Global Infection Rate of Rotavirus C during 1980–2022 and Analysis of Critical Factors in the Host Range Restriction of Virus VP4
Abstract
:1. Introduction
2. Materials and Methods
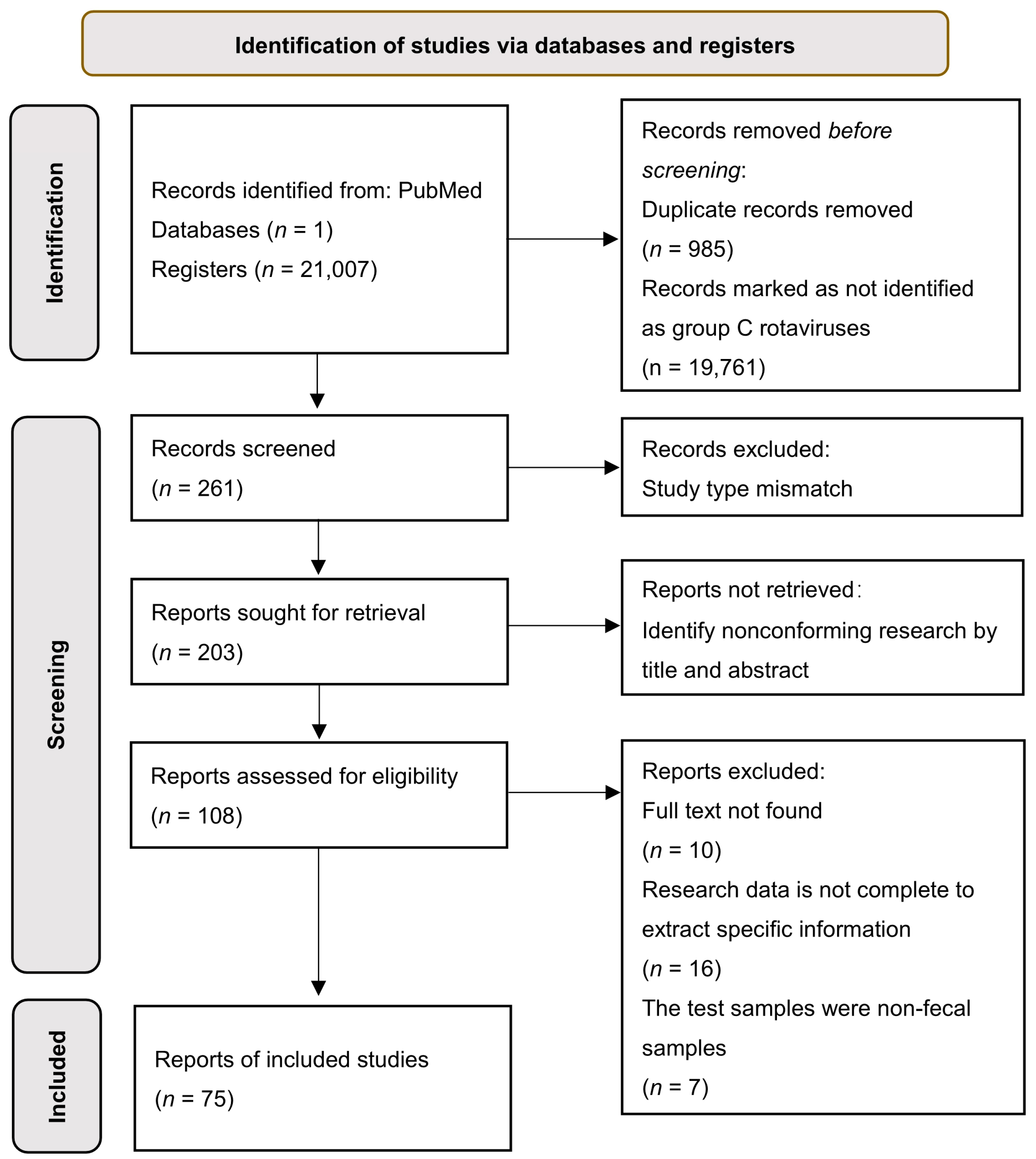
2.1. Systematic Review of Literature Retrieval Strategies and Selection Criteria
2.2. VP4 Gene Retrieval and Sequence Recombination Detection among RVC
2.3. Phylogenetic Tree Construction of VP4 Gene Sequences of RVC and Identification of VP4 Genotypes among Hosts
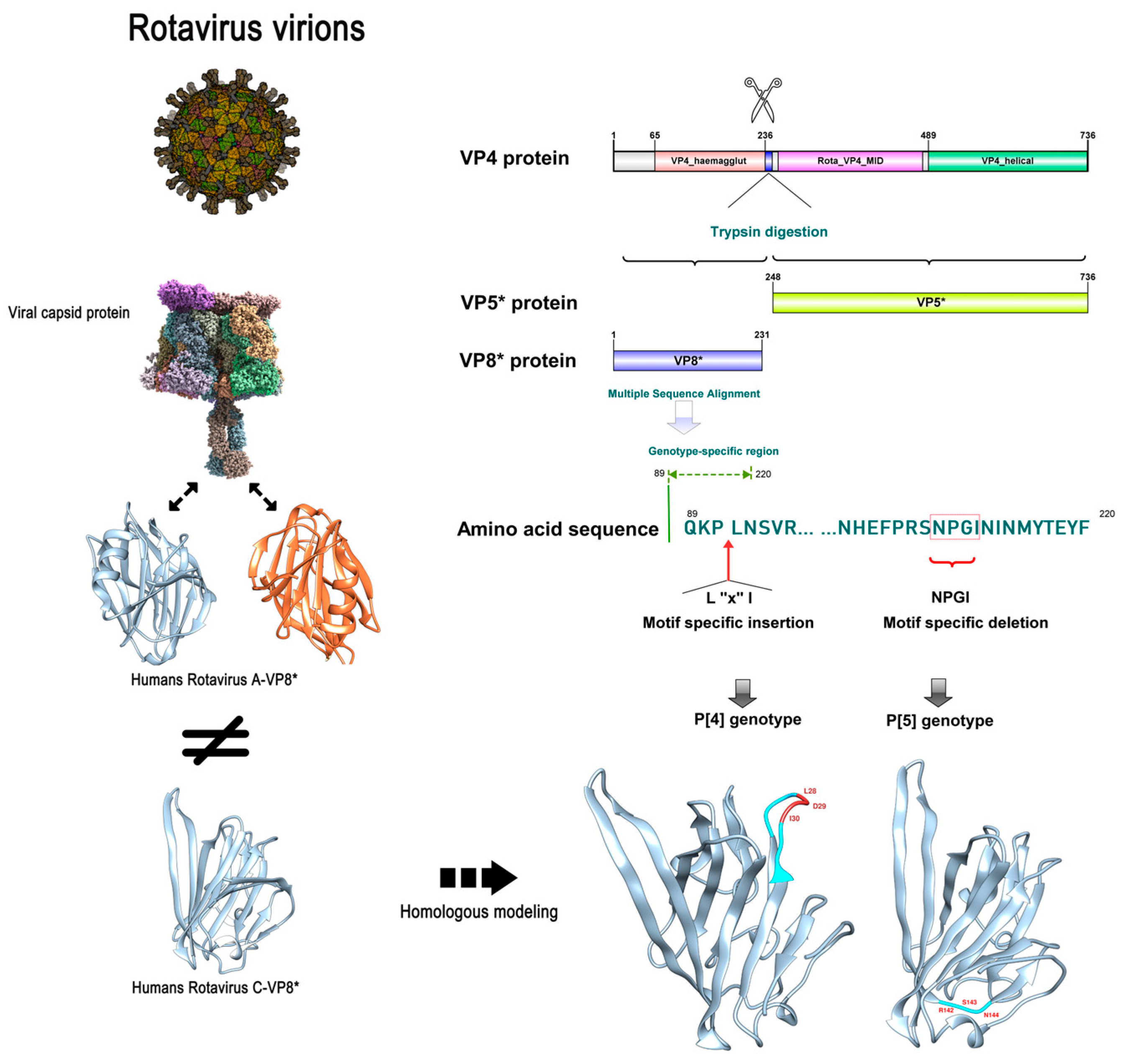
2.4. Amino Acid Sequence Alignment and Specific 3D Structure of RVC Capsid Protein VP4
2.5. Prediction of B-Cell Epitopes of RVC VP4
3. Results
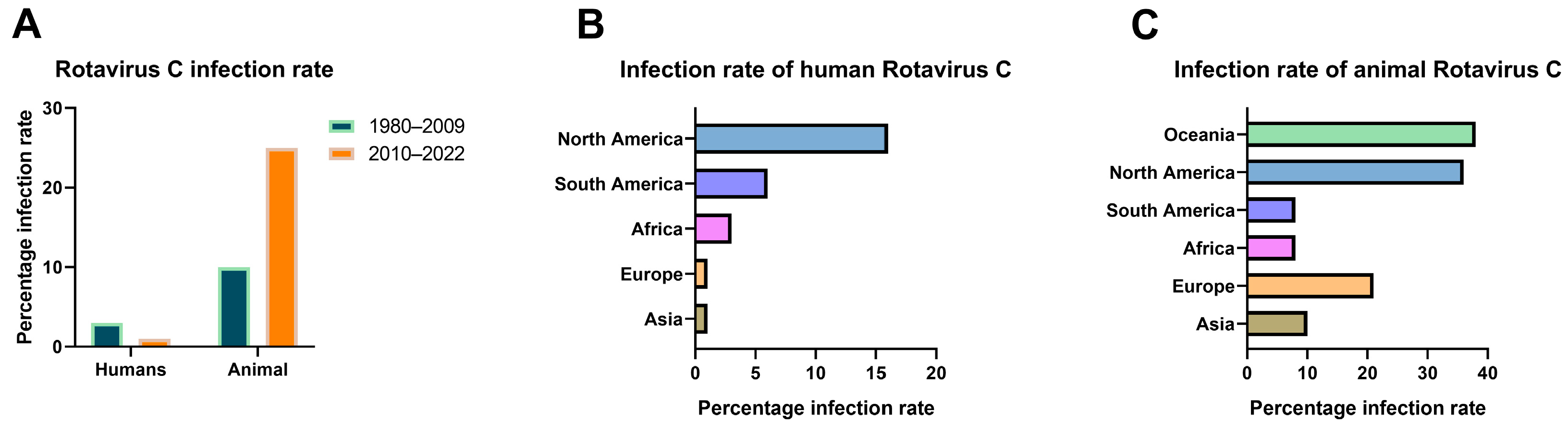
3.1. Current Status of RVC Infections Worldwide
3.2. Evolution of RVC VP4 and the Distribution of P Genotypes
3.3. Specificity Differences of RVC VP4 between Human P [2] and Swine P [4]/P [5]
3.4. Specific Differences in Swine P [4]/P [5] Genotypes Potentially Affecting Viral Invasion into Human Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Caddy, S.; Papa, G.; Borodavka, A.; Desselberger, U. Rotavirus research: 2014–2020. Virus Res. 2021, 304, 198499. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Chen, C.; Zhang, X.; Yan, D.; Jiang, D.; Liu, X.; Yang, M.; Ding, C.; Lan, L.; Hecht, R.; et al. Global burden and trends of rotavirus infection-associated deaths from 1990 to 2019: An observational trend study. Virol. J. 2022, 19, 166. [Google Scholar] [CrossRef] [PubMed]
- Bhuinya, A.; Dass, D.; Banerjee, A.; Mukherjee, A. A tale of antiviral counterattacks in rotavirus infection. Microbiol. Res. 2022, 260, 127046. [Google Scholar] [CrossRef] [PubMed]
- Benedicto-Matambo, P.; Bines, J.E.; Malamba-Banda, C.; Shawa, I.T.; Barnes, K.; Kamng’ona, A.W.; Hungerford, D.; Jambo, K.C.; Iturriza-Gomara, M.; Cunliffe, N.A.; et al. Leveraging Beneficial Off-Target Effects of Live-Attenuated Rotavirus Vaccines. Vaccines 2022, 10, 418. [Google Scholar] [CrossRef]
- Adams, W.R.; Kraft, L.M. Epizootic diarrhea of infant mice: Indentification of the etiologic agent. Science 1963, 141, 359–360. [Google Scholar] [CrossRef]
- Bishop, R.F.; Davidson, G.P.; Holmes, I.H.; Ruck, B.J. Virus particles in epithelial cells of duodenal mucosa from children with acute non-bacterial gastroenteritis. Lancet 1973, 2, 1281–1283. [Google Scholar] [CrossRef]
- Johne, R.; Tausch, S.H.; Grützke, J.; Falkenhagen, A.; Patzina-Mehling, C.; Beer, M.; Höper, D.; Ulrich, R.G. Distantly Related Rotaviruses in Common Shrews, Germany, 2004–2014. Emerg. Infect. Dis. 2019, 25, 2310–2314. [Google Scholar] [CrossRef]
- Sadiq, A.; Bostan, N.; Jadoon, K.; Aziz, A. Effect of rotavirus genetic diversity on vaccine impact. Rev. Med. Virol. 2022, 32, e2259. [Google Scholar] [CrossRef]
- Fischer, T.K.; Gentsch, J.R. Rotavirus typing methods and algorithms. Rev. Med. Virol. 2004, 14, 71–82. [Google Scholar] [CrossRef]
- Da Silva Medeiros, T.N.; Lorenzetti, E.; Alfieri, A.F.; Alfieri, A.A. Phylogenetic analysis of a G6P [5] bovine rotavirus strain isolated in a neonatal diarrhea outbreak in a beef cattle herd vaccinated with G6P [1] and G10P [11] genotypes. Arch. Virol. 2015, 160, 447–451. [Google Scholar] [CrossRef]
- Burke, R.M.; Tate, J.E.; Parashar, U.D. Global Experience with Rotavirus Vaccines. J. Infect. Dis. 2021, 224 (Suppl. S2), S792–S800. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Ciarlet, M.; Heiman, E.; Arijs, I.; Delbeke, T.; McDonald, S.M.; Palombo, E.A.; Iturriza-Gómara, M.; Maes, P.; Patton, J.T.; et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 2008, 82, 3204–3219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suzuki, T.; Hasebe, A. A provisional complete genome-based genotyping system for rotavirus species C from terrestrial mammals. J. Gen. Virol. 2017, 98, 2647–2662. [Google Scholar] [CrossRef] [PubMed]
- Ohseto, M. Epidemiological study of group C rotavirus. Kansenshogaku zasshi. J. Jpn. Assoc. Infect. Dis. 1990, 64, 1264–1274. [Google Scholar] [CrossRef] [Green Version]
- Trovão, N.S.; Shepherd, F.K.; Herzberg, K.; Jarvis, M.C.; Lam, H.C.; Rovira, A.; Culhane, M.R.; Nelson, M.I.; Marthaler, D.G. Evolution of rotavirus C in humans and several domestic animal species. Zoonoses Public Health 2019, 66, 546–557. [Google Scholar] [CrossRef]
- Saif, L.J.; Bohl, E.H.; Theil, K.W.; Cross, R.F.; House, J.A. Rotavirus-like, calicivirus-like, and 23-nm virus-like particles associated with diarrhea in young pigs. J. Clin. Microbiol. 1980, 12, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Soma, J.; Tsunemitsu, H.; Miyamoto, T.; Suzuki, G.; Sasaki, T.; Suzuki, T. Whole-genome analysis of two bovine rotavirus C strains: Shintoku and Toyama. J. Gen. Virol. 2013, 94 Pt 1, 128–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhirakovskaia, E.; Tikunov, A.; Klemesheva, V.; Loginovskikh, N.; Netesov, S.; Tikunova, N. First genetic characterization of rotavirus C in Russia. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2016, 39, 1–8. [Google Scholar] [CrossRef]
- Roczo-Farkas, S.; Dunlop, R.H.; Donato, C.M.; Kirkwood, C.D.; McOrist, S. Rotavirus group C infections in neonatal and grower pigs in Australia. Vet. Rec. 2021, 188, e296. [Google Scholar] [CrossRef]
- Bhat, S.; Kattoor, J.J.; Malik, Y.S.; Sircar, S.; Deol, P.; Rawat, V.; Rakholia, R.; Ghosh, S.; Vlasova, A.N.; Nadia, T.; et al. Species C Rotaviruses in Children with Diarrhea in India, 2010–2013: A Potentially Neglected Cause of Acute Gastroenteritis. Pathogens 2018, 7, 23. [Google Scholar] [CrossRef]
- Rodger, S.M.; Bishop, R.F.; Holmes, I.H. Detection of a rotavirus-like agent associated with diarrhea in an infant. J. Clin. Microbiol. 1982, 16, 724–726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandary, M.B.; Masomian, M.; Poh, C.L. Impact of RNA Virus Evolution on Quasispecies Formation and Virulence. Int. J. Mol. Sci. 2019, 20, 4657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oki, H.; Masuda, T.; Hayashi-Miyamoto, M.; Kawai, M.; Ito, M.; Madarame, H.; Fukase, Y.; Takemae, H.; Sakaguchi, S.; Furuya, T.; et al. Genomic diversity and intragenic recombination of species C rotaviruses. J. Gen. Virol. 2022, 103, 001703. [Google Scholar] [CrossRef] [PubMed]
- Ijaz, M.K.; Sabara, M.I.; Alkarmi, T.; Frenchick, P.J.; Ready, K.F.; Dar, F.K.; Babiuk, L.A. Molecular determinants of rotavirus virulence: Localization of a potential virulence site in a murine rotavirus VP4. Comp. Immunol. Microbiol. Infect. Dis. 1994, 17, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.T.; Diaz, K.; Yang, L.C.; Sharma, A.; Greenberg, H.B.; Smith, J.G. VP4 Is a Determinant of Alpha-Defensin Modulation of Rotaviral Infection. J. Virol. 2022, 96, e0205321. [Google Scholar] [CrossRef]
- Arias, C.F.; Romero, P.; Alvarez, V.; López, S. Trypsin activation pathway of rotavirus infectivity. J. Virol. 1996, 70, 5832–5839. [Google Scholar] [CrossRef] [Green Version]
- Trask, S.D.; Ogden, K.M.; Patton, J.T. Interactions among capsid proteins orchestrate rotavirus particle functions. Curr. Opin. Virol. 2012, 2, 373–379. [Google Scholar] [CrossRef] [Green Version]
- Pang, L.L.; Wang, M.X.; Sun, X.M.; Yuan, Y.; Qing, Y.; Xin, Y.; Zhang, J.Y.; Li, D.D.; Duan, Z.J. Glycan binding patterns of human rotavirus P [10] VP8* protein. Virol. J. 2018, 15, 161. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Huang, P.; Tan, M.; Liu, Y.; Biesiada, J.; Meller, J.; Castello, A.A.; Jiang, B.; Jiang, X. Rotavirus VP8*: Phylogeny, host range, and interaction with histo-blood group antigens. J. Virol. 2012, 86, 9899–9910. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Wang, L.; Qi, J.; Li, D.; Wang, M.; Cong, X.; Peng, R.; Chai, W.; Zhang, Q.; Wang, H.; et al. Human Group C Rotavirus VP8*s Recognize Type A Histo-Blood Group Antigens as Ligands. J. Virol. 2018, 92, e00442-18. [Google Scholar] [CrossRef]
- Hu, L.; Salmen, W.; Sankaran, B.; Lasanajak, Y.; Smith, D.F.; Crawford, S.E.; Estes, M.K.; Prasad, B.V.V. Novel fold of rotavirus glycan-binding domain predicted by AlphaFold2 and determined by X-ray crystallography. Commun. Biol. 2022, 5, 419. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cheng, Z. Cross-Sectional Studies: Strengths, Weaknesses, and Recommendations. Chest 2020, 158, S65–S71. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Settembre, E.C.; Chen, J.Z.; Dormitzer, P.R.; Grigorieff, N.; Harrison, S.C. Atomic model of an infectious rotavirus particle. EMBO J. 2011, 30, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. A Publ. Protein Soc. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Carias, C.; Hu, T.; Chen, Y.T. Burden of rotavirus gastroenteritis on caregivers: Findings from a systematic literature review. Hum. Vaccines Immunother. 2022, 18, 2047545. [Google Scholar] [CrossRef]
- Zhang, L.; Jiang, Z.; Zhou, Z.; Sun, J.; Yan, S.; Gao, W.; Shao, Y.; Bai, Y.; Wu, Y.; Yan, Z.; et al. A TaqMan Probe-Based Multiplex Real-Time PCR for Simultaneous Detection of Porcine Epidemic Diarrhea Virus Subtypes G1 and G2, and Porcine Rotavirus Groups A and C. Viruses 2022, 14, 1819. [Google Scholar] [CrossRef]
- Rajala, E.; Lee, H.S.; Nam, N.H.; Huong, C.T.T.; Son, H.M.; Wieland, B.; Magnusson, U. Skewness in the literature on infectious livestock diseases in an emerging economy—the case of Vietnam. Anim. Health Res. Rev. 2021, 22, 1–13. [Google Scholar] [CrossRef]
- VanderWaal, K.; Deen, J. Global trends in infectious diseases of swine. Proc. Natl. Acad. Sci. USA 2018, 115, 11495–11500. [Google Scholar] [CrossRef] [Green Version]
- Tenthorey, J.L.; Emerman, M.; Malik, H.S. Evolutionary Landscapes of Host-Virus Arms Races. Annu. Rev. Immunol. 2022, 40, 271–294. [Google Scholar] [CrossRef]
- Costa, F.B.; Flores, P.S.; Amorim, A.R.; Mendes, G.D.S.; Santos, N. Porcine rotavirus C strains carrying human-like NSP4 and NSP5. Zoonoses Public Health 2020, 67, 849–861. [Google Scholar] [CrossRef]
- Choudhury, P.R.; Saha, T.; Goel, S.; Shah, J.M.; Ganjewala, D. Cross-species virus transmission and its pandemic potential. Bull. Natl. Res. Cent. 2022, 46, 18. [Google Scholar] [CrossRef] [PubMed]
- Saxena, K.; Blutt, S.E.; Ettayebi, K.; Zeng, X.L.; Broughman, J.R.; Crawford, S.E.; Karandikar, U.C.; Sastri, N.P.; Conner, M.E.; Opekun, A.R.; et al. Human Intestinal Enteroids: A New Model to Study Human Rotavirus Infection, Host Restriction, and Pathophysiology. J. Virol. 2016, 90, 43–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Attar, L.; Dhaliwal, W.; Howard, C.R.; Bridger, J.C. Rotavirus cross-species pathogenicity: Molecular characterization of a bovine rotavirus pathogenic for pigs. Virology 2001, 291, 172–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, K.; Lin, X.D.; Huang, K.Y.; Zhang, B.; Shi, M.; Guo, W.P.; Wang, M.R.; Wang, W.; Xing, J.G.; Li, M.H.; et al. Identification of novel and diverse rotaviruses in rodents and insectivores, and evidence of cross-species transmission into humans. Virology 2016, 494, 168–177. [Google Scholar] [CrossRef]
- Tian, J.; Sun, J.; Li, D.; Wang, N.; Wang, L.; Zhang, C.; Meng, X.; Ji, X.; Suchard, M.A.; Zhang, X.; et al. Emerging viruses: Cross-species transmission of coronaviruses, filoviruses, henipaviruses, and rotaviruses from bats. Cell Rep. 2022, 39, 110969. [Google Scholar] [CrossRef]
- Li, D.; Wang, M.; Qi, J.; Zhang, Q.; Wang, H.; Pang, L.; Sun, X.; Duan, Z. Human group A rotavirus P [25] VP8* specifically binds to A-type histo-blood group antigen. Virology 2021, 555, 56–63. [Google Scholar] [CrossRef]
- Skehel, J.J.; Wiley, D.C. Receptor binding and membrane fusion in virus entry: The influenza hemagglutinin. Annu. Rev. Biochem. 2000, 69, 531–569. [Google Scholar] [CrossRef]
- Böttcher-Friebertshäuser, E.; Garten, W.; Matrosovich, M.; Klenk, H.D. The hemagglutinin: A determinant of pathogenicity. Curr. Top. Microbiol. Immunol. 2014, 385, 3–34. [Google Scholar]
- Gamblin, S.J.; Vachieri, S.G.; Xiong, X.; Zhang, J.; Martin, S.R.; Skehel, J.J. Hemagglutinin Structure and Activities. Cold Spring Harb. Perspect. Med. 2021, 11, a038638. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, S.; Jin, X.; Zang, L.; Liu, Z.; Wen, X.; Ran, X. Global Infection Rate of Rotavirus C during 1980–2022 and Analysis of Critical Factors in the Host Range Restriction of Virus VP4. Viruses 2022, 14, 2826. https://doi.org/10.3390/v14122826
Zhao S, Jin X, Zang L, Liu Z, Wen X, Ran X. Global Infection Rate of Rotavirus C during 1980–2022 and Analysis of Critical Factors in the Host Range Restriction of Virus VP4. Viruses. 2022; 14(12):2826. https://doi.org/10.3390/v14122826
Chicago/Turabian StyleZhao, Simiao, Xinshun Jin, Lingling Zang, Ziwei Liu, Xiaobo Wen, and Xuhua Ran. 2022. "Global Infection Rate of Rotavirus C during 1980–2022 and Analysis of Critical Factors in the Host Range Restriction of Virus VP4" Viruses 14, no. 12: 2826. https://doi.org/10.3390/v14122826





