The Influenza Virus RNA-Polymerase and the Host RNA-Polymerase II: RPB4 Is Targeted by a PB2 Domain That Is Involved in Viral Transcription
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Protein-Protein Interaction Assays
2.2.1. Split-Luciferase Complementation Assay
2.2.2. Bimolecular Fluorescence Complementation (BiFC) Assay
2.3. Minireplicon and Transcription Assays
2.3.1. Minireplicon Assay
2.3.2. Transcription Assay
2.4. Co-Immunoprecipitation Assay
3. Results
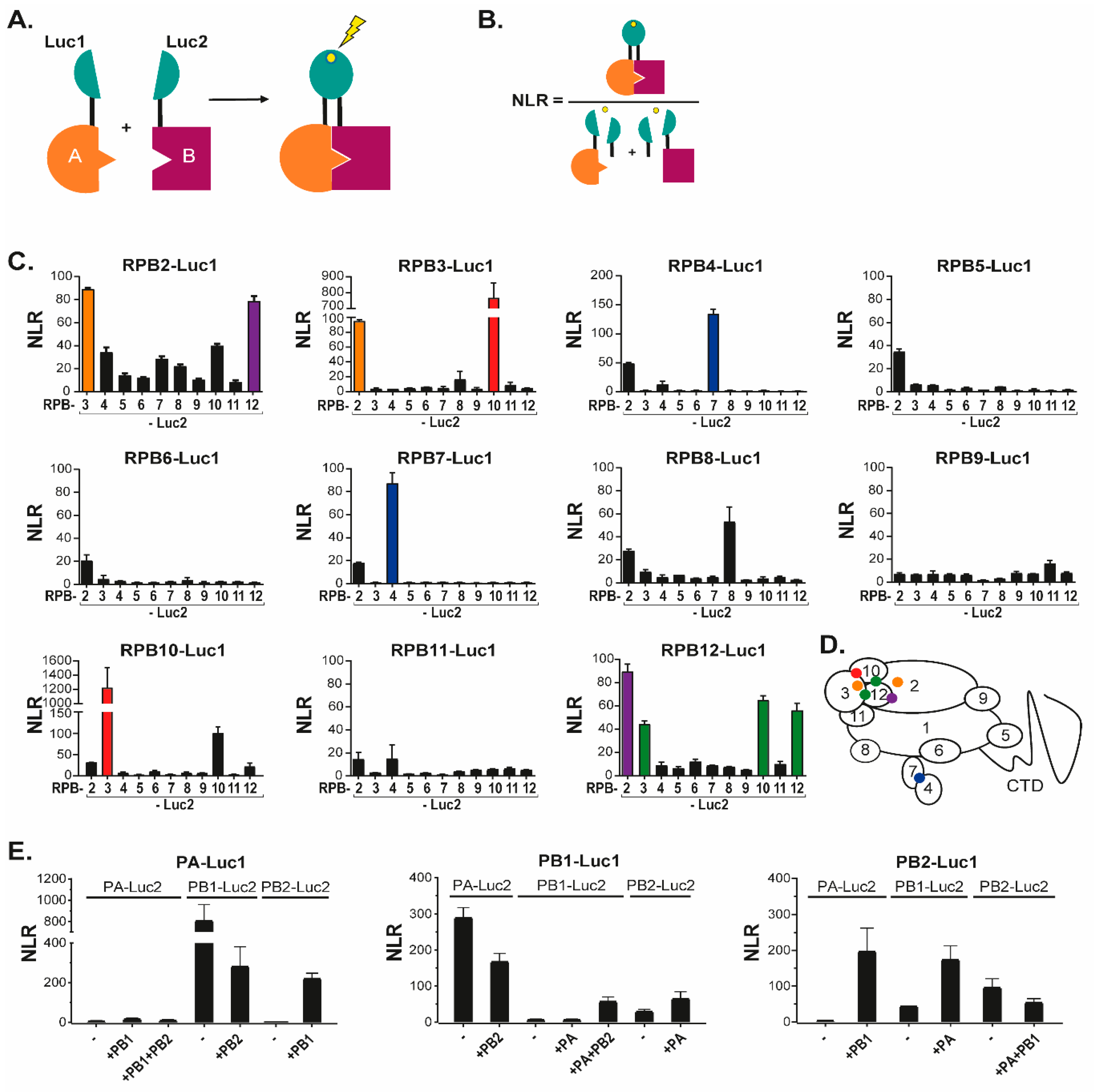
3.1. Novel Interactions between FluPol and Pol II Subunits
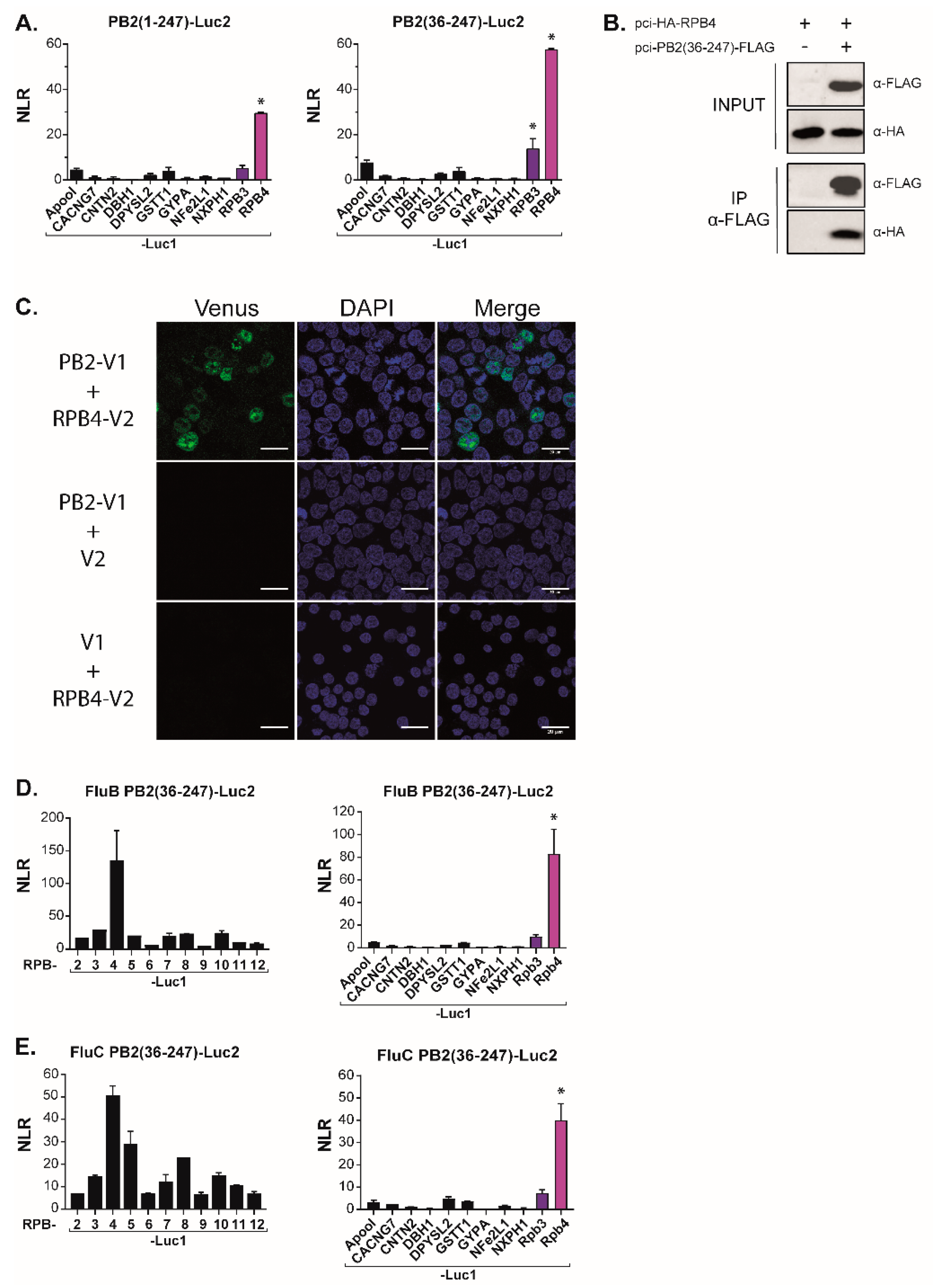
3.2. The N-Terminal Region of PB2 Mediates Interaction with RPB4
3.3. The PB2RPB4 Interaction Domain Is Essential for the Virus Life Cycle
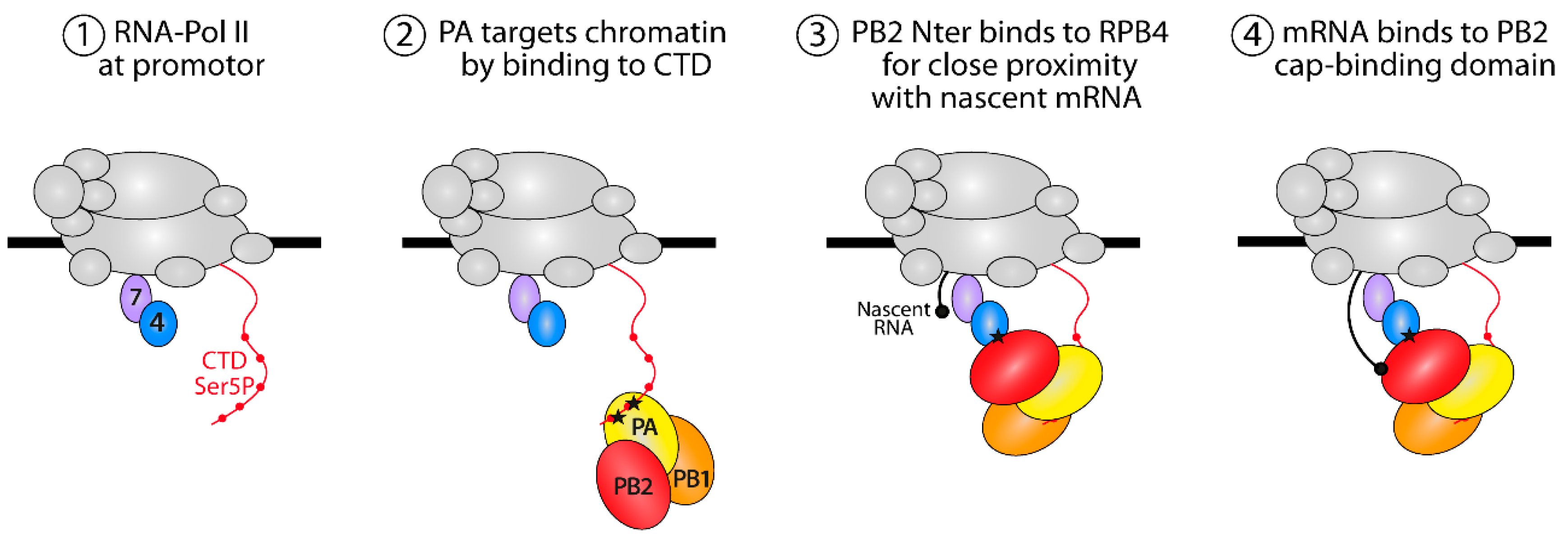
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palese, P.; Shaw, M.L. Orthomyxoviridae: The Viruses and Their Replication. In Fields Virology, 6th ed.; Wolters Kluwer Health/Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; Volume 1, pp. 1151–1185. [Google Scholar]
- Boivin, S.; Cusack, S.; Ruigrok, R.W.H.; Hart, D.J. Influenza A Virus Polymerase: Structural Insights into Replication and Host Adaptation Mechanisms. J. Biol. Chem. 2010, 285, 28411–28417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Te Velthuis, A.J.W.; Fodor, E. Influenza Virus RNA Polymerase: Insights into the Mechanisms of Viral RNA Synthesis. Nat. Rev. Microbiol. 2016, 14, 479–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poon, L.L.; Pritlove, D.C.; Fodor, E.; Brownlee, G.G. Direct Evidence That the Poly(A) Tail of Influenza A Virus MRNA Is Synthesized by Reiterative Copying of a U Track in the Virion RNA Template. J. Virol. 1999, 73, 3473–3476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krischuns, T.; Lukarska, M.; Naffakh, N.; Cusack, S. Influenza Virus RNA-Dependent RNA Polymerase and the Host Transcriptional Apparatus. Annu. Rev. Biochem. 2021, 90, 321–348. [Google Scholar] [CrossRef] [PubMed]
- Wandzik, J.M.; Kouba, T.; Karuppasamy, M.; Pflug, A.; Drncova, P.; Provaznik, J.; Azevedo, N.; Cusack, S. A Structure-Based Model for the Complete Transcription Cycle of Influenza Polymerase. Cell 2020, 181, 877–893.e21. [Google Scholar] [CrossRef] [PubMed]
- Lamb, R.A.; Choppin, P.W. Synthesis of Influenza Virus Polypeptides in Cells Resistant to Alpha-Amanitin: Evidence for the Involvement of Cellular RNA Polymerase II in Virus Replication. J. Virol. 1977, 23, 816–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Engelhardt, O.G.; Smith, M.; Fodor, E. Association of the Influenza A Virus RNA-Dependent RNA Polymerase with Cellular RNA Polymerase II. JVI 2005, 79, 5812–5818. [Google Scholar] [CrossRef] [Green Version]
- Walker, A.P.; Fodor, E. Interplay between Influenza Virus and the Host RNA Polymerase II Transcriptional Machinery. Trends Microbiol. 2019, 27, 398–407. [Google Scholar] [CrossRef] [Green Version]
- Cramer, P.; Bushnell, D.A.; Kornberg, R.D. Structural Basis of Transcription: RNA Polymerase II at 2.8 Angstrom Resolution. Science 2001, 292, 1863–1876. [Google Scholar] [CrossRef] [Green Version]
- Cramer, P. Organization and Regulation of Gene Transcription. Nature 2019, 573, 45–54. [Google Scholar] [CrossRef]
- Nojima, T.; Gomes, T.; Grosso, A.R.F.; Kimura, H.; Dye, M.J.; Dhir, S.; Carmo-Fonseca, M.; Proudfoot, N.J. Mammalian NET-Seq Reveals Genome-Wide Nascent Transcription Coupled to RNA Processing. Cell 2015, 161, 526–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Alonso, M.; Hengrung, N.; Fodor, E. RNA-Free and Ribonucleoprotein-Associated Influenza Virus Polymerases Directly Bind the Serine-5-Phosphorylated Carboxyl-Terminal Domain of Host RNA Polymerase II. J. Virol. 2016, 90, 6014–6021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lukarska, M.; Fournier, G.; Pflug, A.; Resa-Infante, P.; Reich, S.; Naffakh, N.; Cusack, S. Structural Basis of an Essential Interaction between Influenza Polymerase and Pol II CTD. Nature 2017, 541, 117–121. [Google Scholar] [CrossRef]
- Serna Martin, I.; Hengrung, N.; Renner, M.; Sharps, J.; Martínez-Alonso, M.; Masiulis, S.; Grimes, J.M.; Fodor, E. A Mechanism for the Activation of the Influenza Virus Transcriptase. Molecular. Cell 2018, 70, 1101–1110.e4. [Google Scholar] [CrossRef] [Green Version]
- Fodor, E.; Devenish, L.; Engelhardt, O.G.; Palese, P.; Brownlee, G.G.; García-Sastre, A. Rescue of Influenza A Virus from Recombinant DNA. J. Virol. 1999, 73, 9679–9682. [Google Scholar] [CrossRef] [Green Version]
- Le Goffic, R.; Bouguyon, E.; Chevalier, C.; Vidic, J.; Da Costa, B.; Leymarie, O.; Bourdieu, C.; Decamps, L.; Dhorne-Pollet, S.; Delmas, B. Influenza A Virus Protein PB1-F2 Exacerbates IFN-Beta Expression of Human Respiratory Epithelial Cells. J. Immunol. 2010, 185, 4812–4823. [Google Scholar] [CrossRef] [PubMed]
- Cassonnet, P.; Rolloy, C.; Neveu, G.; Vidalain, P.-O.; Chantier, T.; Pellet, J.; Jones, L.; Muller, M.; Demeret, C.; Gaud, G.; et al. Benchmarking a Luciferase Complementation Assay for Detecting Protein Complexes. Nat. Methods 2011, 8, 990–992. [Google Scholar] [CrossRef]
- Da Costa, B.; Sausset, A.; Munier, S.; Ghounaris, A.; Naffakh, N.; Le Goffic, R.; Delmas, B. Temperature-Sensitive Mutants in the Influenza A Virus RNA Polymerase: Alterations in the PA Linker Reduce Nuclear Targeting of the PB1-PA Dimer and Result in Viral Attenuation. J. Virol. 2015, 89, 6376–6390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shyu, Y.J.; Liu, H.; Deng, X.; Hu, C.-D. Identification of New Fluorescent Protein Fragments for Bimolecular Fluorescence Complementation Analysis under Physiological Conditions. Biotechniques 2006, 40, 61–66. [Google Scholar] [CrossRef] [Green Version]
- Leymarie, O.; Jouvion, G.; Hervé, P.-L.; Chevalier, C.; Lorin, V.; Lecardonnel, J.; Da Costa, B.; Delmas, B.; Escriou, N.; Le Goffic, R. Kinetic Characterization of PB1-F2-Mediated Immunopathology during Highly Pathogenic Avian H5N1 Influenza Virus Infection. PLoS ONE 2013, 8, e57894. [Google Scholar] [CrossRef]
- Nilsson-Payant, B.E.; Sharps, J.; Hengrung, N.; Fodor, E. The Surface-Exposed PA51-72-Loop of the Influenza A Virus Polymerase Is Required for Viral Genome Replication. J. Virol. 2018, 92, e00687-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munier, S.; Rolland, T.; Diot, C.; Jacob, Y.; Naffakh, N. Exploration of Binary Virus–Host Interactions Using an Infectious Protein Complementation Assay. Mol. Cell Proteom. 2013, 12, 2845–2855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cramer, P. RNA Polymerase II Structure: From Core to Functional Complexes. Curr. Opin. Genet. Dev. 2004, 14, 218–226. [Google Scholar] [CrossRef]
- Huet, S.; Avilov, S.V.; Ferbitz, L.; Daigle, N.; Cusack, S.; Ellenberg, J. Nuclear Import and Assembly of Influenza A Virus RNA Polymerase Studied in Live Cells by Fluorescence Cross-Correlation Spectroscopy. JVI 2010, 84, 1254–1264. [Google Scholar] [CrossRef] [Green Version]
- Pflug, A.; Guilligay, D.; Reich, S.; Cusack, S. Structure of Influenza A Polymerase Bound to the Viral RNA Promoter. Nature 2014, 516, 355–360. [Google Scholar] [CrossRef]
- Pflug, A.; Lukarska, M.; Resa-Infante, P.; Reich, S.; Cusack, S. Structural Insights into RNA Synthesis by the Influenza Virus Transcription-Replication Machine. Virus Res. 2017, 234, 103–117. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, K.; Obayashi, E.; Kawaguchi, A.; Suzuki, Y.; Tame, J.R.H.; Nagata, K.; Park, S.-Y. Structural Insight into the Essential PB1-PB2 Subunit Contact of the Influenza Virus RNA Polymerase. EMBO J. 2009, 28, 1803–1811. [Google Scholar] [CrossRef] [PubMed]
- Meka, H. Crystal Structure and RNA Binding of the Rpb4/Rpb7 Subunits of Human RNA Polymerase II. Nucleic Acids Res. 2005, 33, 6435–6444. [Google Scholar] [CrossRef] [PubMed]
- Robert, X.; Gouet, P. Deciphering Key Features in Protein Structures with the New ENDscript Server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [Green Version]
- Allepuz-Fuster, P.; Martínez-Fernández, V.; Garrido-Godino, A.I.; Alonso-Aguado, S.; Hanes, S.D.; Navarro, F.; Calvo, O. Rpb4/7 Facilitates RNA Polymerase II CTD Dephosphorylation. Nucleic Acids Res. 2014, 42, 13674–13688. [Google Scholar] [CrossRef] [Green Version]
- Allepuz-Fuster, P.; O’Brien, M.J.; González-Polo, N.; Pereira, B.; Dhoondia, Z.; Ansari, A.; Calvo, O. RNA Polymerase II Plays an Active Role in the Formation of Gene Loops through the Rpb4 Subunit. Nucleic Acids Res. 2019, 47, 8975–8987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, M.; Suzuki, H.; Ishihama, A. Formation of a Carboxy-Terminal Domain Phosphatase (Fcp1)/TFIIF/RNA Polymerase II (Pol II) Complex in Schizosaccharomyces Pombe Involves Direct Interaction between Fcp1 and the Rpb4 Subunit of Pol II. Mol. Cell Biol. 2002, 22, 1577–1588. [Google Scholar] [CrossRef] [Green Version]
- Mitsuzawa, H.; Kanda, E.; Ishihama, A. Rpb7 Subunit of RNA Polymerase II Interacts with an RNA-Binding Protein Involved in Processing of Transcripts. Nucleic Acids Res. 2003, 31, 4696–4701. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Yamaguchi, Y.; Tsugeno, Y.; Yamamoto, J.; Yamada, T.; Nakamura, M.; Hisatake, K.; Handa, H. DSIF, the Paf1 Complex, and Tat-SF1 Have Nonredundant, Cooperative Roles in RNA Polymerase II Elongation. Genes Dev. 2009, 23, 2765–2777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armache, K.-J.; Mitterweger, S.; Meinhart, A.; Cramer, P. Structures of Complete RNA Polymerase II and Its Subcomplex, Rpb4/7. J. Biol. Chem. 2005, 280, 7131–7134. [Google Scholar] [CrossRef] [Green Version]
- Calvo, O. RNA Polymerase II Phosphorylation and Gene Looping: New Roles for the Rpb4/7 Heterodimer in Regulating Gene Expression. Curr. Genet. 2020, 66, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Choder, M. Rpb4 and Rpb7: Subunits of RNA Polymerase II and Beyond. Trends Biochem. Sci. 2004, 29, 674–681. [Google Scholar] [CrossRef]
- Richard, S.; Gross, L.; Fischer, J.; Bendalak, K.; Ziv, T.; Urim, S.; Choder, M. Numerous Post-Translational Modifications of RNA Polymerase II Subunit Rpb4/7 Link Transcription to Post-Transcriptional Mechanisms. Cell Rep. 2021, 34, 108578. [Google Scholar] [CrossRef]
- Vos, S.M.; Pöllmann, D.; Caizzi, L.; Hofmann, K.B.; Rombaut, P.; Zimniak, T.; Herzog, F.; Cramer, P. Architecture and RNA Binding of the Human Negative Elongation Factor. eLife 2016, 5, e14981. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Takagi, T.; Wada, T.; Yano, K.; Furuya, A.; Sugimoto, S.; Hasegawa, J.; Handa, H. NELF, a Multisubunit Complex Containing RD, Cooperates with DSIF to Repress RNA Polymerase II Elongation. Cell 1999, 97, 41–51. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morel, J.; Sedano, L.; Lejal, N.; Da Costa, B.; Batsché, E.; Muchardt, C.; Delmas, B. The Influenza Virus RNA-Polymerase and the Host RNA-Polymerase II: RPB4 Is Targeted by a PB2 Domain That Is Involved in Viral Transcription. Viruses 2022, 14, 518. https://doi.org/10.3390/v14030518
Morel J, Sedano L, Lejal N, Da Costa B, Batsché E, Muchardt C, Delmas B. The Influenza Virus RNA-Polymerase and the Host RNA-Polymerase II: RPB4 Is Targeted by a PB2 Domain That Is Involved in Viral Transcription. Viruses. 2022; 14(3):518. https://doi.org/10.3390/v14030518
Chicago/Turabian StyleMorel, Jessica, Laura Sedano, Nathalie Lejal, Bruno Da Costa, Eric Batsché, Christian Muchardt, and Bernard Delmas. 2022. "The Influenza Virus RNA-Polymerase and the Host RNA-Polymerase II: RPB4 Is Targeted by a PB2 Domain That Is Involved in Viral Transcription" Viruses 14, no. 3: 518. https://doi.org/10.3390/v14030518






