Induction of Innate Immune Response by TLR3 Agonist Protects Mice against SARS-CoV-2 Infection
Abstract
:1. Introduction
2. Methods
2.1. Cells
2.2. Viruses
2.3. Animal Experiments
2.4. Determination of Viral Load in Organs
2.5. Quantitative Real-Time RT-PCR
2.6. General Histopathology
2.7. SARS-CoV-2 Immunohistochemical Staining
2.8. Histological Evaluation
2.9. Percentage of Air Area Calculation
2.10. Flow Cytometry
3. Results
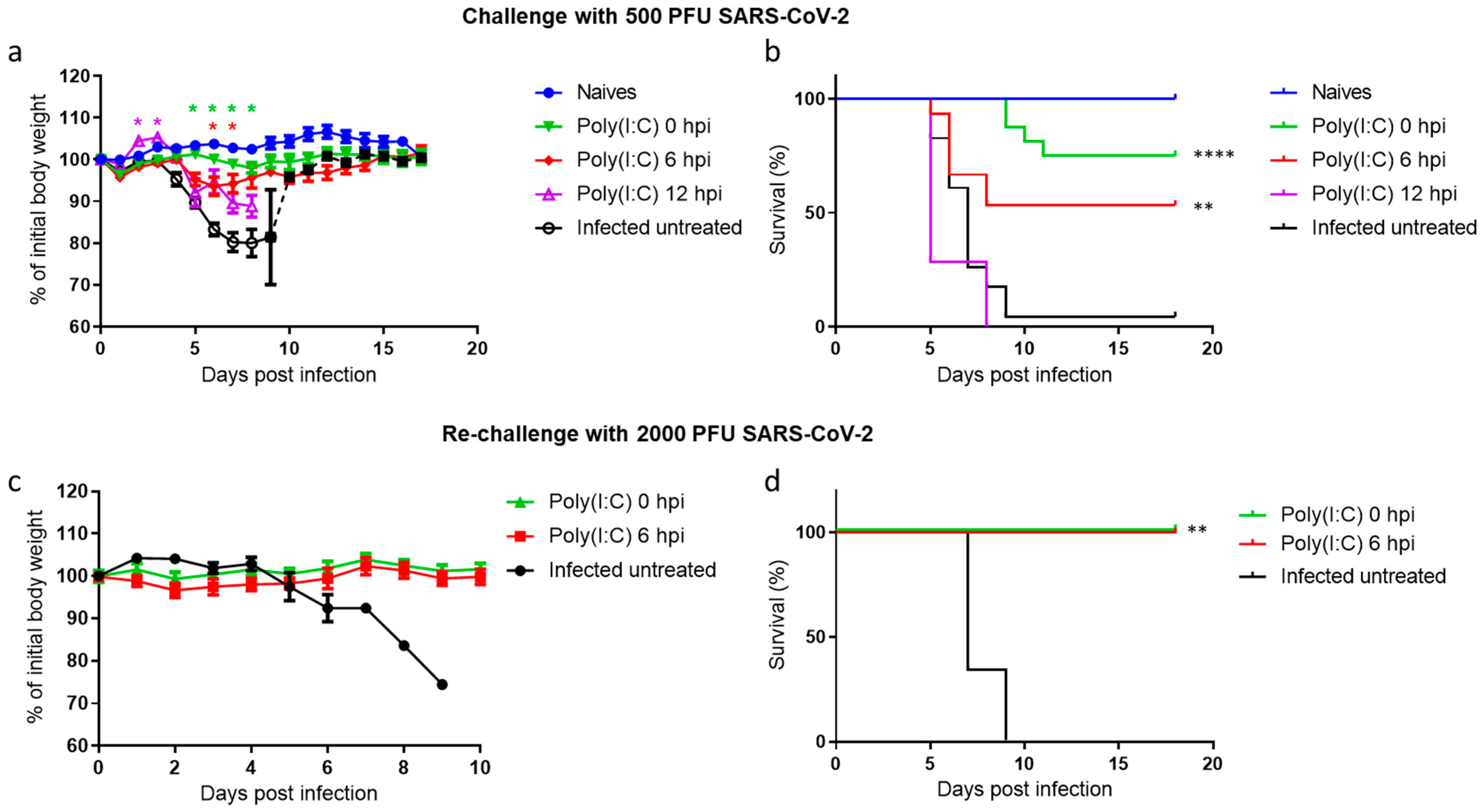
3.1. Poly(I:C) Treatment Protects Mice against a Lethal SARS-CoV-2 Infection
3.2. Poly(I:C) Treatment Reduces Viral Loads in Nasal Turbinates, Lungs and Brains
3.3. Poly(I:C) Alleviates Lung Damage and Reduces SARS-CoV-2 Antigen in Both Lungs and Brains
3.4. Effect of Poly(I:C) Treatment on the Innate Immune Response Following SARS-CoV-2 Infection
3.5. Increased Levels of Alveolar Macrophages and NK Cells in the Lungs Following Poly(I:C) Treatment in SARS-CoV-2 Infected Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Twomey, J.D.; Luo, S.; Dean, A.Q.; Bozza, W.P.; Nalli, A.; Zhang, B. COVID-19 update: The race to therapeutic development. Drug Resist. Updates 2020, 53, 100733. [Google Scholar] [CrossRef]
- Deb, P.; Molla, M.M.A.; Saif-Ur-Rahman, K.M. An update to monoclonal antibody as therapeutic option against COVID-19. Biosaf. Health 2021, 3, 87–91. [Google Scholar] [CrossRef]
- Makdasi, E.; Levy, Y.; Alcalay, R.; Noy-Porat, T.; Zahavy, E.; Mechaly, A.; Epstein, E.; Peretz, E.; Cohen, H.; Bar-On, L.; et al. Neutralizing Monoclonal Anti-SARS-CoV-2 Antibodies Isolated from Immunized Rabbits Define Novel Vulnerable Spike-Protein Epitope. Viruses 2021, 13, 566. [Google Scholar] [CrossRef]
- Noy-Porat, T.; Mechaly, A.; Levy, Y.; Makdasi, E.; Alcalay, R.; Gur, D.; Aftalion, M.; Falach, R.; Ben-Arye, S.L.; Lazar, S.; et al. Therapeutic antibodies, targeting the SARS-CoV-2 spike N-terminal domain, protect lethally infected K18-hACE2 mice. iScience 2021, 24, 102479. [Google Scholar] [CrossRef]
- Rosenfeld, R.; Noy-Porat, T.; Mechaly, A.; Makdasi, E.; Levy, Y.; Alcalay, R.; Falach, R.; Aftalion, M.; Epstein, E.; Gur, D.; et al. Post-exposure protection of SARS-CoV-2 lethal infected K18-hACE2 transgenic mice by neutralizing human monoclonal antibody. Nat. Commun. 2021, 12, 944. [Google Scholar] [CrossRef] [PubMed]
- Musarrat, F.; Chouljenko, V.; Dahal, A.; Nabi, R.; Chouljenko, T.; Jois, S.D.; Kousoulas, K.G. The anti-HIV drug nelfinavir mesylate (Viracept) is a potent inhibitor of cell fusion caused by the SARSCoV-2 spike (S) glycoprotein warranting further evaluation as an antiviral against COVID-19 infections. J. Med. Virol. 2020, 92, 2087–2095. [Google Scholar] [CrossRef]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antivir. Res. 2020, 178, 104787. [Google Scholar] [CrossRef] [PubMed]
- Fischer, W.A.; Eron, J.J.; Holman, W.; Cohen, M.S.; Fang, L.; Szewczyk, L.J.; Sheahan, T.P.; Baric, R.S.; Mollan, K.R.; Wolfe, C.R.; et al. Molnupiravir, an Oral Antiviral Treatment for COVID-19. medRxiv 2021. medRxiv:2021.06.17.21258639. [Google Scholar]
- Bhaskar, S.; Sinha, A.; Banach, M.; Mittoo, S.; Weissert, R.; Kass, J.S.; Rajagopal, S.; Pai, A.R.; Kutty, S. Cytokine Storm in COVID-19-Immunopathological Mechanisms, Clinical Considerations, and Therapeutic Approaches: The REPROGRAM Consortium Position Paper. Front. Immunol. 2020, 11, 1648. [Google Scholar] [CrossRef] [PubMed]
- Yong, H.Y.; Luo, D. RIG-I-Like Receptors as Novel Targets for Pan-Antivirals and Vaccine Adjuvants Against Emerging and Re-Emerging Viral Infections. Front. Immunol. 2018, 9, 1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lester, S.N.; Li, K. Toll-like receptors in antiviral innate immunity. J. Mol. Biol. 2014, 426, 1246–1264. [Google Scholar] [CrossRef]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef] [PubMed]
- Lowery, S.A.; Sariol, A.; Perlman, S. Innate immune and inflammatory responses to SARS-CoV-2: Implications for COVID-19. Cell Host Microbe 2021, 29, 1052–1062. [Google Scholar] [CrossRef]
- Finkel, Y.; Gluck, A.; Nachshon, A.; Winkler, R.; Fisher, T.; Rozman, B.; Mizrahi, O.; Lubelsky, Y.; Zuckerman, B.; Slobodin, B.; et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature 2021, 594, 240–245. [Google Scholar] [CrossRef]
- Rojas, J.M.; Alejo, A.; Martín, V.; Sevilla, N. Viral pathogen-induced mechanisms to antagonize mammalian interferon (IFN) signaling pathway. Cell Mol. Life Sci. 2021, 78, 1423–1444. [Google Scholar] [CrossRef]
- Grandvaux, N.; Servant, M.J.; Hiscott, J. The interferon antiviral response: From viral invasion to evasion. Curr. Opin. Infect. Dis. 2002, 15, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Darazam, I.A.; Shokouhi, S.; Pourhoseingholi, M.A.; Irvani, S.S.N.; Mokhtari, M.; Shabani, M.; Amirdosara, M.; Torabinavid, P.; Golmohammadi, M.; Hashemi, S.; et al. Role of interferon therapy in severe COVID-19: The COVIFERON randomized controlled trial. Sci. Rep. 2021, 11, 8059. [Google Scholar] [CrossRef]
- Nakhlband, A.; Fakhari, A.; Azizi, H. Interferon-beta offers promising avenues to COVID-19 treatment: A systematic review and meta-analysis of clinical trial studies. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2021, 394, 829–838. [Google Scholar] [CrossRef]
- McCartney, S.; Vermi, W.; Gilfillan, S.; Cella, M.; Murphy, T.L.; Schreiber, R.D.; Murphy, K.M.; Colonna, M. Distinct and complementary functions of MDA5 and TLR3 in poly(I:C)-mediated activation of mouse NK cells. J. Exp. Med. 2009, 206, 2967–2976. [Google Scholar] [CrossRef]
- Zhao, J.; Wohlford-Lenane, C.; Zhao, J.; Fleming, E.; Lane, T.E.; McCray, P.B., Jr.; Perlman, S. Intranasal treatment with poly(I*C) protects aged mice from lethal respiratory virus infections. J. Virol. 2012, 86, 11416–11424. [Google Scholar] [CrossRef] [Green Version]
- Salmon, H.; Idoyaga, J.; Rahman, A.; Leboeuf, M.; Remark, R.; Jordan, S.; Casanova-Acebes, M.; Khudoynazarova, M.; Agudo, J.; Tung, N.; et al. Expansion and Activation of CD103(+) Dendritic Cell Progenitors at the Tumor Site Enhances Tumor Responses to Therapeutic PD-L1 and BRAF Inhibition. Immunity 2016, 44, 924–938. [Google Scholar] [CrossRef] [Green Version]
- Boivin, N.; Sergerie, Y.; Rivest, S.; Boivin, G. Effect of pretreatment with toll-like receptor agonists in a mouse model of herpes simplex virus type 1 encephalitis. J. Infect. Dis. 2008, 198, 664–672. [Google Scholar] [CrossRef]
- Kende, M.; Paragas, J.; Salazar, A.M. The efficacy of poly-ICLC against Ebola-Zaire virus (EBOV) infection in mice and cynomolgus monkeys. Antivir. Res. 2019, 163, 179–184. [Google Scholar] [CrossRef]
- Li, Y.G.; Siripanyaphinyo, U.; Tumkosit, U.; Noranate, N.; Atchareeya, A.; Pan, Y.; Kameoka, M.; Kurosu, T.; Ikuta, K.; Takeda, N.; et al. Poly (I:C), an agonist of toll-like receptor-3, inhibits replication of the Chikungunya virus in BEAS-2B cells. Virol. J. 2012, 9, 114. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Huang, S.; Zhao, X.; Chen, M.; Lin, Y.; Xia, Y.; Sun, C.; Yang, X.; Wang, J.; Guo, Y.; et al. Poly(I:C) treatment leads to interferon-dependent clearance of hepatitis B virus in a hydrodynamic injection mouse model. J. Virol. 2014, 88, 10421–11431. [Google Scholar] [CrossRef] [Green Version]
- Israely, T.; Melamed, S.; Achdout, H.; Erez, N.; Politi, B.; Waner, T.; Lustig, S.; Paran, N. TLR3 and TLR9 agonists improve postexposure vaccination efficacy of live smallpox vaccines. PLoS ONE 2014, 9, e110545. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.Y.; Stuart, C.; Takeda, K.; D’Agnillo, F.; Golding, B. Poly(I:C) Induces Human Lung Endothelial Barrier Dysfunction by Disrupting Tight Junction Expression of Claudin-5. PLoS ONE 2016, 11, e0160875. [Google Scholar] [CrossRef]
- National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals, 8th ed.; National Academies Press: Washington, DC, USA, 2011. Available online: https://www.ncbi.nlm.nih.gov/books/NBK54050/ (accessed on 12 January 2022). [CrossRef]
- Kumaki, Y.; Salazar, A.M.; Wandersee, M.K.; Barnard, D.L. Prophylactic and therapeutic intranasal administration with an immunomodulator, Hiltonol((R)) (Poly IC:LC), in a lethal SARS-CoV-infected BALB/c mouse model. Antivir. Res. 2017, 139, 1–12. [Google Scholar] [CrossRef]
- Starkhammar, M.; Kumlien Georén, S.; Swedin, L.; Dahlén, S.E.; Adner, M.; Cardell, L.O. Intranasal administration of poly(I:C) and LPS in BALB/c mice induces airway hyperresponsiveness and inflammation via different pathways. PLoS ONE 2012, 7, e32110. [Google Scholar] [CrossRef]
- Yahalom-Ronen, Y.; Tamir, H.; Melamed, S.; Politi, B.; Shifman, O.; Achdout, H.; Vitner, E.B.; Israeli, O.; Milrot, E.; Stein, D.; et al. A single dose of recombinant VSV-G-spike vaccine provides protection against SARS-CoV-2 challenge. Nat. Commun. 2020, 11, 6402. [Google Scholar] [CrossRef]
- Israely, T.; Paran, N.; Erez, N.; Cherry, L.; Tamir, H.; Achdout, H.; Politi, B.; Israeli, O.; Zaide, G.; Cohen-Gihon, I.; et al. Differential Response Following Infection of Mouse CNS with Virulent and Attenuated Vaccinia Virus Strains. Vaccines 2019, 7, 19. [Google Scholar] [CrossRef] [Green Version]
- Schafer, K.A.; Eighmy, J.; Fikes, J.D.; Halpern, W.G.; Hukkanen, R.R.; Long, G.G.; Meseck, E.K.; Patrick, D.J.; Thibodeau, M.S.; Wood, C.E.; et al. Use of Severity Grades to Characterize Histopathologic Changes. Toxicol. Pathol. 2018, 46, 256–265. [Google Scholar] [CrossRef]
- Winkler, E.S.; Chen, R.E.; Alam, F.; Yildiz, S.; Case, J.B.; Uccellini, M.B.; Holtzman, M.J.; Garcia-Sastre, A.; Schotsaert, M.; Diamond, M.S. SARS-CoV-2 causes lung infection without severe disease in human ACE2 knock-in mice. J. Virol. 2021, 96, JVI0151121. [Google Scholar] [CrossRef]
- Oladunni, F.S.; Park, J.G.; Pino, P.A.; Gonzalez, O.; Akhter, A.; Allué-Guardia, A.; Olmo-Fontánez, A.; Gautam, S.; Garcia-Vilanova, A.; Ye, C.; et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020, 11, 6122. [Google Scholar] [CrossRef]
- Winkler, E.S.; Bailey, A.L.; Kafai, N.M.; Nair, S.; McCune, B.T.; Yu, J.; Fox, J.M.; Chen, R.E.; Earnest, J.T.; Keeler, S.P.; et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020, 21, 1327–1335. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.Q.; Huang, M.; Sun, X.; Deng, F.; Wang, H.; Ning, Y.J. Immune evasion of SARS-CoV-2 from interferon antiviral system. Comput. Struct. Biotechnol. J. 2021, 19, 4217–4225. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, I.; Paakinaho, V.; Baek, S.; Sung, M.H.; Hager, G.L. Synergistic gene expression during the acute phase response is characterized by transcription factor assisted loading. Nat. Commun. 2017, 8, 1849. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Riva, L.; Pu, Y.; Martin-Sancho, L.; Kanamune, J.; Yamamoto, Y.; Sakai, K.; Gotoh, S.; Miorin, L.; De Jesus, P.D.; et al. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021, 34, 108628. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Zhuang, M.W.; Han, L.; Zhang, J.; Nan, M.L.; Zhan, P.; Kang, D.; Liu, X.; Gao, C.; Wang, P.H. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target. Ther. 2020, 5, 299. [Google Scholar] [CrossRef]
- Yang, Z.; Zhang, X.; Wang, F.; Wang, P.; Li, X.; Kuang, E. Suppression of MDA5-mediated antiviral immune responses by NSP8 of SARS-CoV-2. bioRxiv 2020. bioRxiv:2020.08.12.247767. [Google Scholar]
- Beyer, D.K.; Forero, A. Mechanisms of Antiviral Immune Evasion of SARS-CoV-2. J. Mol. Biol. 2021, 10, 167265. [Google Scholar] [CrossRef]
- Liu, G.; Lee, J.H.; Parker, Z.M.; Acharya, D.; Chiang, J.J.; van Gent, M.; Riedl, W.; Davis-Gardner, M.E.; Wies, E.; Chiang, C.; et al. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat. Microbiol. 2021, 6, 467–478. [Google Scholar] [CrossRef]
- Humphries, F.; Shmuel-Galia, L.; Jiang, Z.; Wilson, R.; Landis, P.; Ng, S.L.; Parsi, K.M.; Maehr, R.; Cruz, J.; Morales, A.; et al. A diamidobenzimidazole STING agonist protects against SARS-CoV-2 infection. Sci. Immunol. 2021, 6, eabi9002. [Google Scholar] [CrossRef] [PubMed]
- Croci, S.; Venneri, M.A.; Mantovani, S.; Fallerini, C.; Benetti, E.; Picchiotti, N.; Campolo, F.; Imperatore, F.; Palmieri, M.; Daga, S.; et al. The polymorphism L412F in TLR3 inhibits autophagy and is a marker of severe COVID-19 in males. medRxiv 2021. medRxiv:2021.03.23.21254158. [Google Scholar] [CrossRef]
- Loske, J.; Röhmel, J.; Lukassen, S.; Stricker, S.; Magalhães, V.G.; Liebig, J.; Chua, R.L.; Thürmann, L.; Messingschlager, M.; Seegebarth, A.; et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat. Biotechnol. 2021, 1–6. [Google Scholar] [CrossRef]
- Thompson, M.R.; Kaminski, J.J.; Kurt-Jones, E.A.; Fitzgerald, K.A. Pattern recognition receptors and the innate immune response to viral infection. Viruses 2011, 3, 920–940. [Google Scholar] [CrossRef] [Green Version]
- Zheng, M.; Karki, R.; Williams, E.P.; Yang, D.; Fitzpatrick, E.; Vogel, P.; Jonsson, C.B.; Kanneganti, T.D. TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 2021, 22, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Trinchieri, G.; Sher, A. Cooperation of Toll-like receptor signals in innate immune defence. Nat. Rev. Immunol. 2007, 7, 179–190. [Google Scholar] [CrossRef]
- Pashenkov, M.V.; Murugina, N.E.; Budikhina, A.S.; Pinegin, B.V. Synergistic interactions between NOD receptors and TLRs: Mechanisms and clinical implications. J. Leukoc. Biol. 2019, 105, 669–680. [Google Scholar] [CrossRef]
- Root-Bernstein, R. Innate Receptor Activation Patterns Involving TLR and NLR Synergisms in COVID-19, ALI/ARDS and Sepsis Cytokine Storms: A Review and Model Making Novel Predictions and Therapeutic Suggestions. Int. J. Mol. Sci. 2021, 22, 2108. [Google Scholar] [CrossRef] [PubMed]
- Copaescu, A.; Smibert, O.; Gibson, A.; Philips, E.J.; Trubiano, J.A. The role of IL-6 and other mediators in the cytokine storm associated with SARS-CoV-2 infection. J. Allergy Clin. Immunol. 2020, 146, 518–534. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Pan, M.; Xiao, Z.; Xu, X. Neurological manifestations of the coronavirus (SARS-CoV-2) pandemic 2019–2020. J. Neurol. Neurosurg. Psychiatry 2020, 91, 669–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaw, A.C.; Goldstein, D.R.; Montgomery, R.R. Age-dependent dysregulation of innate immunity. Nat. Rev. Immunol. 2013, 13, 875–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tamir, H.; Melamed, S.; Erez, N.; Politi, B.; Yahalom-Ronen, Y.; Achdout, H.; Lazar, S.; Gutman, H.; Avraham, R.; Weiss, S.; et al. Induction of Innate Immune Response by TLR3 Agonist Protects Mice against SARS-CoV-2 Infection. Viruses 2022, 14, 189. https://doi.org/10.3390/v14020189
Tamir H, Melamed S, Erez N, Politi B, Yahalom-Ronen Y, Achdout H, Lazar S, Gutman H, Avraham R, Weiss S, et al. Induction of Innate Immune Response by TLR3 Agonist Protects Mice against SARS-CoV-2 Infection. Viruses. 2022; 14(2):189. https://doi.org/10.3390/v14020189
Chicago/Turabian StyleTamir, Hadas, Sharon Melamed, Noam Erez, Boaz Politi, Yfat Yahalom-Ronen, Hagit Achdout, Shlomi Lazar, Hila Gutman, Roy Avraham, Shay Weiss, and et al. 2022. "Induction of Innate Immune Response by TLR3 Agonist Protects Mice against SARS-CoV-2 Infection" Viruses 14, no. 2: 189. https://doi.org/10.3390/v14020189
APA StyleTamir, H., Melamed, S., Erez, N., Politi, B., Yahalom-Ronen, Y., Achdout, H., Lazar, S., Gutman, H., Avraham, R., Weiss, S., Paran, N., & Israely, T. (2022). Induction of Innate Immune Response by TLR3 Agonist Protects Mice against SARS-CoV-2 Infection. Viruses, 14(2), 189. https://doi.org/10.3390/v14020189





