Human Adenovirus Type 26 Induced IL-6 Gene Expression in an αvβ3 Integrin- and NF-κB-Dependent Manner
Abstract
:1. Introduction
2. Materials and Methods
3. Results
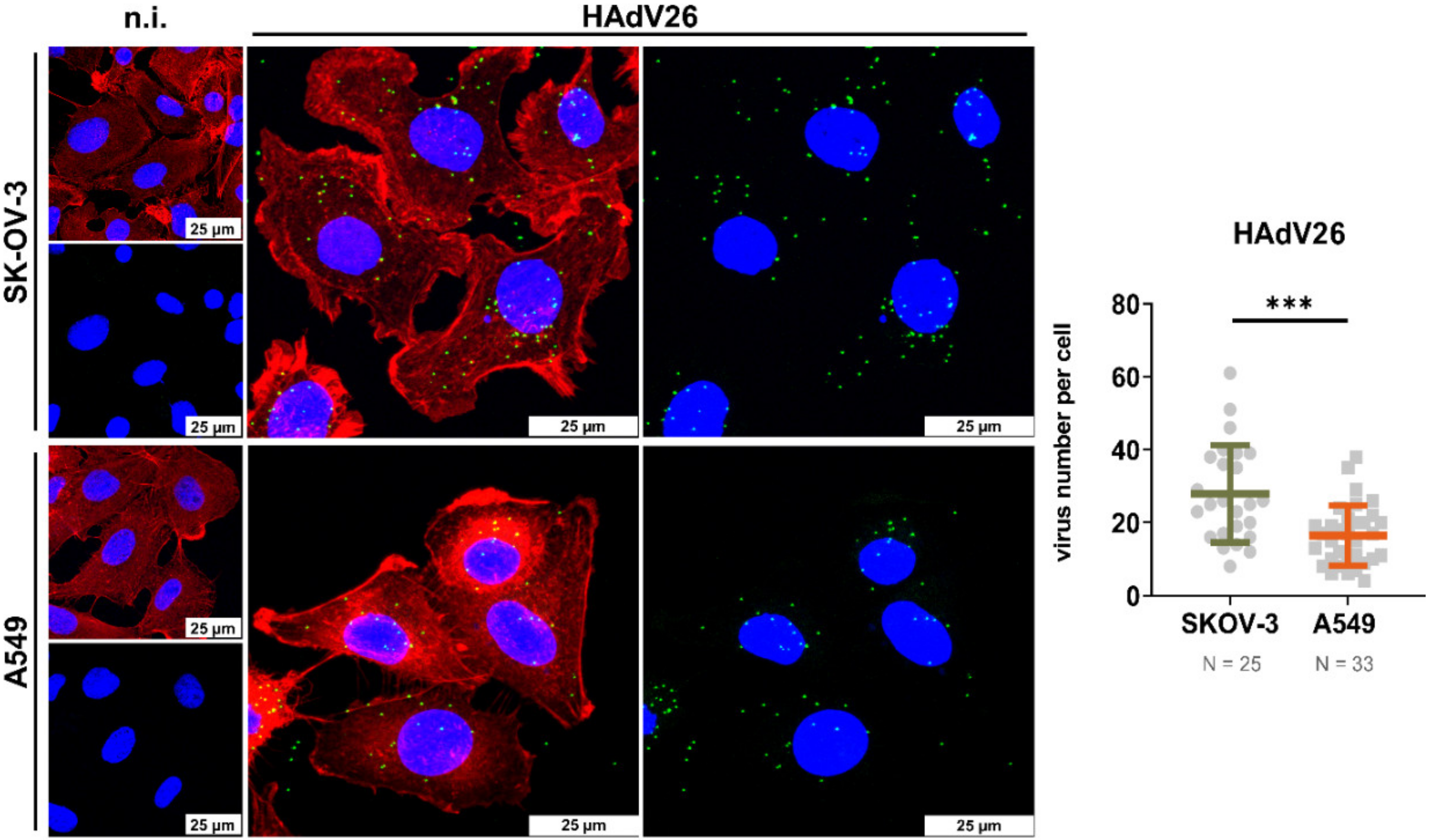
3.1. HAdV26 Infection Triggers Expression of Pro-Inflammatory Cytokines in Human Epithelial Cells In Vitro
3.2. HAdV26 Triggered IL-6 Gene Expression Is αvβ3 Integrin Dependent
3.3. HAdV26-Induced IL-6 Gene Expression Is NF-κB Mediated
3.4. HAdV26-Induced Production of IL-6 Is αvβ3 Integrin Dependent
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Majhen, D.; Calderon, H.; Chandra, N.; Fajardo, C.A.; Rajan, A.; Alemany, R.; Custers, J. Adenovirus-Based Vaccines for Fighting Infectious Diseases and Cancer: Progress in the Field. Hum. Gene Ther. 2014, 25, 301–317. [Google Scholar] [CrossRef] [PubMed]
- Madisch, I.; Harste, G.; Pommer, H.; Heim, A. Phylogenetic Analysis of the Main Neutralization and Hemagglutination Determinants of All Human Adenovirus Prototypes as a Basis for Molecular Classification and Taxonomy. J. Virol. 2005, 79, 15265–15276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, M.P.; Seto, J.; Liu, E.B.; Dehghan, S.; Hudson, N.R.; Lukashev, A.N.; Ivanova, O.; Chodosh, J.; Dyer, D.W.; Jones, M.S.; et al. Computational Analysis of Two Species C Human Adenoviruses Provides Evidence of a Novel Virus. J. Clin. Microbiol. 2011, 49, 3482–3490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, C.M.; Singh, G.; Henquell, C.; Walsh, M.P.; Peigue-Lafeuille, H.; Seto, D.; Jones, M.S.; Dyer, D.W.; Chodosh, J. Computational analysis and identification of an emergent human adenovirus pathogen implicated in a respiratory fatality. Virology 2011, 409, 141–147. [Google Scholar] [CrossRef]
- Gallardo, J.; Pérez-Illana, M.; Martín-González, N.; Martín, C.S. Adenovirus Structure: What Is New? Int. J. Mol. Sci. 2021, 22, 5240. [Google Scholar] [CrossRef]
- Tamanini, A.; Nicolis, E.; Bonizzato, A.; Bezzerri, V.; Melotti, P.; Assael, B.M.; Cabrini, G. Interaction of Adenovirus Type 5 Fiber with the Coxsackievirus and Adenovirus Receptor Activates Inflammatory Response in Human Respiratory Cells. J. Virol. 2006, 80, 11241–11254. [Google Scholar] [CrossRef] [Green Version]
- Wolfrum, N.; Greber, U.F. Adenovirus signalling in entry. Cell. Microbiol. 2012, 15, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Fausther-Bovendo, H.; Kobinger, G.P. Pre-existing immunity against Ad vectors: Humoral, cellular, and innate response, what’s important? Hum. Vaccin. Immunother. 2014, 10, 2875–2884. [Google Scholar]
- Custers, J.; Kim, D.; Leyssen, M.; Gurwith, M.; Tomaka, F.; Robertson, J.; Heijnen, E.; Condit, R.; Shukarev, G.; Heerwegh, D.; et al. Vaccines based on replication incompetent Ad26 viral vectors: Standardized template with key considerations for a risk/benefit assessment. Vaccine 2021, 39, 3081–3101. [Google Scholar] [CrossRef]
- Baden, L.R.; Karita, E.; Mutua, G.; Bekker, L.-G.; Gray, G.; Page-Shipp, L.; Walsh, S.R.; Nyombayire, J.; Anzala, O.; Roux, S.; et al. Assessment of the Safety and Immunogenicity of 2 Novel Vaccine Platforms for HIV-1 Prevention: A Randomized Trial. Ann. Intern. Med. 2016, 164, 313–322. [Google Scholar] [CrossRef]
- Anywaine, Z.; Whitworth, H.; Kaleebu, P.; PrayGod, G.; Shukarev, G.; Manno, D.; Kapiga, S.; Grosskurth, H.; Kalluvya, S.; Bockstal, V.; et al. Safety and Immunogenicity of a 2-Dose Heterologous Vaccination Regimen with Ad26.ZEBOV and MVA-BN-Filo Ebola Vaccines: 12-Month Data From a Phase 1 Randomized Clinical Trial in Uganda and Tanzania. J. Infect. Dis. 2019, 220, 46–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barouch, D.H.; Liu, J.; Peter, L.; Abbink, P.; Iampietro, M.J.; Cheung, A.; Alter, G.; Chung, A.; Dugast, A.-S.; Frahm, N.; et al. Characterization of Humoral and Cellular Immune Responses Elicited by a Recombinant Adenovirus Serotype 26 HIV-1 Env Vaccine in Healthy Adults (IPCAVD 001). J. Infect. Dis. 2012, 207, 248–256. [Google Scholar] [CrossRef] [PubMed]
- van der Fits, L.; Bolder, R.; Heemskerk-van der Meer, M.; Drijver, J.; van Polanen, Y.; Serroyen, J.; Langedijk, J.P.M.; Schuitemaker, H.; Saeland, E.; Zahn, R. Adenovector 26 Encoded Prefusion Conformation Stabilized RSV-F Protein Induces Long-Lasting Th1-Biased Immunity in Neonatal Mice. npj Vaccines 2020, 5, 49. [Google Scholar] [CrossRef]
- Teigler, J.E.; Iampietro, M.J.; Barouch, D.H. Vaccination with Adenovirus Serotypes 35, 26, and 48 Elicits Higher Levels of Innate Cytokine Responses than Adenovirus Serotype 5 in Rhesus Monkeys. J. Virol. 2012, 86, 9590–9598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.-H.; Huang, Y.; Issekutz, A.C.; Griffith, M.; Lin, K.-H.; Anderson, R. Interleukin-1α Released from Epithelial Cells after Adenovirus Type 37 Infection Activates Intercellular Adhesion Molecule 1 Expression on Human Vascular Endothelial Cells. J. Virol. 2002, 76, 427–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbink, P.; Lemckert, A.A.C.; Ewald, B.A.; Lynch, D.M.; Denholtz, M.; Smits, S.; Holterman, L.; Damen, I.; Vogels, R.; Thorner, A.R.; et al. Comparative Seroprevalence and Immunogenicity of Six Rare Serotype Recombinant Adenovirus Vaccine Vectors from Subgroups B and D. J. Virol. 2007, 81, 4654–4663. [Google Scholar] [CrossRef] [Green Version]
- Vogels, R.; Zuijdgeest, D.; van Rijnsoever, R.; Hartkoorn, E.; Damen, I.; de Béthune, M.-P.; Kostense, S.; Penders, G.; Helmus, N.; Koudstaal, W.; et al. Replication-Deficient Human Adenovirus Type 35 Vectors for Gene Transfer and Vaccination: Efficient Human Cell Infection and Bypass of Preexisting Adenovirus Immunity. J. Virol. 2003, 77, 8263–8271. [Google Scholar] [CrossRef] [Green Version]
- Nestić, D.; Uil, T.G.; Ma, J.; Roy, S.; Vellinga, J.; Baker, A.H.; Custers, J.; Majhen, D. Avβ3 Integrin Is Required for Efficient Infection of Epithelial Cells with Human Adenovirus Type 26. J. Virol. 2019, 93, e01474-18. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar]
- Hendrickx, R.; Stichling, N.; Koelen, J.; Kuryk, L.; Lipiec, A.; Greber, U.F. Innate Immunity to Adenovirus. Hum. Gene Ther. 2014, 25, 265–284. [Google Scholar] [CrossRef] [Green Version]
- Gianni, T.; Leoni, V.; Chesnokova, L.S.; Hutt-Fletcher, L.M.; Campadelli-Fiume, G. αvβ3-integrin is a major sensor and activator of innate immunity to herpes simplex virus-1. Proc. Natl. Acad. Sci. USA 2012, 109, 19792–19797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casiraghi, C.; Gianni, T.; Campadelli-Fiume, G. αvβ3 Integrin Boosts the Innate Immune Response Elicited in Epithelial Cells through Plasma Membrane and Endosomal Toll-Like Receptors. J. Virol. 2016, 90, 4243–4248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; A Muruve, D. Molecular basis of the inflammatory response to adenovirus vectors. Gene Ther. 2003, 10, 935–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-ΚB Signaling in Inflammation. Signal Transduct. Target Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Rhee, M.H.; Kim, E.; Cho, J.Y. BAY 11-7082 Is a Broad-Spectrum Inhibitor with Anti-Inflammatory Activity against Multiple Targets. Mediat. Inflamm. 2012, 2012, 416036. [Google Scholar] [CrossRef]
- Mercier, S.; Gahéry-Segard, H.; Monteil, M.; Lengagne, R.; Guillet, J.-G.; Eloit, M.; Denesvre, C. Distinct Roles of Adenovirus Vector-Transduced Dendritic Cells, Myoblasts, and Endothelial Cells in Mediating an Immune Response against a Transgene Product. J. Virol. 2002, 76, 2899–2911. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.H.; Kushwah, R.; Wu, J.; Ng, P.; Palaniyar, N.; Grinstein, S.; Philpott, D.J.; Hu, J. Adenoviral vectors stimulate innate immune responses in macrophages through cross-talk with epithelial cells. Immunol. Lett. 2010, 134, 93–102. [Google Scholar] [CrossRef]
- Coughlan, L. Factors Which Contribute to the Immunogenicity of Non-replicating Adenoviral Vectored Vaccines. Front. Immunol. 2020, 11, 909. [Google Scholar] [CrossRef]
- Yoon, J.-S.; Kim, H.-H.; Lee, Y.; Lee, J.-S. Cytokine induction by respiratory syncytial virus and adenovirus in bronchial epithelial cells. Pediatr. Pulmonol. 2007, 42, 277–282. [Google Scholar] [CrossRef]
- Lam, E.; Ramke, M.; Warnecke, G.; Schrepfer, S.; Kopfnagel, V.; Dobner, T.; Heim, A. Effective Apical Infection of Differentiated Human Bronchial Epithelial Cells and Induction of Proinflammatory Chemokines by the Highly Pneumotropic Human Adenovirus Type 14p1. PLoS ONE 2015, 10, e0131201. [Google Scholar] [CrossRef] [Green Version]
- Booth, J.L.; Metcalf, J.P. Type-Specific Induction of Interleukin-8 by Adenovirus. Am. J. Respir. Cell Mol. Biol. 1999, 21, 521–527. [Google Scholar] [CrossRef]
- Alcorn, M.J.; Booth, J.L.; Coggeshall, K.M.; Metcalf, J.P. Adenovirus Type 7 Induces Interleukin-8 Production via Activation of Extracellular Regulated Kinase 1/2. J. Virol. 2001, 75, 6450–6459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Natarajan, K.; Rajala, M.S.; Chodosh, J. Corneal IL-8 Expression Following Adenovirus Infection Is Mediated by c-Src Activation in Human Corneal Fibroblasts. J. Immunol. 2003, 170, 6234–6243. [Google Scholar] [CrossRef] [PubMed]
- Rajaiya, J.; Xiao, J.; Rajala, R.V.; Chodosh, J. Human adenovirus type 19 infection of corneal cells induces p38 MAPK-dependent interleukin-8 expression. Virol. J. 2008, 5, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Paolo, N.C.; Miao, E.; Iwakura, Y.; Murali-Krishna, K.; Aderem, A.; Flavell, R.A.; Papayannopoulou, T.; Shayakhmetov, D.M. Virus Binding to a Plasma Membrane Receptor Triggers Interleukin-1α-Mediated Proinflammatory Macrophage Response In Vivo. Immunity 2009, 31, 110–121. [Google Scholar] [CrossRef] [Green Version]
- Browne, A.; Tookman, L.A.; Ingemarsdotter, C.K.; Bouwman, R.D.; Pirlo, K.; Wang, Y.; McNeish, I.; Lockley, M. Pharmacological Inhibition of β3 Integrin Reduces the Inflammatory Toxicities Caused by Oncolytic Adenovirus without Compromising Anticancer Activity. Cancer Res. 2015, 75, 2811–2821. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.; Wang, Y.; Wang, H.; Deng, J. Adenovirus 7 Induces Interlukin-6 Expression in Human Airway Epithelial Cells via P38/NF-ΚB Signaling Pathway. Front. Immunol. 2020, 11, 551413. [Google Scholar] [CrossRef]
- Liu, Q.; White, L.R.; Clark, S.A.; Heffner, D.J.; Winston, B.W.; Tibbles, L.A.; Muruve, D.A. Akt/Protein Kinase B Activation by Adenovirus Vectors Contributes to NFkappaB-Dependent CXCL10 Expression. J. Virol. 2005, 79, 14507–14515. [Google Scholar] [CrossRef] [Green Version]
- Machitani, M.; Sakurai, F.; Wakabayashi, K.; Nakatani, K.; Shimizu, K.; Tachibana, M.; Mizuguchi, H. NF-ΚB Promotes Leaky Expression of Adenovirus Genes in a Replication-Incompetent Adenovirus Vector. Sci. Rep. 2016, 6, 19922. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Sun, F.; Han, L.; Qu, Z. Kaposi’s Sarcoma Herpesvirus (KSHV) MicroRNA K12-1 Functions as an Oncogene by Activating NF-ΚB/IL-6/STAT3 Signaling. Oncotarget 2016, 7, 33363–33373. [Google Scholar] [CrossRef]
- Morgan, E.L.; Macdonald, A. Autocrine STAT3 activation in HPV positive cervical cancer through a virus-driven Rac1-NFκB-IL-6 signalling axis. PLoS Pathog. 2019, 15, e1007835. [Google Scholar]







| Gene ID | Primer Sequences |
|---|---|
| IL-6 | F: 5′CAATGAGGAGACTTGCCTGG3′ R: 5′GCACAGCTCTGGCTTGTTCC3′ |
| IL-8 | F: 5′GTTTTTGAAGAGGGCTGAGAATTC3′ R: 5′ATGAAGTGTTGAAGTAGATTTGCTTG3′ |
| IL-1β | F: 5′TGGCAATGAGGATGACTTGTTC3′ R: 5′CTGTAGTGGTGGTCGGAGATT3′ |
| TNF-α | F: 5′AACCTCCTCTCTGCCATCAA3′ R: 5′GGAAGACCCCTCCCAGATAG3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nestić, D.; Božinović, K.; Drašković, I.; Kovačević, A.; van den Bosch, J.; Knežević, J.; Custers, J.; Ambriović-Ristov, A.; Majhen, D. Human Adenovirus Type 26 Induced IL-6 Gene Expression in an αvβ3 Integrin- and NF-κB-Dependent Manner. Viruses 2022, 14, 672. https://doi.org/10.3390/v14040672
Nestić D, Božinović K, Drašković I, Kovačević A, van den Bosch J, Knežević J, Custers J, Ambriović-Ristov A, Majhen D. Human Adenovirus Type 26 Induced IL-6 Gene Expression in an αvβ3 Integrin- and NF-κB-Dependent Manner. Viruses. 2022; 14(4):672. https://doi.org/10.3390/v14040672
Chicago/Turabian StyleNestić, Davor, Ksenija Božinović, Isabela Drašković, Alen Kovačević, Jolien van den Bosch, Jelena Knežević, Jerome Custers, Andreja Ambriović-Ristov, and Dragomira Majhen. 2022. "Human Adenovirus Type 26 Induced IL-6 Gene Expression in an αvβ3 Integrin- and NF-κB-Dependent Manner" Viruses 14, no. 4: 672. https://doi.org/10.3390/v14040672
APA StyleNestić, D., Božinović, K., Drašković, I., Kovačević, A., van den Bosch, J., Knežević, J., Custers, J., Ambriović-Ristov, A., & Majhen, D. (2022). Human Adenovirus Type 26 Induced IL-6 Gene Expression in an αvβ3 Integrin- and NF-κB-Dependent Manner. Viruses, 14(4), 672. https://doi.org/10.3390/v14040672











