HIV-1 gp120 Protein Activates Cyclin-Dependent Kinase 1, a Possible Link to Central Nervous System Cell Death
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Cultivation
2.2. Cell Transfection
2.3. CCK8 Assay
2.4. TUNEL Assay
2.5. Western Blotting Analysis
2.6. RNA Preparation and Transcriptomics Libraries
2.7. Sample Preparation and Mass Spectrometry
2.8. Selection of the Candidate Reference Genes
2.9. Isolation of RNA and cDNA Synthesis
2.10. Bioinformatics Analysis of HIV-Associated Dementia against the Normal Control in GSE35864 Data Set
2.11. Pharmacological Inhibition of CDKI-Gene
2.12. Statistical Analysis
3. Results
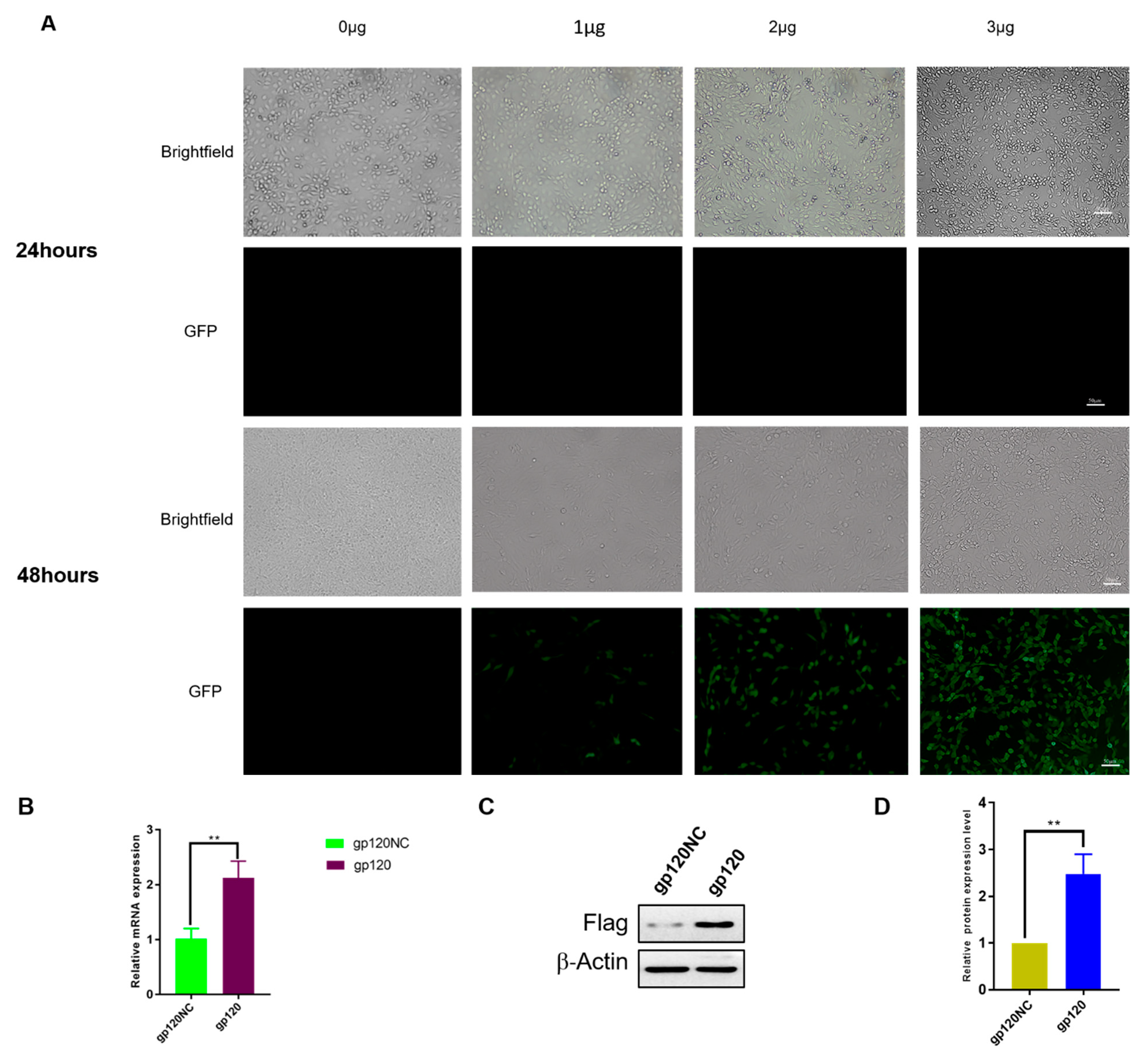
3.1. Transfection of HIV-1 gp120 into PC12 Cells
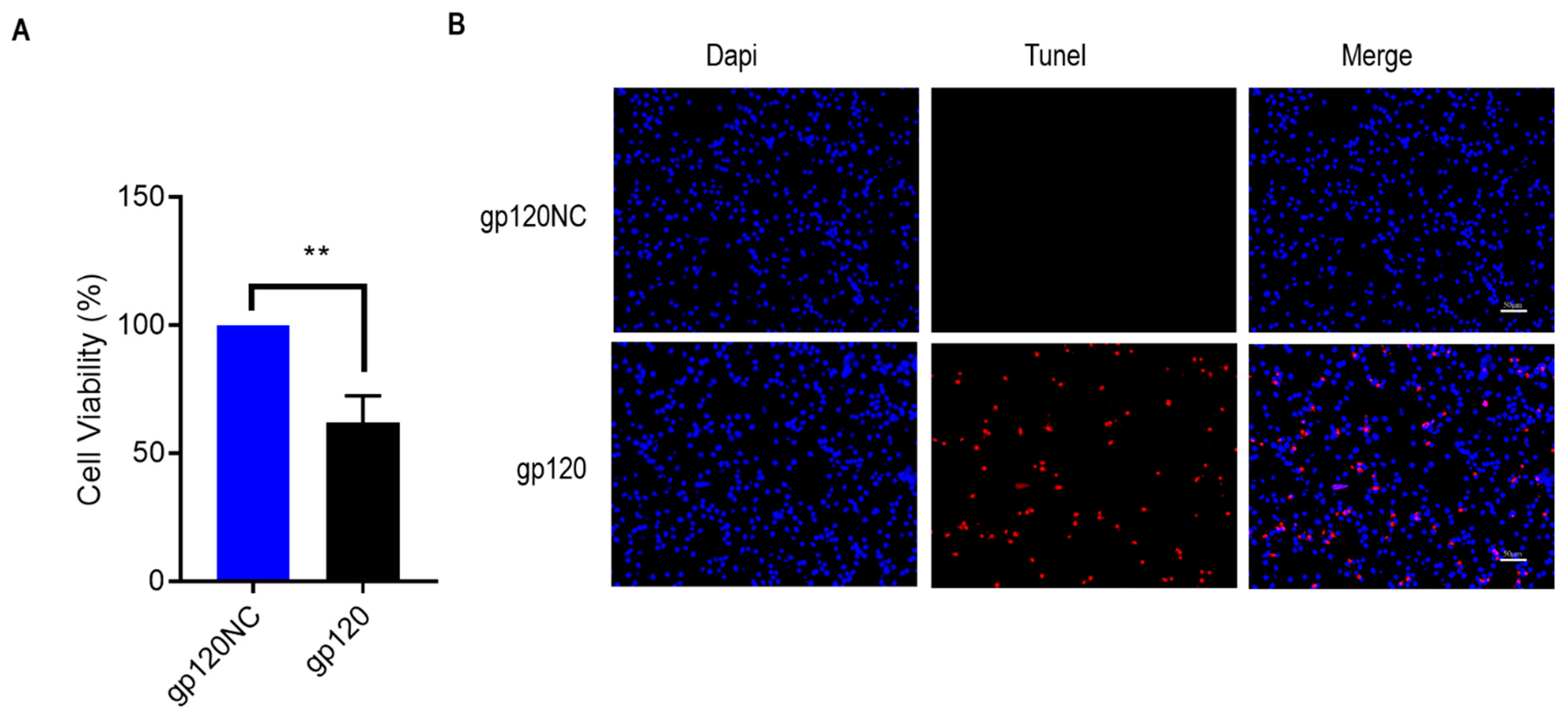
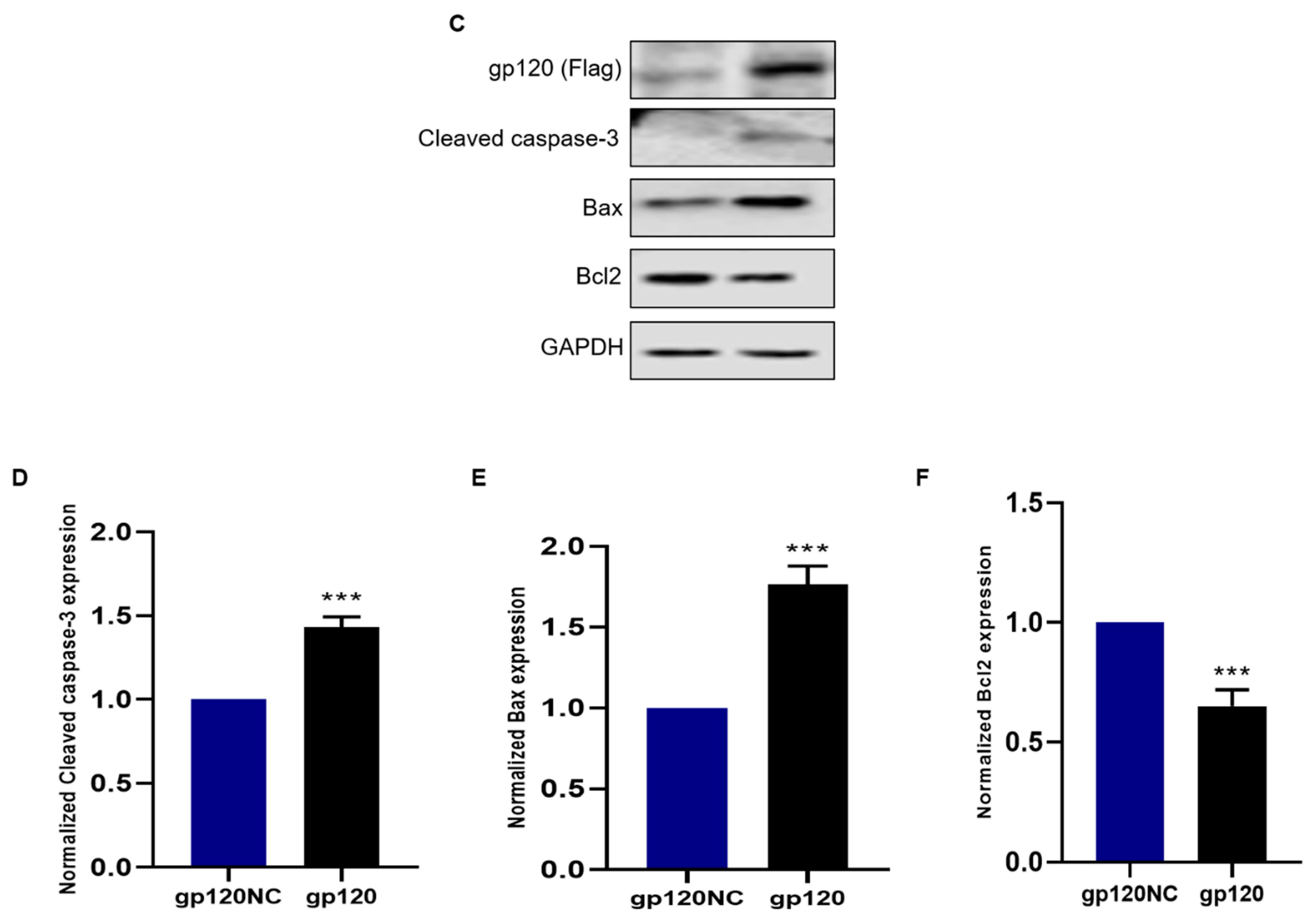
3.2. HIV-1 gp120 Induced Apoptosis of PC12 Cells
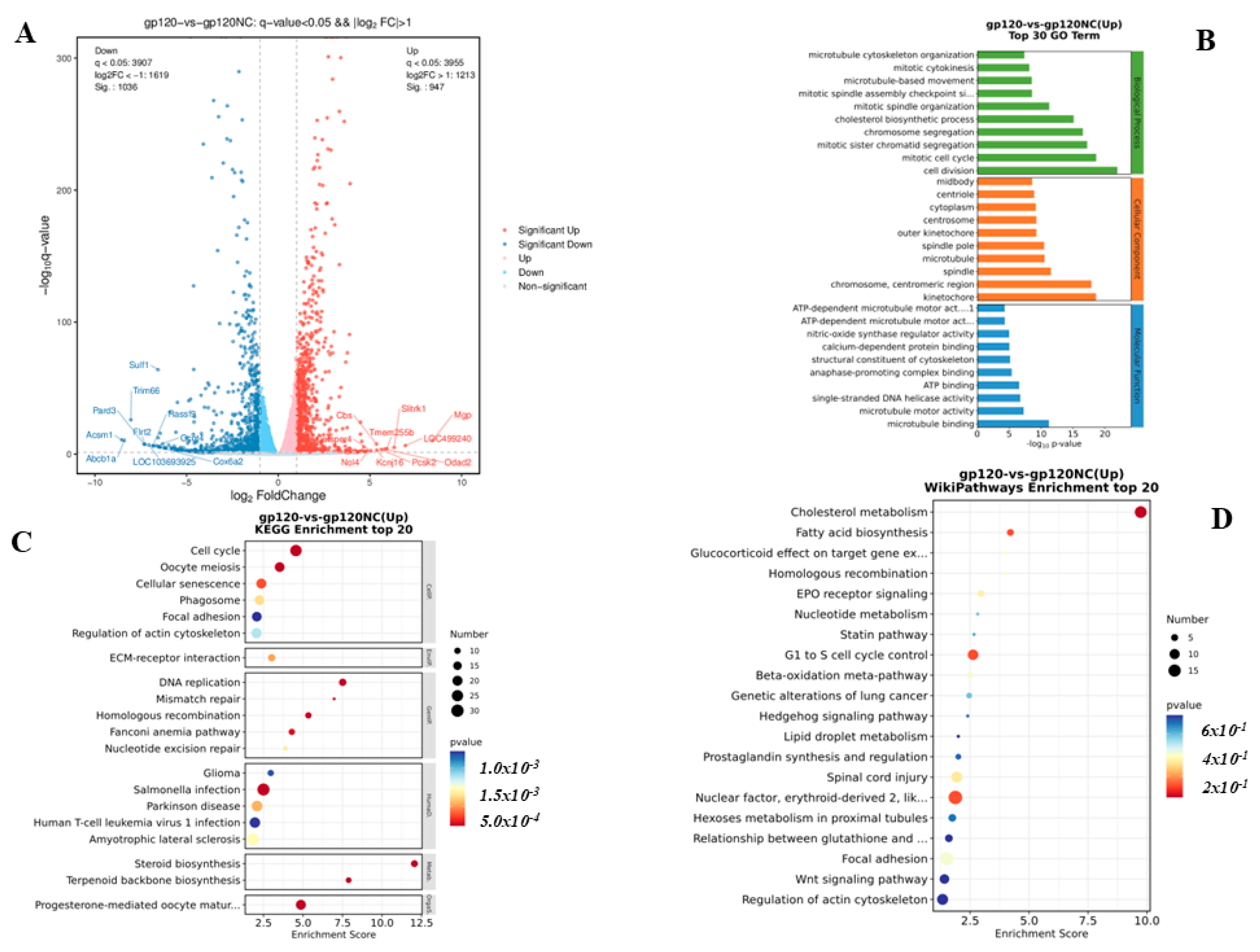
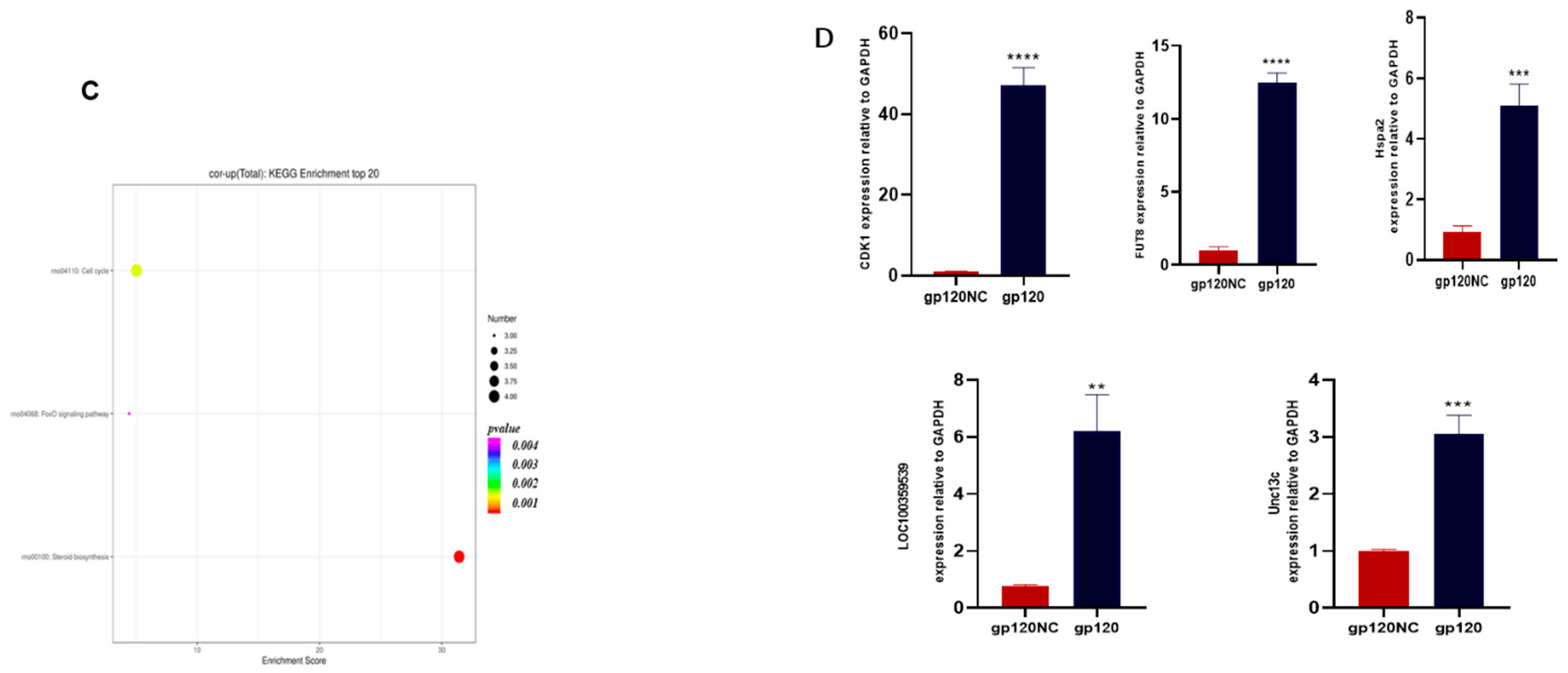
3.3. RNA Transcriptomics Reveals Genes Linked to Neurodegenerative Diseases
3.4. Proteomics Analysis of HIV-1gp120-Expressing Cells
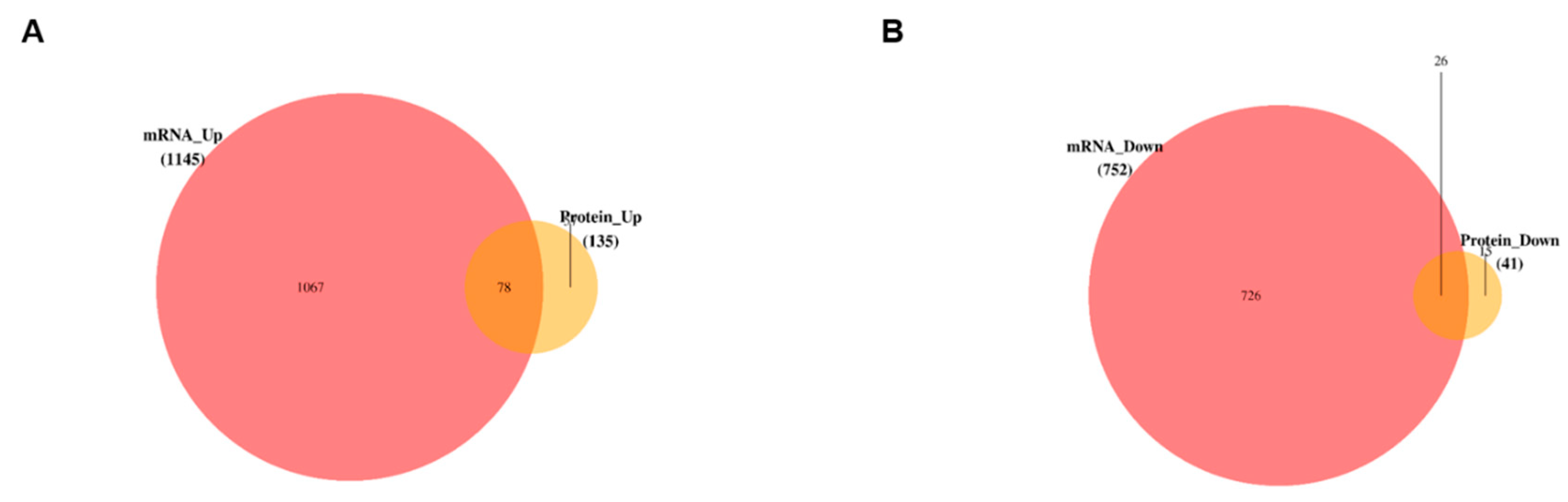
3.5. Data Integration and Validation of Transcriptomics and Proteomics
3.6. CDK1 Is Differentially Upregulated in HIV-Associated Dementia
3.7. A Pharmacological Inhibition of Cdk1 May Attenuate the gp120 Overexpression-Induced Cell Death
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Navia, B.A.; Cho, E.S.; Petito, C.K.; Price, R.W. The AIDS Dementia Complex: II. Neuropathology. Ann. Neurol. 1986, 19, 525–535. [Google Scholar] [CrossRef] [PubMed]
- Saylor, D.; Dickens, A.M.; Sacktor, N.; Haughey, N.; Slusher, B.; Pletnikov, M.; Mankowski, J.L.; Brown, A.; Volsky, D.J.; McArthur, J.C. HIV-Associated Neurocognitive Disorder—Pathogenesis and Prospects for Treatment. Nat. Rev. Neurol. 2016, 12, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Navia, B.A.; Jordan, B.D.; Price, R.W. The AIDS Dementia Complex: I. Clinical Features. Ann. Neurol. 1986, 19, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Borrajo, A.; Spuch, C.; Penedo, M.A.; Olivares, J.M.; Agís-Balboa, R.C. Important Role of Microglia in HIV-1 Associated Neurocognitive Disorders and the Molecular Pathways Implicated in Its Pathogenesis. Ann. Med. 2021, 53, 43–69. [Google Scholar] [CrossRef] [PubMed]
- McArthur, J.C.; Brew, B.J. HIV-Associated Neurocognitive Disorders: Is There a Hidden Epidemic? AIDS 2010, 24, 1367–1370. [Google Scholar] [CrossRef] [PubMed]
- Chai, Q.; Jovasevic, V.; Malikov, V.; Sabo, Y.; Morham, S.; Walsh, D.; Naghavi, M.H. HIV-1 Counteracts an Innate Restriction by Amyloid Precursor Protein Resulting in Neurodegeneration. Nat. Commun. 2017, 8, 1522. [Google Scholar] [CrossRef] [Green Version]
- Albright, A.V.; Shieh, J.T.; Itoh, T.; Lee, B.; Pleasure, D.; O’Connor, M.J.; Doms, R.W.; González-Scarano, F. Microglia Express CCR5, CXCR4, and CCR3, but of These, CCR5 Is the Principal Coreceptor for Human Immunodeficiency Virus Type 1 Dementia Isolates. J. Virol. 1999, 73, 205–213. [Google Scholar] [CrossRef] [Green Version]
- Eugenin, E.A.; D’Aversa, T.G.; Lopez, L.; Calderon, T.M.; Berman, J.W. MCP-1 (CCL2) Protects Human Neurons and Astrocytes from NMDA or HIV-Tat-Induced Apoptosis. J. Neurochem. 2003, 85, 1299–1311. [Google Scholar] [CrossRef]
- András, I.E.; Pu, H.; Tian, J.; Deli, M.A.; Nath, A.; Hennig, B.; Toborek, M. Signaling Mechanisms of HIV-1 Tat-Induced Alterations of Claudin-5 Expression in Brain Endothelial Cells. J. Cereb. Blood Flow Metab. Off. J. Int. Soc. Cereb. Blood Flow Metab. 2005, 25, 1159–1170. [Google Scholar] [CrossRef] [Green Version]
- Torres-Muñoz, J.E.; Núñez, M.; Petito, C.K. Successful Application of Hyperbranched Multidisplacement Genomic Amplification to Detect HIV-1 Sequences in Single Neurons Removed from Autopsy Brain Sections by Laser Capture Microdissection. J. Mol. Diagn. 2008, 10, 317–324. [Google Scholar] [CrossRef]
- Chi, X.; Amet, T.; Byrd, D.; Chang, K.-H.; Shah, K.; Hu, N.; Grantham, A.; Hu, S.; Duan, J.; Tao, F.; et al. Direct Effects of HIV-1 Tat on Excitability and Survival of Primary Dorsal Root Ganglion Neurons: Possible Contribution to HIV-1-Associated Pain. PLoS ONE 2011, 6, e24412. [Google Scholar] [CrossRef] [PubMed]
- Rao, V.R.; Ruiz, A.P.; Prasad, V.R. Viral and Cellular Factors Underlying Neuropathogenesis in HIV Associated Neurocognitive Disorders (HAND). AIDS Res. Ther. 2014, 11, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cantó-Nogués, C.; Sánchez-Ramón, S.; Álvarez, S.; Lacruz, C.; Muñóz-Fernánde, M.Á. HIV-1 Infection of Neurons Might Account for Progressive HIV-1-Associated Encephalopathy in Children. J. Mol. Neurosci. 2005, 27, 079–090. [Google Scholar] [CrossRef] [PubMed]
- Kaul, M.; Garden, G.A.; Lipton, S.A. Pathways to Neuronal Injury and Apoptosis in HIV-Associated Dementia. Nature 2001, 410, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Langer, S.; Sauter, D. Unusual Fusion Proteins of HIV-1. Front. Microbiol. 2016, 7, 2152. [Google Scholar] [CrossRef] [Green Version]
- Wallace, V.C.J.; Blackbeard, J.; Segerdahl, A.R.; Hasnie, F.; Pheby, T.; McMahon, S.B.; Rice, A.S.C. Characterization of Rodent Models of HIV-Gp120 and Anti-Retroviral-Associated Neuropathic Pain. Brain 2007, 130, 2688–2702. [Google Scholar] [CrossRef] [Green Version]
- Silverstein, P.S.; Shah, A.; Weemhoff, J.; Kumar, S.; Singh, D.P.; Kumar, A. HIV-1 Gp120 and Drugs of Abuse: Interactions in the Central Nervous System. Curr. HIV Res. 2012, 10, 369–383. [Google Scholar] [CrossRef] [Green Version]
- Yoon, V.; Fridkis-Hareli, M.; Munisamy, S.; Lee, J.; Anastasiades, D.; Stevceva, L. The GP120 Molecule of HIV-1 and Its Interaction with T Cells. Curr. Med. Chem. 2010, 17, 741–749. [Google Scholar] [CrossRef]
- Alirezaei, M.; Watry, D.D.; Flynn, C.F.; Kiosses, W.B.; Masliah, E.; Williams, B.R.G.; Kaul, M.; Lipton, S.A.; Fox, H.S. Human Immunodeficiency Virus-1/Surface Glycoprotein 120 Induces Apoptosis through RNA-Activated Protein Kinase Signaling in Neurons. J. Neurosci. 2007, 27, 11047–11055. [Google Scholar] [CrossRef] [Green Version]
- Shah, A.; Vaidya, N.K.; Bhat, H.K.; Kumar, A. HIV-1 Gp120 Induces Type-1 Programmed Cell Death through ER Stress Employing IRE1α, JNK and AP-1 Pathway. Sci. Rep. 2016, 6, 18929. [Google Scholar] [CrossRef] [Green Version]
- Shah, A.; Verma, A.S.; Patel, K.H.; Noel, R.; Rivera-Amill, V.; Silverstein, P.S.; Chaudhary, S.; Bhat, H.K.; Stamatatos, L.; Singh, D.P.; et al. HIV-1 Gp120 Induces Expression of IL-6 through a Nuclear Factor-Kappa B-Dependent Mechanism: Suppression by Gp120 Specific Small Interfering RNA. PLoS ONE 2011, 6, e21261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI Gene Expression and Hybridization Array Data Repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, J.; Kharebava, G.; Piao, C.; Stoica, B.A.; Dinizo, M.; Sabirzhanov, B.; Hanscom, M.; Guanciale, K.; Faden, A.I. Inhibition of E2F1/CDK1 Pathway Attenuates Neuronal Apoptosis in Vitro and Confers Neuroprotection after Spinal Cord Injury in Vivo. PLoS ONE 2012, 7, e42129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiatrak, B.; Kubis-Kubiak, A.; Piwowar, A.; Barg, E. PC12 Cell Line: Cell Types, Coating of Culture Vessels, Differentiation and Other Culture Conditions. Cells 2020, 9, 958. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Sulcove, J.; Frank, I.; Jaffer, S.; Ozdener, H.; Kolson, D.L. Development of a Human Neuronal Cell Model for Human Immunodeficiency Virus (HIV)-Infected Macrophage-Induced Neurotoxicity: Apoptosis Induced by HIV Type 1 Primary Isolates and Evidence for Involvement of the Bcl-2/Bcl-XL-Sensitive Intrinsic Apoptosis P. J. Virol. 2002, 76, 9407–9419. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Liu, J.; Xu, C.; Keblesh, J.; Zang, W.; Xiong, H. HIV-1gp120 Induces Neuronal Apoptosis through Enhancement of 4-Aminopyridine-Senstive Outward K+ Currents. PLoS ONE 2011, 6, e25994. [Google Scholar] [CrossRef]
- Yang, Y.; Mufson, E.J.; Herrup, K. Neuronal Cell Death Is Preceded by Cell Cycle Events at All Stages of Alzheimer’s Disease. J. Neurosci. Off. J. Soc. Neurosci. 2003, 23, 2557–2563. [Google Scholar] [CrossRef] [Green Version]
- Herrup, K.; Yang, Y. Cell Cycle Regulation in the Postmitotic Neuron: Oxymoron or New Biology? Nat. Rev. Neurosci. 2007, 8, 368–378. [Google Scholar] [CrossRef]
- Andreau, K.; Perfettini, J.-L.; Castedo, M.; Métivier, D.; Scott, V.; Pierron, G.; Kroemer, G. Contagious Apoptosis Facilitated by the HIV-1 Envelope: Fusion-Induced Cell-to-Cell Transmission of a Lethal Signal. J. Cell Sci. 2004, 117, 5643–5653. [Google Scholar] [CrossRef] [Green Version]
- Potapova, T.A.; Daum, J.R.; Byrd, K.S.; Gorbsky, G.J. Fine Tuning the Cell Cycle: Activation of the Cdk1 Inhibitory Phosphorylation Pathway during Mitotic Exit. Mol. Biol. Cell 2009, 20, 1737–1748. [Google Scholar] [CrossRef]
- Nigg, E.A.; Blangy, A.; Lane, H.A. Dynamic Changes in Nuclear Architecture during Mitosis: On the Role of Protein Phosphorylation in Spindle Assembly and Chromosome Segregation. Exp. Cell Res. 1996, 229, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.J.; Zhu, B.T. Upregulated Cyclin B1/CDK1 Mediates Apoptosis Following 2-Methoxyestradiol-Induced Mitotic Catastrophe: Role of Bcl-X(L) Phosphorylation. Steroids 2019, 150, 108381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Shi, H.; Ren, F.; Zhang, M.; Ji, P.; Wang, W.; Liu, C. The Aberrant Upstream Pathway Regulations of CDK1 Protein Were Implicated in the Proliferation and Apoptosis of Ovarian Cancer Cells. J. Ovarian Res. 2017, 10, 60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fields, J.A.; Dumaop, W.; Crews, L.; Adame, A.; Spencer, B.; Metcalf, J.; He, J.; Rockenstein, E.; Masliah, E. Mechanisms of HIV-1 Tat Neurotoxicity via CDK5 Translocation and Hyper-Activation: Role in HIV-Associated Neurocognitive Disorders. Curr. HIV Res. 2015, 13, 43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castedo, M.; Perfettini, J.L.; Roumier, T.; Kroemer, G. Cyclin-Dependent Kinase-1: Linking Apoptosis to Cell Cycle and Mitotic Catastrophe. Cell Death Differ. 2002, 9, 1287–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castedo, M.; Roumier, T.; Blanco, J.; Ferri, K.F.; Barretina, J.; Tintignac, L.A.; Andreau, K.; Perfettini, J.-L.; Amendola, A.; Nardacci, R.; et al. Sequential Involvement of Cdk1, MTOR and P53 in Apoptosis Induced by the HIV-1 Envelope. EMBO J. 2002, 21, 4070–4080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hilton, G.D.; Stoica, B.A.; Byrnes, K.R.; Faden, A.I. Roscovitine Reduces Neuronal Loss, Glial Activation, and Neurologic Deficits after Brain Trauma. J. Cereb. Blood Flow Metab. 2008, 28, 1845–1859. [Google Scholar] [CrossRef] [Green Version]
- Di Giovanni, S.; Movsesyan, V.; Ahmed, F.; Cernak, I.; Schinelli, S.; Stoica, B.; Faden, A.I. Cell Cycle Inhibition Provides Neuroprotection and Reduces Glial Proliferation and Scar Formation after Traumatic Brain Injury. Proc. Natl. Acad. Sci. USA 2005, 102, 8333–8338. [Google Scholar] [CrossRef]



| GENE | Sequences | Product Size (bp) |
|---|---|---|
| HIV-1gp120 F | TAGAGCTAGCGAATTCATGAGAGTGACGGGGATCAGG | 37 |
| HIV-1gp120 R | CACCTCCACCGGATCCGCGCTTTTCTCTCTCCACC | 35 |
| Fut8 F | ACGTGGTTCGTTGACAGACA | 20 |
| Fut8 R | TACTGTGCATGGGCTTGAGG | 20 |
| Unc13c F | GACTGCCTACACCCCTGTTC | 20 |
| Unc13c R | CCCGCAGTTGTTGGATGTTG | 20 |
| Cdk1 F | CCGGTTGACATCTGGAGCAT | 20 |
| Cdk1 R | TAAACGCCACGATCTTCCCC | 20 |
| LOC100359539 F | GCAGCCCCATTCAGAGTCTT | 20 |
| LOC100359539 R | GTGAACTACCCCAGGGAACG | 20 |
| Hspa2 F | CCATCGCTTATGGCCTGGAT | 20 |
| Hspa2 R | TGCTGGAGGGATCCCAGTTA | 20 |
| Gapdh F | CGTCCCGTAGACAAAATGGTGAA | 23 |
| Gapdh R | GCCGTGAGTGGAGTCATACTGGAACA | 26 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saro, A.; Gao, Z.; Kambey, P.A.; Li, M.; Huang, J. HIV-1 gp120 Protein Activates Cyclin-Dependent Kinase 1, a Possible Link to Central Nervous System Cell Death. Viruses 2022, 14, 2793. https://doi.org/10.3390/v14122793
Saro A, Gao Z, Kambey PA, Li M, Huang J. HIV-1 gp120 Protein Activates Cyclin-Dependent Kinase 1, a Possible Link to Central Nervous System Cell Death. Viruses. 2022; 14(12):2793. https://doi.org/10.3390/v14122793
Chicago/Turabian StyleSaro, Adonira, Zhaolin Gao, Piniel Alphayo Kambey, Min Li, and Jufang Huang. 2022. "HIV-1 gp120 Protein Activates Cyclin-Dependent Kinase 1, a Possible Link to Central Nervous System Cell Death" Viruses 14, no. 12: 2793. https://doi.org/10.3390/v14122793
APA StyleSaro, A., Gao, Z., Kambey, P. A., Li, M., & Huang, J. (2022). HIV-1 gp120 Protein Activates Cyclin-Dependent Kinase 1, a Possible Link to Central Nervous System Cell Death. Viruses, 14(12), 2793. https://doi.org/10.3390/v14122793






