Spike Gene Evolution and Immune Escape Mutations in Patients with Mild or Moderate Forms of COVID-19 and Treated with Monoclonal Antibodies Therapies
Abstract
:1. Background
2. Methods
Ethics Statement
3. Results
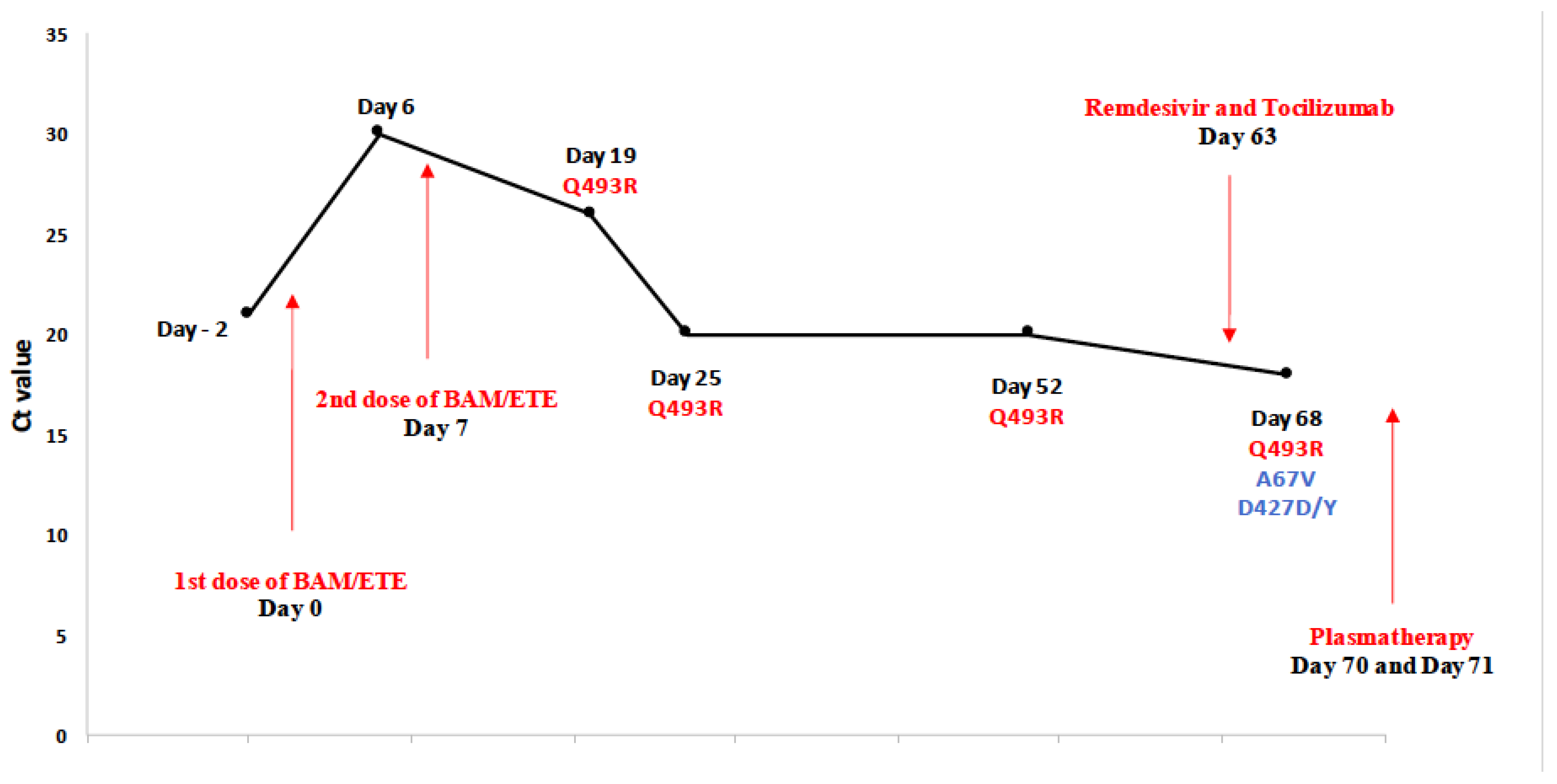
Description of Patient P5
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical Course and Risk Factors for Mortality of Adult Inpatients with COVID-19 in Wuhan, China: A Retrospective Cohort Study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Rabinowich, L.; Grupper, A.; Baruch, R.; Ben-Yehoyada, M.; Halperin, T.; Turner, D.; Katchman, E.; Levi, S.; Houri, I.; Lubezky, N.; et al. Low Immunogenicity to SARS-CoV-2 Vaccination among Liver Transplant Recipients. J. Hepatol. 2021, 75, 435–438. [Google Scholar] [CrossRef] [PubMed]
- Mazzola, A.; Todesco, E.; Drouin, S.; Hazan, F.; Marot, S.; Thabut, D.; Varnous, S.; Soulié, C.; Barrou, B.; Marcelin, A.-G.; et al. Poor Antibody Response after Two Doses of SARS-CoV-2 Vaccine in Transplant Recipients. Clin. Infect. Dis. 2021, ciab580. [Google Scholar] [CrossRef] [PubMed]
- Beigel, J.H.; Tomashek, K.M.; Dodd, L.E.; Mehta, A.K.; Zingman, B.S.; Kalil, A.C.; Hohmann, E.; Chu, H.Y.; Luetkemeyer, A.; Kline, S.; et al. Remdesivir for the Treatment of COVID-19—Final Report. N. Engl. J. Med. 2020, 383, 1813–1826. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, D.; Du, G.; Du, R.; Zhao, J.; Jin, Y.; Fu, S.; Gao, L.; Cheng, Z.; Lu, Q.; et al. Remdesivir in Adults with Severe COVID-19: A Randomised, Double-Blind, Placebo-Controlled, Multicentre Trial. Lancet 2020, 395, 1569–1578. [Google Scholar] [CrossRef]
- Salama, C.; Han, J.; Yau, L.; Reiss, W.G.; Kramer, B.; Neidhart, J.D.; Criner, G.J.; Kaplan-Lewis, E.; Baden, R.; Pandit, L.; et al. Tocilizumab in Patients Hospitalized with COVID-19 Pneumonia. N. Engl. J. Med. 2021, 384, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Joyner, M.J.; Carter, R.E.; Senefeld, J.W.; Klassen, S.A.; Mills, J.R.; Johnson, P.W.; Theel, E.S.; Wiggins, C.C.; Bruno, K.A.; Klompas, A.M.; et al. Convalescent Plasma Antibody Levels and the Risk of Death from COVID-19. N. Engl. J. Med. 2021, 384, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Dougan, M.; Nirula, A.; Azizad, M.; Mocherla, B.; Gottlieb, R.L.; Chen, P.; Hebert, C.; Perry, R.; Boscia, J.; Heller, B.; et al. Bamlanivimab plus Etesevimab in Mild or Moderate COVID-19. N. Engl. J. Med. 2021, 385, 1382–1392. [Google Scholar] [CrossRef] [PubMed]
- Jensen, B.; Luebke, N.; Feldt, T.; Keitel, V.; Brandenburger, T.; Kindgen-Milles, D.; Lutterbeck, M.; Freise, N.F.; Schoeler, D.; Haas, R.; et al. Emergence of the E484K Mutation in SARS-CoV-2-Infected Immunocompromised Patients Treated with Bamlanivimab in Germany. Lancet Reg. Health Eur. 2021, 8, 100164. [Google Scholar] [CrossRef] [PubMed]
- Guigon, A.; Faure, E.; Lemaire, C.; Chopin, M.-C.; Tinez, C.; Assaf, A.; Lazrek, M.; Hober, D.; Bocket, L.; Engelmann, I.; et al. Emergence of Q493R Mutation in SARS-CoV-2 Spike Protein during Bamlanivimab/Etesevimab Treatment and Resistance to Viral Clearance. J. Infect. 2021. [Google Scholar] [CrossRef] [PubMed]
- Focosi, D.; Novazzi, F.; Genoni, A.; Dentali, F.; Gasperina, D.D.; Baj, A.; Maggi, F. Emergence of SARS-CoV-2 Spike Protein Escape Mutation Q493R after Treatment for COVID-19. Emerg. Infect. Dis. 2021, 27, 2728–2731. [Google Scholar] [CrossRef] [PubMed]
- Pommeret, F.; Colomba, J.; Bigenwald, C.; Laparra, A.; Bockel, S.; Bayle, A.; Michot, J.-M.; Hueso, T.; Albiges, L.; Tiberghien, P.; et al. Bamlanivimab+ Etesevimab Therapy Induces SARS-CoV-2 Immune Escape Mutations and Secondary Clinical Deterioration in COVID-19 Patients with B-Cell Malignancies. Ann. Oncol. 2021, 32, 1445–1447. [Google Scholar] [CrossRef] [PubMed]
- Vellas, C.; Del Bello, A.; Debard, A.; Steinmeyer, Z.; Tribaudeau, L.; Ranger, N.; Jeanne, N.; Martin-Blondel, G.; Delobel, P.; Kamar, N.; et al. Influence of Treatment with Neutralizing Monoclonal Antibodies on the SARS-CoV-2 Nasopharyngeal Load and Quasispecies. Clin. Microbiol. Infect. 2021. [Google Scholar] [CrossRef] [PubMed]
- Weisblum, Y.; Schmidt, F.; Zhang, F.; DaSilva, J.; Poston, D.; Lorenzi, J.C.; Muecksch, F.; Rutkowska, M.; Hoffmann, H.-H.; Michailidis, E.; et al. Escape from Neutralizing Antibodies by SARS-CoV-2 Spike Protein Variants. eLife 2020, 9, e61312. [Google Scholar] [CrossRef] [PubMed]
- Baum, A.; Fulton, B.O.; Wloga, E.; Copin, R.; Pascal, K.E.; Russo, V.; Giordano, S.; Lanza, K.; Negron, N.; Ni, M.; et al. Antibody Cocktail to SARS-CoV-2 Spike Protein Prevents Rapid Mutational Escape Seen with Individual Antibodies. Science 2020, 369, 1014–1018. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, M. Omicron Overpowers Key COVID Antibody Treatments in Early Tests. Nature 2021. [Google Scholar] [CrossRef] [PubMed]
| Monoclonal Antibodies | |||||||
|---|---|---|---|---|---|---|---|
| Risk Factors of Severe COVID-19 Form | mABS | Time of Sampling | Ct | Nextstrain Clade | Mutation of the Spike Gene | ||
| P1 | Heart transplantation Corticosteroid-induced diabetes | BAM | D0 | 17 | 20B (B1.214.2) | Ins213TDR (ACAGATCGA), Q414K, N450K, D614G, T716I | |
| D4 | 20 | 20B (B1.214.2) | Ins213TDR (ACAGATCGA), Q414K, N450K, D614G, T716I | ||||
| D8 | 29 | - | Missing sample | ||||
| D9 | 30 | - | Low viral load | ||||
| P2 | Necrotizing myopathy | BAM | Pre-administration | 33 | - | Low viral load | |
| D0 | 17 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D4 | 28 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D6 | 22 | 20I (Alpha, V1) | delH69, delV70, delY144, Q493Q/R, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| P3 | Diabetes type 2 Dyslipidaemia | BAM/ETE | D0 | 17 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | |
| D3 | 24 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D5 | 32 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H, G1204G/E | ||||
| D7 | 27 | 20I (Alpha, V1) 1 | N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| P4 | Myelodysplastic syndrome Trisomy 8 Corticosteroids | BAM/ETE | D0 | 21 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | |
| D2 | 19 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D4 | 24 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D8 | 33 | - | Low viral load | ||||
| P5 | Bi-phenotypic acute leukemia GVHD treated with systemic corticosteroids | BAM/ETE | D0 | 21 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | |
| D6 | 30 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D19 | 26 | 20I (Alpha, V1) 2 | Q493R, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D25 | 20 | 20I (Alpha, V1) | delH69, delV70, delY144, Q493R, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D52 | 20 | 20I (Alpha, V1) | delH69, delV70, delY144, Q493R, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D68 | 18 | 20I (Alpha, V1) | A67V, delH69, delV70, delY144, D427D/Y, Q493R, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| P6 | Arterial hypertension, obesity, dyslipidemia, renal cancer under chemotherapy | CAS/IMD | D0 | 13.5 | 20J (Gamma, V3) | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | |
| D2 | 31 | 20J (Gamma, V3) | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | ||||
| D5 | 32 | 20J (Gamma, V3) | L18F, T20N, P26S, D138Y, R190S, E406E/G, K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | ||||
| D7 | 33 | 20J (Gamma, V3) 3 | K417T, E484K, N501Y, D614G, H655Y, T1027I, V1176F | ||||
| P7 | Multiple myeloma under chemotherapy | CAS/IMD | D0 | 16 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | |
| D4 | 29 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D6 | 30 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D8 | 21 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D13 | 36 | - | Low viral load | ||||
| P8 | Multiple myeloma under chemotherapy | CAS/IMD | D0 | 21 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | |
| D0Bis | 18 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D1 | 16.5 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D9 | 33 | - | Low viral load | ||||
| P9 | Age > 80 years, obesity | CAS/IMD | D0 | 17 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | |
| P10 | Diabetes type 2 Arterial hypertension Metabolic syndrome Obesity (BMC = 35.8) | CAS/IMD | D0 | 16 | - | Missing sample | |
| D4 | 25 | 20I (Alpha, V1) 4 | N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D6 | 30 | 20I (Alpha, V1) | T20I, delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| P11 | Age > 80 years Arterial hypertension Chronic obstructive bronchopneumopathy | CAS/IMD | D0 | 25 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | |
| D3 | 31 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D5 | 30 | 20I (Alpha, V1) | delH69, delV70, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D7 | 20I (Alpha, V1) 5 | delH69, delV70, delY144, N501Y, A570D, D614G | |||||
| P12 | Diabetes type 2 Vascularitis Renal transplantation | CAS/IMD | D0 | 14 | 20I (Alpha, V1) | delH69, delV70, delY144, K182R, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | |
| D4 | 25 | 20I (Alpha, V1) | delH69, delV70, delY144, K182R, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D8 | 22 | 20I (Alpha, V1) | delH69, delV70, delY144, K182R, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| P13 | Multiple sclerosis under immunosuppressor treatment (Ocrelizumab) | CAS/IMD | pre-administration | 17 | 20I (Alpha, V1) | delH69, delV70, S98F, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | |
| D0 | 20 | 20I (Alpha, V1) | delH69, delV70, S98F, delY144, N501Y, A570D, D614G, P681H, T716I, S982A, D1118H | ||||
| D7 | 33 | - | Low viral load | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jary, A.; Marot, S.; Faycal, A.; Leon, S.; Sayon, S.; Zafilaza, K.; Ghidaoui, E.; Quoc, S.N.; Nemlaghi, S.; Choquet, S.; et al. Spike Gene Evolution and Immune Escape Mutations in Patients with Mild or Moderate Forms of COVID-19 and Treated with Monoclonal Antibodies Therapies. Viruses 2022, 14, 226. https://doi.org/10.3390/v14020226
Jary A, Marot S, Faycal A, Leon S, Sayon S, Zafilaza K, Ghidaoui E, Quoc SN, Nemlaghi S, Choquet S, et al. Spike Gene Evolution and Immune Escape Mutations in Patients with Mild or Moderate Forms of COVID-19 and Treated with Monoclonal Antibodies Therapies. Viruses. 2022; 14(2):226. https://doi.org/10.3390/v14020226
Chicago/Turabian StyleJary, Aude, Stéphane Marot, Antoine Faycal, Sacha Leon, Sophie Sayon, Karen Zafilaza, Emna Ghidaoui, Stéphanie Nguyen Quoc, Safaa Nemlaghi, Sylvain Choquet, and et al. 2022. "Spike Gene Evolution and Immune Escape Mutations in Patients with Mild or Moderate Forms of COVID-19 and Treated with Monoclonal Antibodies Therapies" Viruses 14, no. 2: 226. https://doi.org/10.3390/v14020226
APA StyleJary, A., Marot, S., Faycal, A., Leon, S., Sayon, S., Zafilaza, K., Ghidaoui, E., Quoc, S. N., Nemlaghi, S., Choquet, S., Dres, M., Pourcher, V., Calvez, V., Junot, H., Marcelin, A.-G., & Soulié, C. (2022). Spike Gene Evolution and Immune Escape Mutations in Patients with Mild or Moderate Forms of COVID-19 and Treated with Monoclonal Antibodies Therapies. Viruses, 14(2), 226. https://doi.org/10.3390/v14020226






