A Community Study of SARS-CoV-2 Detection by RT-PCR in Saliva: A Reliable and Effective Method
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Collection
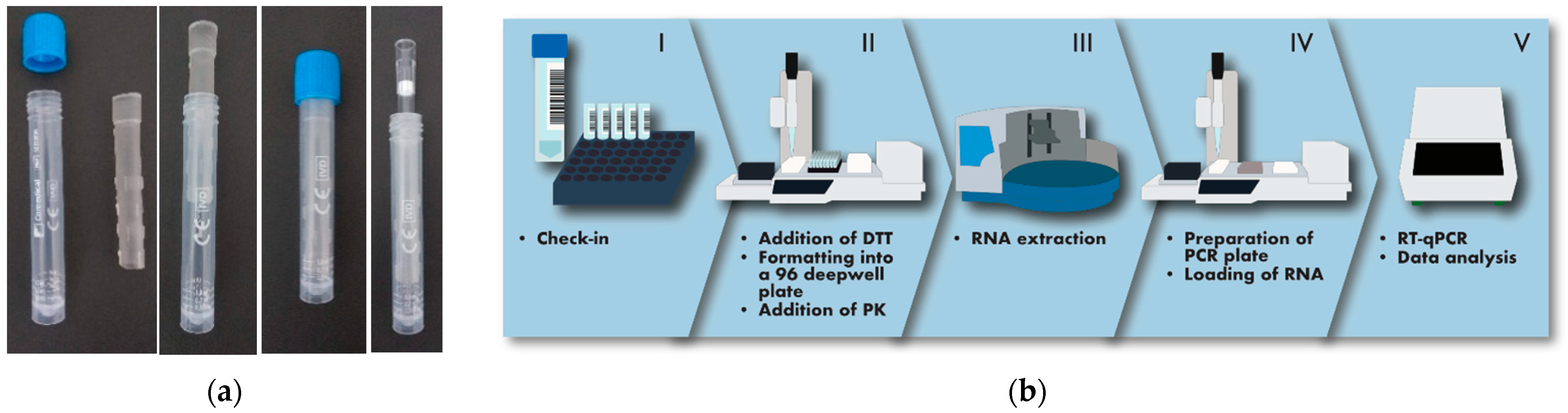
2.3. The Analytical Process
2.4. Statistical Analysis
3. Results
3.1. Design of an Analytical Workflow Allowing Large-Scale Screening of SARS-CoV-2 in Saliva
3.2. Global Comparison of SARS-CoV-2 Detection in Saliva and NP Swabs
3.3. Saliva and NP Swabs Give Concordant Results in Symptomatic Subjects and in Individuals Recruited from Contact Tracing
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mercer, T.R.; Salit, M. Testing at Scale during the COVID-19 Pandemic. Nat. Rev. Genet. 2021, 22, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, O.; Martiny, D.; Rochas, O.; Belkum, A.; van Kozlakidis, Z. Considerations for Diagnostic COVID-19 Tests. Nat. Rev. Microbiol. 2020, 19, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Marty, F.M.; Chen, K.; Verrill, K.A. How to Obtain a Nasopharyngeal Swab Specimen. N. Engl. J. Med. 2020, 382, e76. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.-W.; Tsang, O.T.-Y.; Yip, C.C.-Y.; Chan, K.-H.; Wu, T.-C.; Chan, J.M.-C.; Leung, W.-S.; Chik, T.S.-H.; Choi, C.Y.-C.; Kandamby, D.H.; et al. Consistent Detection of 2019 Novel Coronavirus in Saliva. Clin. Infect. Dis. 2020, 71, 841–843. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, P.; Chowdhry, A.; Kharbanda, O.P.; Popli, D.B.; Gautam, K.; Saini, V. Exploring Salivary Diagnostics in COVID-19: A Scoping Review and Research Suggestions. BDJ Open 2021, 7, 1–10. [Google Scholar] [CrossRef]
- Tan, S.H.; Allicock, O.; Armstrong-Hough, M.; Wyllie, A.L. Saliva as a Gold-Standard Sample for SARS-CoV-2 Detection. Lancet Respir. Med. 2021, 9, 562–564. [Google Scholar] [CrossRef]
- Warsi, I.; Khurshid, Z.; Shazam, H.; Umer, M.F.; Imran, E.; Khan, M.O.; Slowey, P.D.; Goodson, J.M. Saliva Exhibits High Sensitivity and Specificity for the Detection of SARS-COV-2. Diseases 2021, 9, 38. [Google Scholar] [CrossRef]
- Atieh, M.A.; Guirguis, M.; Alsabeeha, N.H.M.; Cannon, R.D. The Diagnostic Accuracy of Saliva Testing for SARS-CoV-2: A Systematic Review and Meta-Analysis. Oral Dis. 2021, 00, 1–15. [Google Scholar] [CrossRef]
- Allicock, O.M.; Petrone, M.E.; Yolda-Carr, D.; Breban, M.; Watkins, A.E.; Rothman, J.E.; Farhadian, S.F.; Wyllie, A.L. Evaluation of Saliva Self-Collection Devices for SARS-CoV-2 Diagnostics 1. Available online: https://ssrn.com/abstract=3877563 (accessed on 1 January 2022). [CrossRef]
- Matic, N.; Stefanovic, A.; Leung, V.; Lawson, T.; Ritchie, G.; Li, L.; Champagne, S.; Romney, M.G.; Lowe, C.F. Practical Challenges to the Clinical Implementation of Saliva for SARS-CoV-2 Detection. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 447–450. [Google Scholar] [CrossRef]
- Ott, I.M.; Strine, M.S.; Watkins, A.E.; Boot, M.; Kalinich, C.C.; Harden, C.A.; Vogels, C.B.F.; Casanovas-Massana, A.; Moore, A.J.; Muenker, M.C.; et al. Stability of SARS-CoV-2 RNA in Nonsupplemented Saliva. Emerg. Infect. Dis. 2021, 27, 1146. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, D.; Yang, P.; Poon, L.L.M.; Wang, Q. Viral Load of SARS-CoV-2 in Clinical Samples. Lancet Infect. Dis. 2020, 20, 411–412. [Google Scholar] [CrossRef]
- Zhu, J.; Guo, J.; Xu, Y.; Chen, X. Viral Dynamics of SARS-CoV-2 in Saliva from Infected Patients. J. Infect. 2020, 81, e48–e50. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Matuck, B.F.; Dolhnikoff, M.; Duarte-Neto, A.N.; Maia, G.; Gomes, S.C.; Sendyk, D.I.; Zarpellon, A.; de Andrade, N.P.; Monteiro, R.A.; Pinho, J.R.R.; et al. Salivary Glands Are a Target for SARS-CoV-2: A Source for Saliva Contamination. J. Pathol. 2021, 254, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Pascolo, L.; Zupin, L.; Melato, M.; Tricarico, P.M.; Crovella, S. TMPRSS2 and ACE2 Coexpression in SARS-CoV-2 Salivary Glands Infection. J. Dent. Res. 2020, 99, 1120–1121. [Google Scholar] [CrossRef] [PubMed]
- Teo, A.K.J.; Choudhury, Y.; Tan, I.B.; Cher, C.Y.; Chew, S.H.; Wan, Z.Y.; Cheng, L.T.E.; Oon, L.L.E.; Tan, M.H.; Chan, K.S.; et al. Saliva Is More Sensitive than Nasopharyngeal or Nasal Swabs for Diagnosis of Asymptomatic and Mild COVID-19 Infection. Sci. Rep. 2021, 11, 3134. [Google Scholar] [CrossRef]
- Lu, J.; Peng, J.; Xiong, Q.; Liu, Z.; Lin, H.; Tan, X.; Kang, M.; Yuan, R.; Zeng, L.; Zhou, P.; et al. Clinical, Immunological and Virological Characterization of COVID-19 Patients That Test Re-Positive for SARS-CoV-2 by RT-PCR. EBioMedicine 2020, 59, 102960. [Google Scholar] [CrossRef] [PubMed]
- Piralla, A.; Ricchi, M.; Cusi, M.G.; Prati, P.; Vicari, N.; Scarsi, G.; Gandolfo, C.; Anichini, G.; Terrosi, C.; Percivalle, E.; et al. Residual SARS-CoV-2 RNA in Nasal Swabs of Convalescent COVID-19 Patients: Is Prolonged Quarantine Always Justified? Int. J. Infect. Dis. 2021, 102, 299–302. [Google Scholar] [CrossRef]
- Zhou, L.; Ayeh, S.K.; Chidambaram, V.; Karakousis, P.C. Modes of Transmission of SARS-CoV-2 and Evidence for Preventive Behavioral Interventions. BMC Infect. Dis. 2021, 21, 1–9. [Google Scholar] [CrossRef]
- Halfmann, P.; Iida, S.; Iwatsuki-Horimoto, K.; Maemura, T.; Kiso, M.; Scheaffer, S.M.; Darling, T.; Joshi, A.; Loeber, S.; Singh, G.; et al. The SARS-CoV-2 B.1.1.529 Omicron Virus Causes Attenuated Infection and Disease in Mice and Hamsters. Nature 2022. Available online: https://www.nature.com/articles/s41586-022-04441-6#citeas (accessed on 1 January 2022). [CrossRef]
- McMahan, K.; Giffin, V.; Tostanoski, L.H.; Chung, B.; Siamatu, M.; Suthar, M.S.; Halfmann, P.; Kawaoka, Y.; Piedra-Mora, C.; Martinot, A.J.; et al. Reduced Pathogenicity of the SARS-CoV-2 Omicron Variant in Hamsters. BioRxiv 2022. [Google Scholar] [CrossRef]
- Bentley, E.G.; Kirby, A.; Sharma, P.; Kipar, A.; Mega, D.F.; Bramwell, C.; Penrice-Randal, R.; Prince, T.; Brown, J.C.; Zhou, J.; et al. SARS-CoV-2 Omicron-B.1.1.529 Variant Leads to Less Severe Disease than Pango B and Delta Variants Strains in a Mouse Model of Severe COVID-19. BioRxiv 2021. [Google Scholar] [CrossRef]
- Peacock, T.P.; Brown, J.C.; Zhou, J.; Thakur, N.; Newman, J.; Kugathasan, R.; Sukhova, K.; Kaforou, M.; Bailey, D.; Barclay, W.S. The SARS-CoV-2 Variant, Omicron, Shows Rapid Replication in Human Primary Nasal Epithelial Cultures and Efficiently Uses the Endosomal Route of Entry. BioRxiv 2022, 2021-12. [Google Scholar] [CrossRef]
- Marais, G.; Hsiao, N.; Iranzadeh, A.; Doolabh, D.; Enoch, A.; Chu, C.; Williamson, C.; Brink, A.; Hardie, D. Saliva Swabs Are the Preferred Sample for Omicron Detection. MedRxiv 2021. [Google Scholar] [CrossRef]
- Schrom, J.; Marquez, C.; Pilarowski, G.; Wang, G.; Mitchell, A.; Puccinelli, R.; Black, D.; Rojas, S.; Ribeiro, S.; Martinez, J.; et al. Direct Comparison of SARS-CoV-2 Nasal RT-PCR and Rapid Antigen Test (BinaxNOWTM) at a Community Testing Site During an Omicron Surge. MedRxiv 2022. [Google Scholar] [CrossRef]


| n. of Individuals | Positive NP Swabs | Positive Saliva Samples | |||
|---|---|---|---|---|---|
| n | % (95% CI) | n | % (95% CI) | ||
| Total | 1003 | 344 | 34.3 (31.4–37.3) | 312 | 31.1 (28.3–34.0) |
| Males | 434 | 175 | 40 (35.8–45) | 158 | 36.4 (32.0–41.0) |
| Females | 569 | 169 | 29.7 (26.1–33.6) | 154 | 27.1 (23.6–30.9) |
| Contact tracing | 193 | 56 | 29.0 (22.7–36.0) | 65 | 33.7 (27.4–40.6) |
| Convalescents | 442 | 276 | 62.4 (57.8–66.8) | 235 | 53.2% (48.5–57.8) |
| Screening | 351 | 4 | 1.1 (0.3–3.0) | 4 | 1.1 (0.3–3.0) |
| Suspected | 17 | 8 | 47.1 (26.2–69.0) | 8 | 47.1 (26.2–69.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fronza, F.; Groff, N.; Martinelli, A.; Passerini, B.Z.; Rensi, N.; Cortelletti, I.; Vivori, N.; Adami, V.; Helander, A.; Bridi, S.; et al. A Community Study of SARS-CoV-2 Detection by RT-PCR in Saliva: A Reliable and Effective Method. Viruses 2022, 14, 313. https://doi.org/10.3390/v14020313
Fronza F, Groff N, Martinelli A, Passerini BZ, Rensi N, Cortelletti I, Vivori N, Adami V, Helander A, Bridi S, et al. A Community Study of SARS-CoV-2 Detection by RT-PCR in Saliva: A Reliable and Effective Method. Viruses. 2022; 14(2):313. https://doi.org/10.3390/v14020313
Chicago/Turabian StyleFronza, Filippo, Nelli Groff, Angela Martinelli, Beatrice Zita Passerini, Nicolò Rensi, Irene Cortelletti, Nicolò Vivori, Valentina Adami, Anna Helander, Simone Bridi, and et al. 2022. "A Community Study of SARS-CoV-2 Detection by RT-PCR in Saliva: A Reliable and Effective Method" Viruses 14, no. 2: 313. https://doi.org/10.3390/v14020313









