A Polyvalent Broad-Spectrum Escherichia Phage Tequatrovirus EP01 Capable of Controlling Salmonella and Escherichia coli Contamination in Foods
Abstract
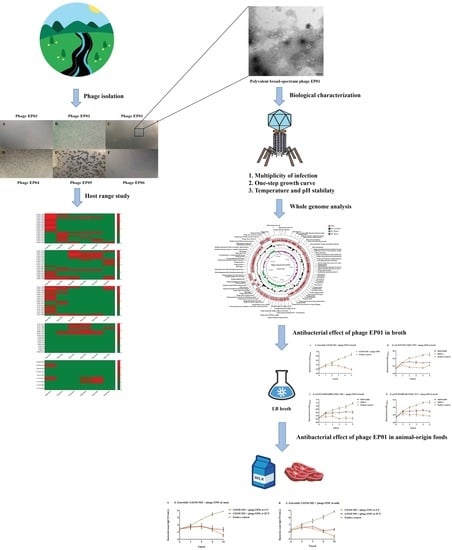
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Phages and Growth Conditions
2.2. Host Range of Isolated Phages
2.3. Morphological Analysis
2.4. Optimal Multiplicity of Infection Test
2.5. One-Step Growth Test
2.6. Thermal and pH Stability
2.7. Extraction and Analysis of Phage Tequatrovirus EP01 Genome
2.8. Bacterial Challenge Assay against S. Enteritidis, E. coli O157:H7, E. coli O114:K90 (B90), and E. coli O142:K86 (B) Contamination in LB Broth
2.9. Bacterial Challenge Assay against S. Enteritidis, E. coli O157:H7, E. coli O114:K90 (B90), and E. coli O142:K86 (B) Contamination on the Surface of Raw Meat and in Fresh Milk
2.9.1. Food Samples Preparation
2.9.2. Application of Phage Tequatrovirus EP01 on the Surface of Raw Meat and in Fresh Milk
2.10. Statistical Analysis
3. Results
3.1. Phage Tequatrovirus EP01 Exhibited Polyvalent Broad Spectrum
3.2. Biological Characteristics Analysis
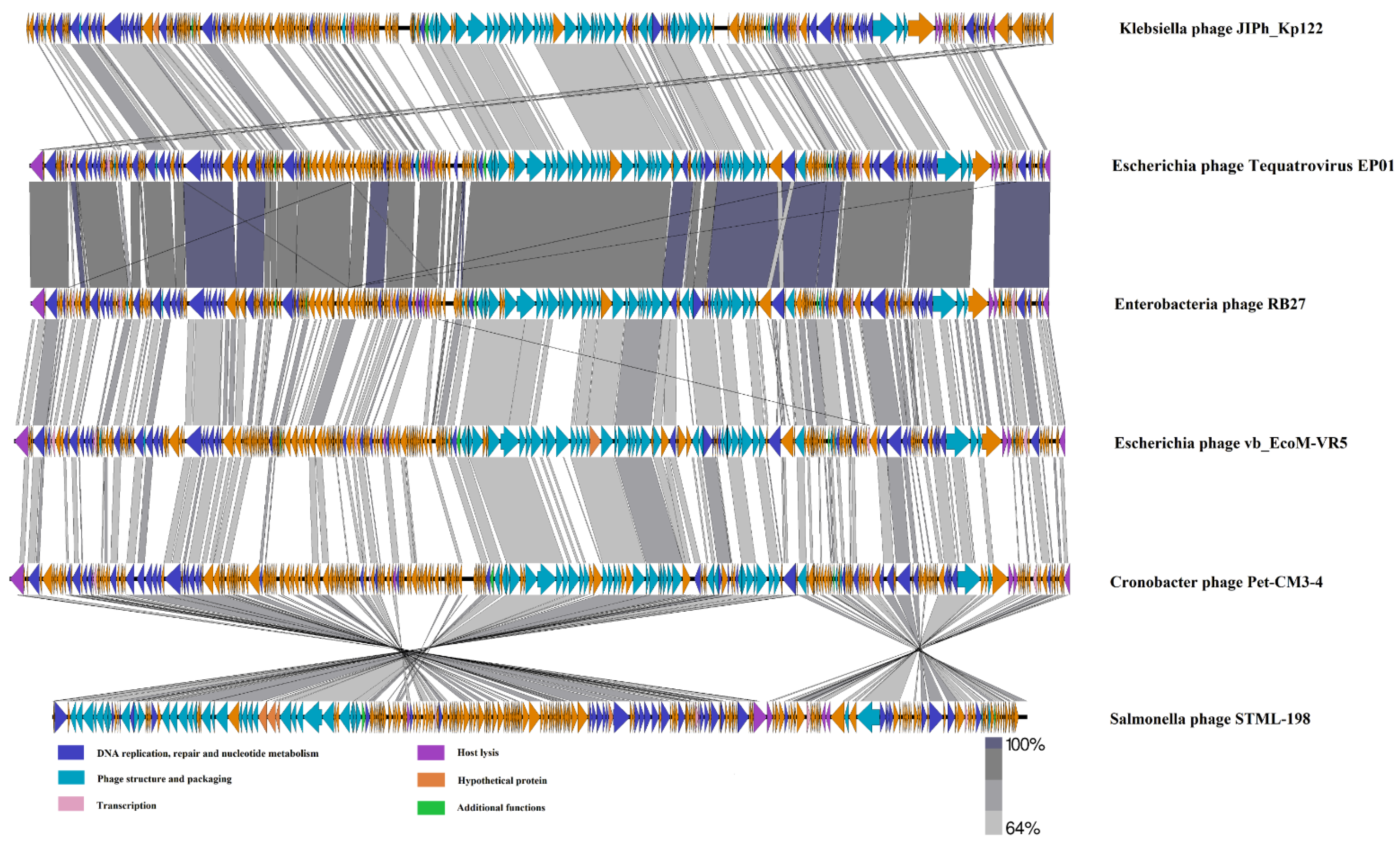
3.3. Phage Tequatrovirus EP01 Genomic Analysis
3.4. Phage Tequatrovirus EP01 Could Inhibit the Growth of S. Enteritidis, E. coli O157:H7, E. coli O114:K90 (B90), and E. coli O142:K86 (B) in LB Broth
3.5. Phage Tequatrovirus EP01 Could Control S. Enteritidis, E. coli O157:H7, E. coli O114:K90 (B90), and E. coli O142:K86 (B) Contamination on the Surface of Raw Meat and in Fresh Milk
3.5.1. Application of Phage Tequatrovirus EP01 Controlling S. Enteritidis
3.5.2. Application of Phage Tequatrovirus EP01 Controlling E. coli O157:H7
3.5.3. Application of Phage Tequatrovirus EP01 Controlling E. coli O114:K90 (B90)
3.5.4. Application of Phage Tequatrovirus EP01 Controlling E. coli O142:K86 (B)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Staes, I.; Passaris, I.; Cambre, A.; Aertsen, A. Population heterogeneity tactics as driving force in Salmonella virulence and survival. Food Res. Int. 2019, 125, 108560. [Google Scholar] [CrossRef] [PubMed]
- Fatica, M.K.; Schneider, K.R. Salmonella and produce: Survival in the plant environment and implications in food safety. Virulence 2011, 2, 573–579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; McGee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T.; et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Rangel, J.M.; Sparling, P.H.; Crowe, C. Epidemiology of Escherichia coli O157:H7 Outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 2005, 11, 603. [Google Scholar] [CrossRef] [PubMed]
- Ahn, D.U.; Lee, E.J. Mechanisms and prevention of off-odor production and color changes in irradiated meat. In Irradiation of Food and Packaging: Recent Developments; Komolprasert, V., Morehouse, K.M., Eds.; Acs Symposium Series; American Chemical Society: Washington, DC, USA, 2004; Volume 875, pp. 43–76. [Google Scholar]
- Richardson, S.D. Disinfection by-products and other emerging contaminants in drinking water. Trac-Trends Anal. Chem. 2003, 22, 666–684. [Google Scholar] [CrossRef]
- Mokgatla, R.M.; Gouws, P.A.; Brozel, V.S. Mechanisms contributing to hypochlorous acid resistance of a Salmonella isolate from a poultry-processing plant. J. Appl. Microbiol. 2002, 92, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Bajpai, V.K.; Baek, K.H.; Kang, S.C. Control of Salmonella in foods by using essential oils: A review. Food Res. Int. 2012, 45, 722–734. [Google Scholar] [CrossRef]
- Li, Z.; Ma, W.; Li, W.; Ding, Y.; Zhang, Y.; Yang, Q.; Wang, J.; Wang, X. A broad-spectrum phage controls multidrug-resistant Salmonella in liquid eggs. Food Res. Int. 2020, 132, 109011. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Sharma, M.; Millner, P.; Calaway, T.; Singh, M. Inactivation of Escherichia coli O157:H7 Attached to Spinach Harvester Blade Using Bacteriophage. Foodborne Pathog. Dis. 2011, 8, 541–546. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.; Shi, J.; Ma, W.; Li, Z.; Wang, J.; Li, J.; Wang, X. Isolation, characterization, and application of a novel specific Salmonella bacteriophage in different food matrices. Food Res. Int. 2018, 111, 631–641. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, Z.; Zhou, Y.; Bao, H.; Wang, R.; Li, T.; Pang, M.; Sun, L.; Zhou, X. Application of a phage in decontaminating Vibrio parahaemolyticus in oysters. Int. J. Food Microbiol. 2018, 275, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Magnone, J.P.; Marek, P.J.; Sulakvelidze, A.; Senecal, A.G. Additive Approach for Inactivation of Escherichia coli O157:H7, Salmonella, and Shigella spp. on Contaminated Fresh Fruits and Vegetables Using Bacteriophage Cocktail and Produce Wash. J. Food Prot. 2013, 76, 1336–1341. [Google Scholar] [CrossRef] [PubMed]
- Duc, H.M.; Son, H.M.; Yi, H.P.S.; Sato, J.; Ngan, P.H.; Masuda, Y.; Honjoh, K.I.; Miyamoto, T. Isolation, characterization and application of a polyvalent phage capable of controlling Salmonella and Escherichia coli O157:H7 in different food matrices. Food Res. Int. 2020, 131, 108977. [Google Scholar] [CrossRef]
- Yoon, H.; Yun, J.; Lim, J.A.; Roh, E.; Jung, K.S.; Chang, Y.; Ryu, S.; Heu, S. Characterization and genomic analysis of two Staphylococcus aureus bacteriophages isolated from poultry/livestock farms. J. Gen. Virol. 2013, 94, 2569–2576. [Google Scholar] [CrossRef] [PubMed]
- Kasman, L.M.; Kasman, A.; Westwater, C.; Dolan, J.; Schmidt, M.G.; Norris, J.S. Overcoming the phage replication threshold: A mathematical model with implications for phage therapy. J. Virol. 2002, 76, 5557–5564. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Yuan, J.; Guo, C.; Ge, C.; Wang, X.; Wei, D.; Li, X.; Si, H.; Hu, C. Identification and complete genome of lytic “Kp34likevirus” phage vB_KpnP_Bp5 and therapeutic potency in the treatment of lethal Klebsiella pneumoniae infections in mice. Virus Res. 2021, 297, 198348. [Google Scholar] [CrossRef] [PubMed]
- Shende, R.K.; Hirpurkar, S.D.; Sannat, C.; Rawat, N.; Pandey, V. Isolation and characterization of bacteriophages with lytic activity against common bacterial pathogens. Vet. World 2017, 10, 973–978. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.L.; Bao, H.D.; Wei, C.R.; Zhang, H.; Zhou, Y.; Wang, R. Characterization and partial genomic analysis of a lytic Myoviridae bacteriophage against Staphylococcus aureus isolated from dairy cows with mastitis in Mid-east of China. Virus Genes 2015, 50, 111–117. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2019, 47, D687–D692. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Dereeper, A.; Audic, S.; Claverie, J.M.; Blanc, G. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol. Biol. 2010, 10, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K.; Battistuzzi, F.U. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S.R.; Thuman-Commike, P.A. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology 2011, 411, 393–415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorensen, A.N.; Woudstra, C.; Sorensen, M.C.H.; Brondsted, L. Subtypes of tail spike proteins predicts the host range of Ackermannviridae phages. Comput. Struct. Biotechnol. J. 2021, 19, 4854–4867. [Google Scholar] [CrossRef]
- Suga, A.; Kawaguchi, M.; Yonesaki, T.; Otsuka, Y. Manipulating Interactions between T4 Phage Long Tail Fibers and Escherichia coli Receptors. Appl. Environ. Microbiol. 2021, 87. [Google Scholar] [CrossRef]
- Bao, H.; Zhang, P.; Zhang, H.; Zhou, Y.; Zhang, L.; Wang, R. Bio-Control of Salmonella Enteritidis in Foods Using Bacteriophages. Viruses 2015, 7, 4836–4853. [Google Scholar] [CrossRef]
- Park, M.; Lee, J.H.; Shin, H.; Kim, M.; Choi, J.; Kang, D.H.; Heu, S.; Ryu, S. Characterization and comparative genomic analysis of a novel bacteriophage, SFP10, simultaneously inhibiting both Salmonella enterica and Escherichia coli O157:H7. Appl. Environ. Microbiol. 2012, 78, 58–69. [Google Scholar] [CrossRef] [Green Version]









Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Li, L.; Han, K.; Wang, L.; Cao, Y.; Ma, D.; Wang, X. A Polyvalent Broad-Spectrum Escherichia Phage Tequatrovirus EP01 Capable of Controlling Salmonella and Escherichia coli Contamination in Foods. Viruses 2022, 14, 286. https://doi.org/10.3390/v14020286
Zhou Y, Li L, Han K, Wang L, Cao Y, Ma D, Wang X. A Polyvalent Broad-Spectrum Escherichia Phage Tequatrovirus EP01 Capable of Controlling Salmonella and Escherichia coli Contamination in Foods. Viruses. 2022; 14(2):286. https://doi.org/10.3390/v14020286
Chicago/Turabian StyleZhou, Yuqing, Lei Li, Kaiou Han, Leping Wang, Yajie Cao, Dongxin Ma, and Xiaoye Wang. 2022. "A Polyvalent Broad-Spectrum Escherichia Phage Tequatrovirus EP01 Capable of Controlling Salmonella and Escherichia coli Contamination in Foods" Viruses 14, no. 2: 286. https://doi.org/10.3390/v14020286