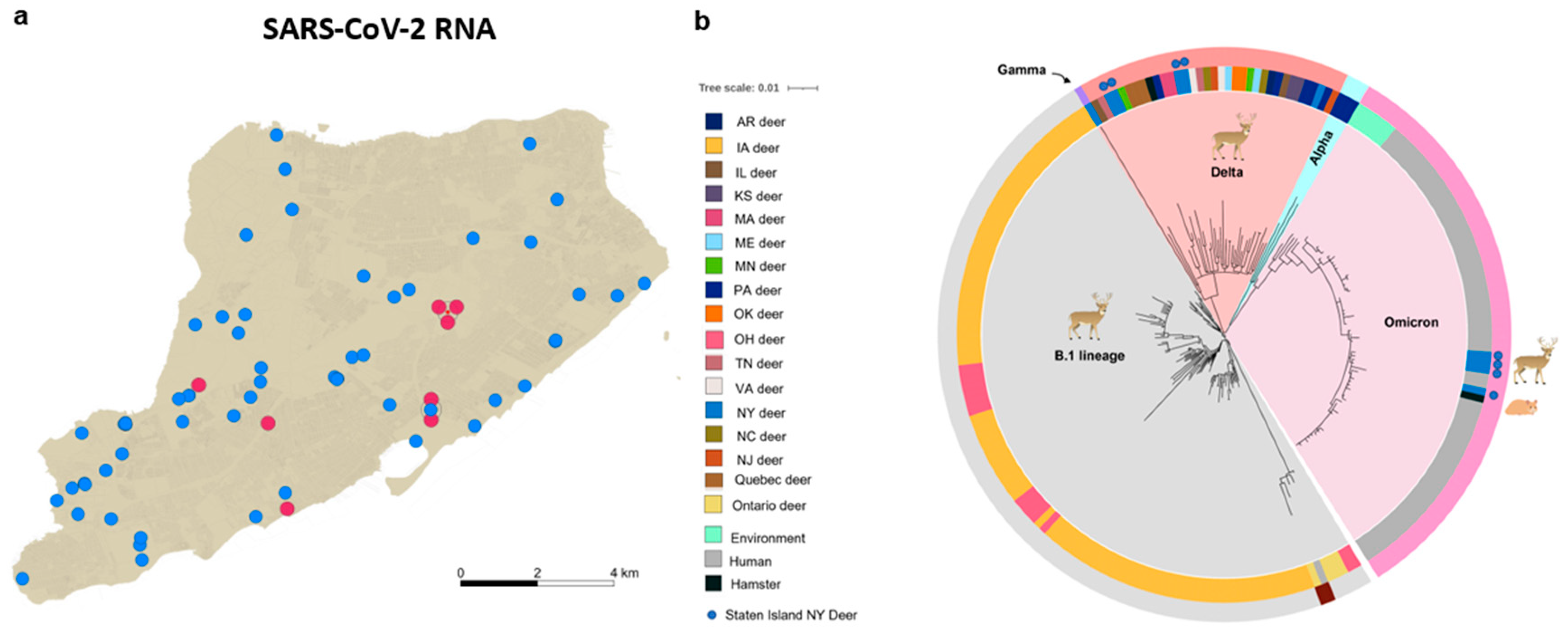
3.1. Molecular and Genetic Identification of SARS-CoV-2 Delta and Omicron Variants in Nasal and Tonsillar Swabs from White-Tailed Deer on Staten Island, New York
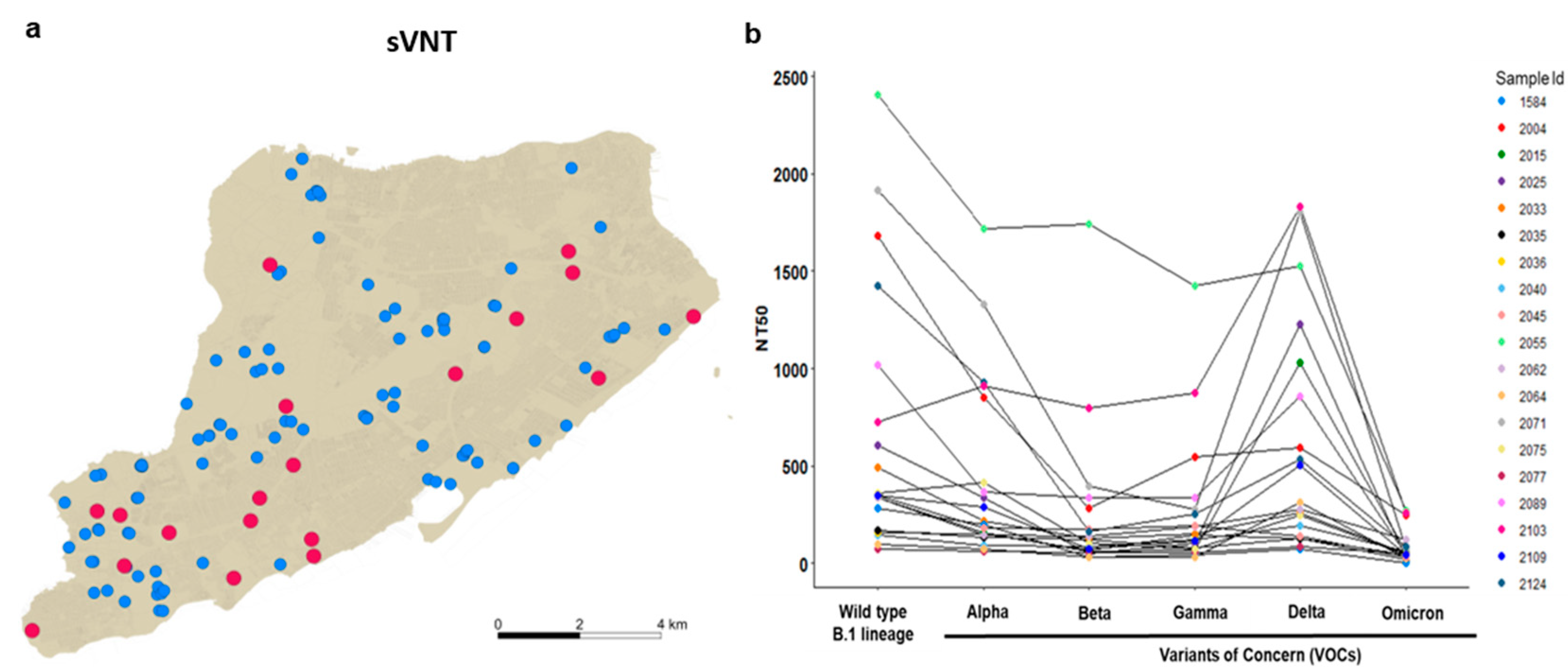
3.2. Delta and Omicron Infections Occur among Staten Island Deer despite Serological Evidence of Prior SARS-CoV-2 Exposure
3.3. Nasal Shedding of SARS-CoV-2 RNA in White-Tailed Deer with High Levels of Neutralizing Antibodies without Clinical Signs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Damas, J.; Hughes, G.M.; Keough, K.C.; Painter, C.A.; Persky, N.S.; Corbo, M.; Hiller, M.; Koepfli, K.P.; Pfenning, A.R.; Zhao, H.; et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl. Acad. Sci. USA 2020, 117, 22311–22322. [Google Scholar] [CrossRef]
- Hobbs, E.C.; Reid, T.J. Animals and SARS-CoV-2: Species susceptibility and viral transmission in experimental and natural conditions, and the potential implications for community transmission. Transbound. Emerg. Dis. 2021, 68, 1850–1867. [Google Scholar] [CrossRef] [PubMed]
- Meekins, D.A.; Gaudreault, N.N.; Richt, J.A. Natural and Experimental SARS-CoV-2 Infection in Domestic and Wild Animals. Viruses 2021, 13, 1993. [Google Scholar] [CrossRef] [PubMed]
- Oreshkova, N.; Molenaar, R.J.; Vreman, S.; Harders, F.; Oude Munnink, B.B.; Hakze-van der Honing, R.W.; Gerhards, N.; Tolsma, P.; Bouwstra, R.; Sikkema, R.S.; et al. SARS-CoV-2 infection in farmed minks, the Netherlands, April and May 2020. Eurosurveillance 2020, 25, 210325c. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.R.; Navas-Martin, S. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 2005, 69, 635–664. [Google Scholar] [CrossRef] [Green Version]
- Cool, K.; Gaudreault, N.N.; Morozov, I.; Trujillo, J.D.; Meekins, D.A.; McDowell, C.; Carossino, M.; Bold, D.; Mitzel, D.; Kwon, T.; et al. Infection and transmission of ancestral SARS-CoV-2 and its alpha variant in pregnant white-tailed deer. Emerg. Microbes Infect. 2022, 11, 95–112. [Google Scholar] [CrossRef]
- Palmer, M.V.; Martins, M.; Falkenberg, S.; Buckley, A.; Caserta, L.C.; Mitchell, P.K.; Cassmann, E.D.; Rollins, A.; Zylich, N.C.; Renshaw, R.W.; et al. Susceptibility of white-tailed deer (Odocoileus virginianus) to SARS-CoV-2. J. Virol. 2021, 95, e00083-21. [Google Scholar] [CrossRef]
- Kuchipudi, S.V.; Surendran-Nair, M.; Ruden, R.M.; Yon, M.; Nissly, R.H.; Vandegrift, K.J.; Nelli, R.K.; Li, L.; Jayarao, B.M.; Maranas, C.D.; et al. Multiple spillovers from humans and onward transmission of SARS-CoV-2 in white-tailed deer. Proc. Natl. Acad. Sci. USA 2022, 119, e2121644119. [Google Scholar] [CrossRef]
- Hale, V.L.; Dennis, P.M.; McBride, D.S.; Nolting, J.M.; Madden, C.; Huey, D.; Ehrlich, M.; Grieser, J.; Winston, J.; Lombardi, D.; et al. SARS-CoV-2 infection in free-ranging white-tailed deer. Nature 2022, 602, 481–486. [Google Scholar] [CrossRef]
- Marques, A.D.; Sherrill-Mix, S.; Everett, J.K.; Adhikari, H.; Reddy, S.; Ellis, J.C.; Zeliff, H.; Greening, S.S.; Cannuscio, C.C.; Strelau, K.M. Multiple Introductions of SARS-CoV-2 Alpha and Delta Variants into White-Tailed Deer in Pennsylvania. MBio 2022, 13, e0210122. [Google Scholar] [CrossRef]
- USDA Confirmed Cases of SARS-CoV-2 in Animals in the United States. Available online: https://www.aphis.usda.gov/aphis/dashboards/tableau/sars-dashboard (accessed on 1 February 2022).
- Pickering, B.; Lung, O.; Maguire, F.; Kruczkiewicz, P.; Kotwa, J.D.; Buchanan, T.; Gagnier, M.; Guthrie, J.L.; Jardine, C.M.; Marchand-Austin, A.; et al. Divergent SARS-CoV-2 variant emerges in white-tailed deer with deer-to-human transmission. Nat. Microbiol. 2022, 7, 2011–2024. [Google Scholar] [CrossRef] [PubMed]
- Farinholt, T.; Doddapaneni, H.; Qin, X.; Menon, V.; Meng, Q.; Metcalf, G.; Chao, H.; Gingras, M.C.; Avadhanula, V.; Farinholt, P.; et al. Transmission event of SARS-CoV-2 delta variant reveals multiple vaccine breakthrough infections. BMC Med. 2021, 19, 255. [Google Scholar] [CrossRef] [PubMed]
- Riediker, M.; Briceno-Ayala, L.; Ichihara, G.; Albani, D.; Poffet, D.; Tsai, D.H.; Iff, S.; Monn, C. Higher viral load and infectivity increase risk of aerosol transmission for Delta and Omicron variants of SARS-CoV-2. Swiss Med. Wkly. 2022, 152, w30133. [Google Scholar] [PubMed]
- Ito, K.; Piantham, C.; Nishiura, H. Relative instantaneous reproduction number of Omicron SARS-CoV-2 variant with respect to the Delta variant in Denmark. J. Med. Virol. 2021, 94, 2265–2268. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, K.N.; Mendes, A.; Strydom, A.; Rotherham, L.; Mulumba, M.; Venter, M. SARS-CoV-2 Reverse Zoonoses to Pumas and Lions, South Africa. Viruses 2022, 14, 120. [Google Scholar] [CrossRef]
- Yen, H.-L.; Sit, T.H.C.; Brackman, C.J.; Chuk, S.S.Y.; Gu, H.; Tam, K.W.S.; Law, P.Y.T.; Leung, G.M.; Peiris, M.; Poon, L.L.M.; et al. Transmission of SARS-CoV-2 delta variant (AY.127) from pet hamsters to humans, leading to onward human-to-human transmission: A case study. Lancet 2022, 399, 1070–1078. [Google Scholar] [CrossRef] [PubMed]
- Mallapaty, S. Where did Omicron come from? Three key theories. Nature 2022, 602, 26–28. [Google Scholar] [CrossRef]
- Kotwa, J.D.; Massé, A.; Gagnier, M.; Aftanas, P.; Blais-Savoie, J.; Bowman, J.; Buchanan, T.; Chee, H.-Y.; Dibernardo, A.; Kruczkiewicz, P.; et al. First detection of SARS-CoV-2 infection in Canadian wildlife identified in free-ranging white-tailed deer (Odocoileus virginianus) from southern Québec, Canada. bioRxiv 2022, 476458. [Google Scholar] [CrossRef]
- Zhang, J.; Ding, N.; Ren, L.; Song, R.; Chen, D.; Zhao, X.; Chen, B.; Han, J.; Li, J.; Song, Y.; et al. COVID-19 reinfection in the presence of neutralizing antibodies. Natl. Sci. Rev. 2021, 8, nwab006. [Google Scholar] [CrossRef]
- Chandler, J.C.; Bevins, S.N.; Ellis, J.W.; Linder, T.J.; Tell, R.M.; Jenkins-Moore, M.; Root, J.J.; Lenoch, J.B.; Robbe-Austerman, S.; DeLiberto, T.J.; et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc. Natl. Acad. Sci. USA 2021, 118, e2114828118. [Google Scholar] [CrossRef]
- Tan, C.W.; Chia, W.N.; Qin, X.; Liu, P.; Chen, M.I.; Tiu, C.; Hu, Z.; Chen, V.C.; Young, B.E.; Sia, W.R.; et al. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020, 38, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Neerukonda, S.N.; Vassell, R.; Herrup, R.; Liu, S.; Wang, T.; Takeda, K.; Yang, Y.; Lin, T.L.; Wang, W.; Weiss, C.D. Establishment of a well-characterized SARS-CoV-2 lentiviral pseudovirus neutralization assay using 293T cells with stable expression of ACE2 and TMPRSS2. PLoS ONE 2021, 16, e0248348. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.J.; Lapedes, A.S.; de Jong, J.C.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.; Fouchier, R.A. Mapping the antigenic and genetic evolution of influenza virus. Science 2004, 305, 371–376. [Google Scholar] [CrossRef] [Green Version]
- Kendra, J.A.; Tohma, K.; Ford-Siltz, L.A.; Lepore, C.J.; Parra, G.I. Antigenic cartography reveals complexities of genetic determinants that lead to antigenic differences among pandemic GII.4 noroviruses. Proc. Natl. Acad. Sci. USA 2021, 118, e2015874118. [Google Scholar] [CrossRef] [PubMed]
- Smithgall, M.C.; Dowlatshahi, M.; Spitalnik, S.L.; Hod, E.A.; Rai, A.J. Types of Assays for SARS-CoV-2 Testing: A Review. Lab. Med. 2020, 51, e59–e65. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Patel, D.; Pandya, A. Emerging diagnostic tools for detection of COVID-19 and perspective. Biomed. Microdevices 2020, 22, 83. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.K.; Tham, J.W.M.; Png, S.; Chai, C.N.; Ng, S.C.; Tan, E.J.M.; Ng, L.J.; Chua, R.P.; Sani, M.; Seow, Y.; et al. Clinical performance of Roche cobas 6800, Luminex ARIES, MiRXES Fortitude Kit 2.1, Altona RealStar, and Applied Biosystems TaqPath for SARS-CoV-2 detection in nasopharyngeal swabs. J. Med. Virol. 2021, 93, 4603–4607. [Google Scholar] [CrossRef]
- Long, S.W.; Olsen, R.J.; Christensen, P.A.; Subedi, S.; Olson, R.; Davis, J.J.; Saavedra, M.O.; Yerramilli, P.; Pruitt, L.; Reppond, K.; et al. Sequence Analysis of 20,453 Severe Acute Respiratory Syndrome Coronavirus 2 Genomes from the Houston Metropolitan Area Identifies the Emergence and Widespread Distribution of Multiple Isolates of All Major Variants of Concern. Am. J. Pathol. 2021, 191, 983–992. [Google Scholar] [CrossRef]
- Christensen, P.A.; Olsen, R.J.; Long, S.W.; Subedi, S.; Davis, J.J.; Hodjat, P.; Walley, D.R.; Kinskey, J.C.; Ojeda Saavedra, M.; Pruitt, L.; et al. Delta Variants of SARS-CoV-2 Cause Significantly Increased Vaccine Breakthrough COVID-19 Cases in Houston, Texas. Am. J. Pathol. 2022, 192, 320–331. [Google Scholar] [CrossRef]
- O’Toole, A.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef]
- Olsen, R.J.; Christensen, P.A.; Long, S.W.; Subedi, S.; Hodjat, P.; Olson, R.; Nguyen, M.; Davis, J.J.; Yerramilli, P.; Saavedra, M.O.; et al. Trajectory of Growth of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Variants in Houston, Texas, January through May 2021, Based on 12,476 Genome Sequences. Am. J. Pathol. 2021, 191, 1754–1773. [Google Scholar] [CrossRef]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, C. SARS-CoV-2 Variant Calling and Consensus Assembly Pipeline. Available online: https://github.com/onecodex/sars-cov-2.git (accessed on 25 February 2022).
- Li, H. Toolkit for Processing Sequences in FASTA/Q Formats. Available online: https://github.com/lh3/seqtk (accessed on 25 February 2022).
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [Green Version]
- Wu, F.; Zhao, S.; Yu, B.; Chen, Y.-M.; Wang, W.; Song, Z.-G.; Hu, Y.; Tao, Z.-W.; Tian, J.-H.; Pei, Y.-Y. A new coronavirus associated with human respiratory disease in China. Nature 2020, 579, 265–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, K.G. iVar Version 1.2.2. Available online: https://github.com/andersen-lab/ivar/releases (accessed on 25 February 2021).
- Grubaugh, N.D.; Gangavarapu, K.; Quick, J.; Matteson, N.L.; De Jesus, J.G.; Main, B.J.; Tan, A.L.; Paul, L.M.; Brackney, D.E.; Grewal, S.; et al. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019, 20, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, S.W.; Olsen, R.J.; Christensen, P.A.; Bernard, D.W.; Davis, J.J.; Shukla, M.; Nguyen, M.; Saavedra, M.O.; Yerramilli, P.; Pruitt, L.; et al. Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area. MBio 2020, 11, e0270720. [Google Scholar] [CrossRef]
- NYCHealth Omicron Variant: NYC Report for 13 January 2022. Available online: https://www1.nyc.gov/assets/doh/downloads/pdf/covid/omicron-variant-report-jan-13–22.pdf (accessed on 1 February 2022).
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef] [Green Version]
- Van der Straten, K.; Guerra, D.; van Gils, M.; Bontjer, I.; Caniels, T.G.; van Willigen, H.D.; Wynberg, E.; Poniman, M.; Burger, J.A.; Bouhuijs, J.H.; et al. Mapping the antigenic diversification of SARS-CoV-2. medRxiv 2022. [Google Scholar] [CrossRef]
- Sheehan, M.M.; Reddy, A.J.; Rothberg, M.B. Reinfection Rates among Patients Who Previously Tested Positive for Coronavirus Disease 2019: A Retrospective Cohort Study. Clin. Infect. Dis. 2021, 73, 1882–1886. [Google Scholar] [CrossRef]
- Agarwal, V.; Venkatakrishnan, A.J.; Puranik, A.; Kirkup, C.; Lopez-Marquez, A.; Challener, D.W.; Theel, E.S.; O’Horo, J.C.; Binnicker, M.J.; Kremers, W.K.; et al. Long-term SARS-CoV-2 RNA shedding and its temporal association to IgG seropositivity. Cell Death Discov. 2020, 6, 138. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.C.S.; Lam, S.D.; Richard, D.; Owen, C.; Berchtold, D.; Orengo, C.; Nair, M.S.; Kuchipudi, S.V.; Kapur, V.; van Dorp, L.; et al. Transmission of SARS-CoV-2 from humans to animals and potential host adaptation. Nat. Commun. 2022, 13, 2988. [Google Scholar] [CrossRef] [PubMed]
- Bashor, L.; Gagne, R.B.; Bosco-Lauth, A.M.; Bowen, R.A.; Stenglein, M.; VandeWoude, S. SARS-CoV-2 evolution in animals suggests mechanisms for rapid variant selection. Proc. Natl. Acad. Sci. USA 2021, 118, e2105253118. [Google Scholar] [CrossRef] [PubMed]
- Fagre, A.; Lewis, J.; Eckley, M.; Zhan, S.; Rocha, S.M.; Sexton, N.R.; Burke, B.; Geiss, B.; Peersen, O.; Bass, T.; et al. SARS-CoV-2 infection, neuropathogenesis and transmission among deer mice: Implications for spillback to New World rodents. PLoS Pathog. 2021, 17, e1009585. [Google Scholar] [CrossRef]
- Francisco, R.; Hernandez, S.M.; Mead, D.G.; Adcock, K.G.; Burke, S.C.; Nemeth, N.M.; Yabsley, M.J. Experimental Susceptibility of North American Raccoons (Procyon lotor) and Striped Skunks (Mephitis mephitis) to SARS-CoV-2. Front. Vet. Sci. 2021, 8, 715307. [Google Scholar] [CrossRef]
- Griffin, B.D.; Chan, M.; Tailor, N.; Mendoza, E.J.; Leung, A.; Warner, B.M.; Duggan, A.T.; Moffat, E.; He, S.; Garnett, L.; et al. SARS-CoV-2 infection and transmission in the North American deer mouse. Nat. Commun. 2021, 12, 3612. [Google Scholar] [CrossRef]
- Moreira-Soto, A.; Walzer, C.; Czirják, G.Á.; Richter, M.; Marino, S.F.; Posautz, A.; de Yebra, P.R.; McEwen, G.K.; Drexler, J.F.; Greenwood, A.D. SARS-CoV-2 has not emerged in roe, red or fallow deer in Germany or Austria during the COVID 19 pandemic. bioRxiv 2022. [Google Scholar] [CrossRef]
- Chen, C.; Boorla, V.S.; Banerjee, D.; Chowdhury, R.; Cavener, V.S.; Nissly, R.H.; Gontu, A.; Boyle, N.R.; Vandegrift, K.; Nair, M.S.; et al. Computational prediction of the effect of amino acid changes on the binding affinity between SARS-CoV-2 spike RBD and human ACE2. Proc. Natl. Acad. Sci. USA 2021, 118, e2106480118. [Google Scholar] [CrossRef]
- Fischhoff, I.R.; Castellanos, A.A.; Rodrigues, J.; Varsani, A.; Han, B.A. Predicting the zoonotic capacity of mammals to transmit SARS-CoV-2. Proc. Royal Soc. B 2021, 288, 20211651. [Google Scholar] [CrossRef]
- Roberts, M.G.; Heesterbeek, J.A.P. Characterizing reservoirs of infection and the maintenance of pathogens in ecosystems. J. R. Soc. Interface 2020, 17, 20190540. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vandegrift, K.J.; Yon, M.; Surendran Nair, M.; Gontu, A.; Ramasamy, S.; Amirthalingam, S.; Neerukonda, S.; Nissly, R.H.; Chothe, S.K.; Jakka, P.; et al. SARS-CoV-2 Omicron (B.1.1.529) Infection of Wild White-Tailed Deer in New York City. Viruses 2022, 14, 2770. https://doi.org/10.3390/v14122770
Vandegrift KJ, Yon M, Surendran Nair M, Gontu A, Ramasamy S, Amirthalingam S, Neerukonda S, Nissly RH, Chothe SK, Jakka P, et al. SARS-CoV-2 Omicron (B.1.1.529) Infection of Wild White-Tailed Deer in New York City. Viruses. 2022; 14(12):2770. https://doi.org/10.3390/v14122770
Chicago/Turabian StyleVandegrift, Kurt J., Michele Yon, Meera Surendran Nair, Abhinay Gontu, Santhamani Ramasamy, Saranya Amirthalingam, Sabarinath Neerukonda, Ruth H. Nissly, Shubhada K. Chothe, Padmaja Jakka, and et al. 2022. "SARS-CoV-2 Omicron (B.1.1.529) Infection of Wild White-Tailed Deer in New York City" Viruses 14, no. 12: 2770. https://doi.org/10.3390/v14122770
APA StyleVandegrift, K. J., Yon, M., Surendran Nair, M., Gontu, A., Ramasamy, S., Amirthalingam, S., Neerukonda, S., Nissly, R. H., Chothe, S. K., Jakka, P., LaBella, L., Levine, N., Rodriguez, S., Chen, C., Sheersh Boorla, V., Stuber, T., Boulanger, J. R., Kotschwar, N., Aucoin, S. G., ... Kuchipudi, S. V. (2022). SARS-CoV-2 Omicron (B.1.1.529) Infection of Wild White-Tailed Deer in New York City. Viruses, 14(12), 2770. https://doi.org/10.3390/v14122770









