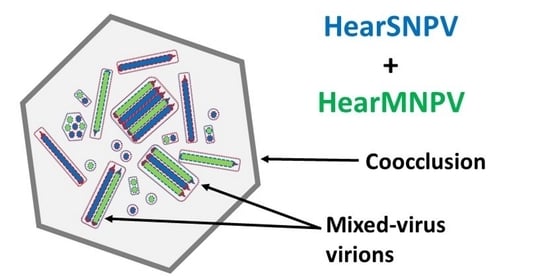
Coocclusion of Helicoverpa armigera Single Nucleopolyhedrovirus (HearSNPV) and Helicoverpa armigera Multiple Nucleopolyhedrovirus (HearMNPV): Pathogenicity and Stability in Homologous and Heterologous Hosts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Insect Colonies, Viruses and Cell Line
2.2. Biological Activity of HearSNPV and HearMNPV OBs
2.3. Superinfection with HearMNPV and HearSNPV at Different Time Intervals
2.4. Quantification of HearMNPV and HearSNPV Genomes in OB Samples
2.5. Analysis of Mixed-Virus ODVs
2.6. Biological Activity of Cooccluded Mixed-Virus OBs
2.7. Stability of Mixed-Virus Preparations during Serial Passage in Larvae
3. Results
3.1. Insecticidal Characteristics of HearSNPV and HearMNPV OBs
3.2. Relative Prevalence of HearSNPV and HearMNPV Genomes in Mixtures
3.3. HearSNPV and HearMNPV Genomes Are Present within the Same ODV
3.4. Biological Activity of Cooccluded Mixtures
3.5. Host Range and Stability of Mixed-Virus Preparations in Serial Passage
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arnemann, J.A.; Roxburgh, S.; Walsh, T.; Guedes, J.; Gordon, K.; Smagghe, G.; Tay, W.T. Multiple incursion pathways for Helicoverpa armigera in Brazil show its genetic diversity spreading in a connected world. Sci. Rep. 2019, 9, 19380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mironidis, G.K.; Kapantaidaki, D.; Bentila, M.; Morou, E.; Savopoulou-Soultani, M.; Vontas, J. Resurgence of the cotton bollworm Helicoverpa armigera in northern Greece associated with insecticide resistance. Insect Sci. 2013, 20, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Sun, X. History and current status of development and use of viral insecticides in China. Viruses 2015, 7, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Grzywacz, D.; Moore, S. Production, formulation, and bioassay of baculoviruses for pest control. In Microbial Control of Insect and Mite Pests; Lacey, L.A., Ed.; Academic Press: San Diego, CA, USA, 2017; pp. 109–124. [Google Scholar]
- Harrison, R.L.; Herniou, E.A.; Jehle, J.A.; Theilmann, D.A.; Burand, J.P.; Krell, P.J.; van Oers, M.M.; Mowery, J.D. ICTV virus taxonomy profile: Baculoviridae. J. Gen. Virol. 2019, 99, 1185–1186. [Google Scholar] [CrossRef] [PubMed]
- Sajjan, D.B.; Hinchigeri, S.B. Structural organization of baculovirus occlusion bodies and protective role of multilayered polyhedron envelope protein. Food Environ. Virol. 2016, 8, 86–100. [Google Scholar] [CrossRef]
- Rohrmann, G.F. Baculovirus Molecular Biology, 4th ed.; National Center for Biotechnology Information: Bethesda, MD, USA, 2019. Available online: https://www.ncbi.nlm.nih.gov/books/NBK543458/ (accessed on 21 December 2021).
- Erlandson, M.A.; Toprak, U.; Hegedus, D.D. Role of the peritrophic matrix in insect-pathogen interactions. J. Ins. Physiol. 2019, 117, 103894. [Google Scholar] [CrossRef]
- Passarelli, A.L. Barriers to success: How baculoviruses establish efficient systemic infections. Virology 2011, 411, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Williams, T. Viruses. In Ecology of Invertebrate Diseases; Hajek, A.E., Shapiro-Ilan, D.I., Eds.; Wiley: Chichester, UK, 2018; pp. 215–285. [Google Scholar]
- Erlandson, M. Genetic variation in field populations of baculoviruses: Mechanisms for generating variation and its potential role in baculovirus epizootiology. Virol. Sin. 2009, 24, 458–469. [Google Scholar] [CrossRef]
- Masson, T.; Fabre, M.L.; Pidre, M.L.; Niz, J.M.; Berretta, M.F.; Romanowski, V.; Ferrelli, M.L. Genomic diversity in a population of Spodoptera frugiperda nucleopolyhedrovirus. Infect. Genet. Evol. 2021, 90, 104749. [Google Scholar] [CrossRef]
- Clavijo, G.; Williams, T.; Muñoz, D.; Caballero, P.; López-Ferber, M. Mixed genotype transmission bodies and virions contribute to the maintenance of diversity in an insect virus. Proc. R. Soc. B Biol. Sci. 2010, 277, 943–951. [Google Scholar] [CrossRef] [Green Version]
- Bernal, A.; Simón, O.; Williams, T.; Muñoz, D.; Caballero, P. A Chrysodeixis chalcites single nucleopolyhedrovirus population from the Canary Islands is genotypically structured to maximize survival. Appl. Environ. Microbiol. 2013, 79, 7709–7718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beperet, I.; Simón, O.; López-Ferber, M.; van Lent, J.; Williams, T.; Caballero, P. Mixtures of insect pathogenic viruses in a single virion: Towards the development of custom designed insecticides. Appl. Environ. Microbiol. 2021, 87, e02180-20A. [Google Scholar] [CrossRef] [PubMed]
- Bull, J.C.; Godfray, H.C.J.; O’Reilly, D.R. Persistence of an occlusion-negative recombinant nucleopolyhedrovirus in Trichoplusia ni indicates high multiplicity of cellular infection. Appl. Environ. Microbiol. 2001, 67, 5204–5209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cory, J.S.; Green, B.M.; Paul, R.K.; Hunter-Fujita, F. Genotypic and phenotypic diversity of a baculovirus population within an individual insect host. J. Invertebr. Pathol. 2005, 89, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Baillie, V.L.; Bouwer, G. The effect of inoculum dose on the genetic diversity detected within Helicoverpa armigera nucleopolyhedrovirus populations. J. Gen. Virol. 2013, 94, 2524–2529. [Google Scholar] [CrossRef] [PubMed]
- Chateigner, A.; Bézier, A.; Labrousse, C.; Jiolle, D.; Barbe, V.; Herniou, E.A. Ultra deep sequencing of a baculovirus population reveals widespread genomic variations. Viruses 2015, 7, 3625–3646. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Chen, Y.; Zhao, Y.; Liu, X.; Dong, B.; Jones, I.M.; Chen, H. Baculovirus superinfection: A probable restriction factor on the surface display of proteins for library screening. PLoS ONE 2013, 8, e54631. [Google Scholar] [CrossRef] [Green Version]
- Beperet, I.; Irons, S.; Simón, O.; King, L.A.; Williams, T.; Possee, R.D.; López-Ferber, M.; Caballero, P. Superinfection exclusion in alphabaculovirus infections is concomitant with actin reorganization. J. Virol. 2014, 88, 3548–3556. [Google Scholar] [CrossRef] [Green Version]
- Gettig, R.R.; McCarthy, W.J. Genotypic variation among isolates of Heliothis spp. nuclear polyhedrosis viruses from different geographical regions. Virology 1982, 117, 245–251. [Google Scholar] [CrossRef]
- Figueiredo, E.; Muñoz, D.; Escribano, A.; Mexia, A.; Vlak, J.M.; Caballero, P. Biochemical identification and comparative insecticidal activity of nucleopolyhedrovirus pathogenic for Heliothis armigera (Lep. Noctuidae) larvae. J. Appl. Entomol. 1999, 123, 165–169. [Google Scholar] [CrossRef]
- Ogembo, J.G.; Kunjeku, E.C.; Sithanantham, S. A preliminary study on the pathogenicity of two isolates of nucleopolyhedroviruses infecting the African bollworm, Helicoverpa armigera (Lepidoptera: Noctuidae). Int. J. Trop. Ins. Sci. 2005, 25, 218–222. [Google Scholar] [CrossRef]
- Zhang, C.X.; Ma, X.C.; Guo, Z.J. Comparison of complete genome sequence between C1 and G4 isolates of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus. Virology 2005, 333, 190–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, C.F.; Payne, C.C. The susceptibility of Heliothis armigera to three nuclear polyhedrosis viruses. Ann. Appl. Biol. 1984, 104, 405–412. [Google Scholar] [CrossRef]
- Sun, X.L.; Zhang, G.Y. Comparison of four Heliothis armigera NPV isolates. Virol. Sin. 1994, 12, 309–318. [Google Scholar]
- Figueiredo, E.; Muñoz, D.; Murillo, R.; Mexia, A.; Caballero, P. Diversity of Iberian nucleopolyhedrovirus wild-type isolates infecting Helicoverpa armigera (Lepidoptera: Noctuidae). Biol. Control 2009, 50, 43–49. [Google Scholar] [CrossRef] [Green Version]
- Ogembo, J.G.; Caoili, B.L.; Shikata, M.; Chaeychomsri, S.; Kobayashi, M.; Ikeda, M. Comparative genomic sequence analysis of novel Helicoverpa armigera nucleopolyhedrovirus (NPV) isolated from Kenya and three other previously sequenced Helicoverpa spp. NPV. Virus Genes 2009, 39, 261–272. [Google Scholar] [CrossRef]
- Rowley, D.L.; Popham, H.J.; Harrison, R.L. Genetic variation and virulence of nucleopolyhedroviruses isolated worldwide from the heliothine pests Helicoverpa armigera, Helicoverpa zea, and Heliothis virescens. J. Invertebr. Pathol. 2011, 107, 112–126. [Google Scholar] [CrossRef]
- Arrizubieta, M.; Simón, O.; Williams, T.; Caballero, P. Genomic sequence of five Helicoverpa armigera nucleopolyhedrovirus genotypes from Spain that differ in their insecticidal properties. Genome Anounc. 2015, 3, e00548-15. [Google Scholar] [CrossRef] [Green Version]
- Raghavendra, A.T.; Jalali, S.K.; Ojha, R.; Shivalingaswamy, T.M.; Bhatnagar, R. Whole genome sequence and comparative genomic sequence analysis of Helicoverpa armigera nucleopolyhedrovirus (HearNPV-L1) isolated from India. VirusDisease 2017, 28, 61–68. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Jagadish, K.S.; Tak, K.R.; Peter, A. Genetic diversity among different geographical isolates of the gram pod borer, Helicoverpa armigera (Hübner)(Lepidoptera: Noctuidae) nucleopolyhedrosis virus (Hear NPV). Egypt. J. Biol. Pest Contr. 2019, 29, 61. [Google Scholar] [CrossRef] [Green Version]
- Eroglu, G.B.; Inan, C.; Nalcacioglu, R.; Demirbag, Z. Genome sequence analysis of a Helicoverpa armigera single nucleopolyhedrovirus (HearNPV-TR) isolated from Heliothis peltigera in Turkey. PLoS ONE 2020, 15, e0234635. [Google Scholar] [CrossRef] [PubMed]
- Gröner, A. Specificity and safety of baculoviruses, In The Biology of Baculoviruses: Biological Properties and Molecular Biology; Federici, B.B., Granados, R.R., Eds.; CRC: Boca Ratón, FL, USA, 1986; Volume 1, pp. 177–202. [Google Scholar]
- Tang, P.; Zhang, H.; Li, Y.; Han, B.; Wang, G.; Qin, Q.; Zhang, Z. Genomic sequencing and analyses of HearMNPV—A new multinucleocapsid nucleopolyhedrovirus isolated from Helicoverpa armigera. Virol. J. 2012, 9, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rovesti, L.; Crook, N.E.; Winstanley, D. Biological and biochemical relationships between the nucleopolyhedroviruses of Mamestra brassicae and Heliothis armigera. J. Invertebr. Pathol. 2000, 75, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Tompkins, G.J.; Dougherty, E.M.; Adams, J.R.; Diggs, D. Changes in the virulence of nuclear polyhedrosis viruses when propagated in alternate noctuid (Lepidoptera: Noctuidae) cell lines and hosts. J. Econ. Entomol. 1988, 81, 1027–1032. [Google Scholar] [CrossRef]
- Belda, I.M.; Beperet, I.; Williams, T.; Caballero, P. Genetic variation and biological stability of two closely related alphabaculoviruses during serial passage in permissive and semi-permissive heterologous hosts. Viruses 2019, 11, 660. [Google Scholar] [CrossRef] [Green Version]
- Moore, S.D.; Bouwer, G.; Pittway, T.M. Evaluation of Helicoverpa armigera nucleopolyhedrosis virus (HearNPV) for control of Helicoverpa armigera (Lepidoptera: Noctuidae) on citrus in South Africa. Biocontrol Sci. Technol. 2004, 14, 239–250. [Google Scholar] [CrossRef]
- Arrizubieta, M.; Williams, T.; Caballero, P.; Simón, O. Selection of a nucleopolyhedrovirus isolate from Helicoverpa armigera as the basis for a biological insecticide. Pest Manag. Sci. 2014, 70, 967–976. [Google Scholar] [CrossRef]
- Greene, G.L.; Leppla, N.C.; Dickerson, W.A. Velvetbean caterpillar: A rearing procedure and artificial medium. J. Econ. Entomol. 1976, 69, 487–488. [Google Scholar] [CrossRef]
- Mclntosh, A.H.; Ignoffo, C.M. Replication and infectivity of the single-embedded nuclear polyhedrosis virus, Baculovirus heliothis, in homologous cell lines. J. Invertebr. Pathol. 1981, 37, 258–264. [Google Scholar] [CrossRef]
- Hughes, P.R.; Wood, H.A. A synchronous peroral technique for the bioassay of insect viruses. J. Invertebr. Pathol. 1981, 37, 154–159. [Google Scholar] [CrossRef]
- Le Ora Software. POLO-PC a User’s Guide to Probit or Logit Analysis; LeOra Software: Berkeley, CA, USA, 1987. [Google Scholar]
- Crawley, M.J. GLIM for Ecologists; Blackwell: Oxford, UK, 1993. [Google Scholar]
- Guo, Z.J.; An, S.H.; Wang, D.; Liu, Y.H.; Kumar, V.S.; Zhang, C.X. Characterization of Ha29, a specific gene for Helicoverpa armigera single-nucleocapsid nucleopolyhedrovirus. J. Biochem. Mol. Biol. 2005, 38, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Jamovi Statistical Software 2021. Jamovi Project v.2.3.0.0. Available online: https://www.jamovi.org (accessed on 14 January 2022).
- Tabashnik, B.E. Evaluation of synergism among Bacillus thuringiensis toxins. Appl. Env. Microbiol. 1992, 58, 3343–3346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beperet-Arive, I. Regulation of Multiple Infection in Alphabaculoviruses: Critical Factors that Determine Success. Doctoral Thesis, Universidad Pública de Navarra, Pamplona, Spain, 2014. Available online: https://hdl.handle.net/2454/17404 (accessed on 23 December 2021).
- Williams, T.; López-Ferber, M.; Caballero, P. Nucleopolyhedrovirus coocclusion technology: A new concept in the development of biological insecticides. Front. Microbiol. 2022, 12, 810026. [Google Scholar] [CrossRef] [PubMed]
- Makalliwa, G.A.; Wang, X.; Zhang, H.; Zhang, N.; Chen, C.; Li, J.; Deng, F.; Wang, H.; Wang, M.; Hu, Z. HearNPV pseudotyped with PIF1, 2, and 3 from MabrNPV: Infectivity and complex stability. Virol. Sin. 2018, 33, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Torres-Vila, L.M.; Rodríguez-Molina, M.C.; Lacasa-Plasencia, A. Impact of Helicoverpa armigera larval density and crop phenology on yield and quality losses in processing tomato: Developing fruit count-based damage thresholds for IPM decision-making. Crop Protec. 2003, 22, 521–532. [Google Scholar] [CrossRef]
- Rojas, J.C.; Wyatt, T.D.; Birch, M.C. Flight and oviposition behavior toward different host plant species by the cabbage moth, Mamestra brassicae (L.) (Lepidoptera: Noctuidae). J. Insect Behav. 2000, 13, 247–254. [Google Scholar] [CrossRef]
- Cartea, M.E.; Francisco, M.; Lema, M.; Soengas, P.; Velasco, P. Resistance of cabbage (Brassica oleracea capitata group) crops to Mamestra brassicae. J. Econ. Entomol. 2010, 103, 1866–1874. [Google Scholar] [CrossRef] [PubMed]
- CABI. Invasive Species Compendium. Datasheet Helicoverpa armigera (Cotton Bollworm). 2021. Available online: https://www.cabi.org/isc/datasheet/26757#tohostsOrSpeciesAffected (accessed on 21 December 2021).
- Acharya, R.; Akintola, A.A.; Malekera, M.J.; Kamulegeya, P.; Nyakunga, K.B.; Mutimbu, M.K.; Shrestha, Y.K.; Hemayet, J.S.; Hoat, T.X.; Dao, H.T.; et al. Genetic relationship of fall armyworm (Spodoptera frugiperda) populations that invaded Africa and Asia. Insects 2021, 12, 439. [Google Scholar] [CrossRef]
- Sanches, M.M.; Guimarães, G.C.; Sihler, W.; Souza, M.L. Successful co-infection of two different baculovirus species in the same cell line reveals a potential strategy for large in vitro production. Braz. J. Microbiol. 2021, 52, 1835–1843. [Google Scholar] [CrossRef]
- Sanches, M.M.; Sihler, W.; Silva, C.E.P.; Guimarães, G.C.; Benito, N.P.; Sosa-Gómez, D.R.; de Souza, M.L. Characterization of a Chrysodeixis includens nucleopolyhedrovirus isolate from Brazilian cerrado and assessment of its co-infection with Anticarsia gemmatalis multiple nucleopolyhedrovirus. Braz. Arch. Biol. Technol. 2019, 62, e19180688. [Google Scholar] [CrossRef]
- Anonymous. AgBiTech launches lepidopteran biocontrol options. Outlooks Pest Manag. 2019, 30, 277–281. [Google Scholar]
- Pavan, O.H.O.; Ribeiro, H.C.T. Selection of a baculovirus strain with a bivalent insecticidal activity. Mem. Inst. Oswaldo Cruz 1989, 84, 63–65. [Google Scholar] [CrossRef] [Green Version]
- Kolodny-Hirsch, D.M.; Van Beek, N.A.M. Selection of a morphological variant of Autographa californica nuclear polyhedrosis virus with increased virulence following serial passage in Plutella xylostella. J. Invertebr. Pathol. 1997, 69, 205–211. [Google Scholar] [CrossRef]
- Graillot, B.; Blachère-López, C.; Besse, S.; Siegwart, M.; López-Ferber, M. Host range extension of Cydia pomonella granulovirus: Adaptation to oriental fruit moth, Grapholita molesta. BioControl 2016, 62, 19–27. [Google Scholar] [CrossRef]
- Shapiro, M.; Martignoni, M.E.; Cunningham, J.C.; Goodwin, R.H. Potential use of the saltmarsh caterpillar as a production host for nucleopolyhedrosis viruses. J. Econ. Entomol. 1982, 75, 69–71. [Google Scholar] [CrossRef]
- Kitchin, D.; Bouwer, G. Significant differences in the intra-host genetic diversity of Helicoverpa armigera nucleopolyhedrovirus dnapol after serial in vivo passages in the same insect population. Arch. Virol. 2018, 163, 713–718. [Google Scholar] [CrossRef] [PubMed]
- Arrizubieta, M.; Simón, O.; Williams, T.; Caballero, P. A novel binary mixture of Helicoverpa armigera single nucleopolyhedrovirus genotypic variants has improved insecticidal characteristics for control of cotton bollworms. Appl. Environ. Microbiol. 2015, 81, 3984–3993. [Google Scholar] [CrossRef] [Green Version]
- Simón, O.; Williams, T.; Caballero, P.; López-Ferber, M. Dynamics of deletion genotypes in an experimental insect virus population. Proc. R. Soc. B Biol. Sci. 2006, 273, 783–790. [Google Scholar] [CrossRef] [Green Version]
- Zwart, M.P.; Pijlman, G.P.; Sardanyés, J.; Duarte, J.; Januário, C.; Elena, S.F. Complex dynamics of defective interfering baculoviruses during serial passage in insect cells. J. Biol. Phys. 2013, 39, 327–342. [Google Scholar] [CrossRef] [Green Version]
- Hitchman, R.B.; Hodgson, D.J.; King, L.A.; Hails, R.S.; Cory, J.S.; Possee, R.D. Host mediated selection of pathogen genotypes as a mechanism for the maintenance of baculovirus diversity in the field. J. Invertebr. Pathol. 2007, 94, 153–162. [Google Scholar] [CrossRef]
- Hou, D.; Chen, X.; Zhang, L.K. Proteomic analysis of Mamestra brassicae nucleopolyhedrovirus progeny virions from two different hosts. PLoS ONE 2016, 11, e0153365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aguirre, E.; Beperet, I.; Williams, T.; Caballero, P. Generation of variability in Chrysodeixis includens nucleopolyhedrovirus (ChinNPV): The role of a single variant. Viruses 2021, 13, 1895. [Google Scholar] [CrossRef] [PubMed]
- Clavijo, G.; Williams, T.; Muñoz, D.; López-Ferber, M.; Caballero, P. Entry into midgut epithelial cells is a key step in the selection of genotypes in a nucleopolyhedrovirus. Virol. Sin. 2009, 24, 350–358. [Google Scholar] [CrossRef]
- Zwart, M.P.; Hemerik, L.; Cory, J.S.; de Visser, J.A.G.M.; Bianchi, F.J.J.A.; Van Oers, M.M.; Vlak, J.M.; Hoekstra, R.F.; Van der Werf, W. An experimental test of the independent action hypothesis in virus–insect pathosystems. Proc. R. Soc. B Biol. Sci. 2009, 276, 2233–2242. [Google Scholar] [CrossRef] [Green Version]
- Mu, J.; van Lent, J.W.; Smagghe, G.; Wang, Y.; Chen, X.; Vlak, J.M.; van Oers, M.M. Live imaging of baculovirus infection of midgut epithelium cells: A functional assay of per os infectivity factors. J. Gen. Virol. 2014, 95, 2531–2539. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Shang, Y.; Chen, C.; Liu, S.; Chang, M.; Zhang, N.; Hu, H.; Zhang, F.; Zhang, T.; Wang, Z.; et al. Baculovirus per os infectivity factor complex: Components and assembly. J. Virol. 2019, 93, e02053-18. [Google Scholar] [CrossRef] [Green Version]
- López-Ferber, M.; Simón, O.; Williams, T.; Caballero, P. Defective or effective? Mutualistic interactions between virus genotypes. Proc. R. Soc. B Biol. Sci. 2003, 270, 2249–2255. [Google Scholar] [CrossRef] [Green Version]
- Zwart, M.P.; Elena, S.F. Matters of size: Genetic bottlenecks in virus infection and their potential impact on evolution. Annu. Rev. Virol. 2015, 2, 161–179. [Google Scholar] [CrossRef]
- Washburn, J.O.; Trudeau, D.; Wong, J.F.; Volkman, L.E. Early pathogenesis of Autographa californica multiple nucleopolyhedrovirus and Helicoverpa zea single nucleopolyhedrovirus in Heliothis virescens: A comparison of the ‘M’ and ‘S’ strategies for establishing fatal infection. J. Gen. Virol. 2003, 84, 343–351. [Google Scholar] [CrossRef]





| Virus | LC50 (OBs/mL) | Potency | Range of 95% C.I. | MTD (h) | Range of 95% C.I. | Mean OB Production (±SE) (×107) |
|---|---|---|---|---|---|---|
| HearMNPV | 1.8 × 105 a | 1 | - | 143.4a | 140.4–146.5 | 6.1 ± 1.2 a |
| HearSNPV | 2.9 × 104 b | 6.2 | 4.0–10.3 | 130.0b | 127.3–132.7 | 11 ± 2.4 b |
| Single Nucleocapsid ODVs | Multinucleocapsid ODVs 1 | |||
|---|---|---|---|---|
| Sample | HearSNPV | HearMNPV | HearSNPV | HearMNPV |
| T0 | 99.78 | 0.22 | - | - |
| T12 | 97.09 | 2.91 | 0.47 | 99.53 |
| T24 | 98.73 | 1.27 | 0.88 | 99.12 |
| T48 | 55.77 | 44.23 | 0.00 | 100.00 |
| T72 | 47.73 | 52.27 | 0.00 | 100.00 |
| Sample | Replicate | Log[Copy Number] (mean ± SD) | Cq Value (mean ± SD) |
|---|---|---|---|
| Upper band | Replicate 1 | 6.686 ± 0.096 | 11.16 ± 0.21 |
| Replicate 2 | 6.743 ± 0.050 | 11.01 ± 0.13 | |
| Replicate 3 | 6.019 ± 0.027 | 12.83 ± 0.07 | |
| Replicate 4 | 6.041 ± 0.090 | 12.77 ± 0.23 | |
| Mean ± SD | 6.372 ± 0.396 | 11.94 ± 0.99 | |
| Lower bands | Replicate 1 | 1.355 ± 0.062 | 24.49 ± 0.16 |
| Replicate 2 | 1.462 ± 0.037 | 24.22 ± 0.09 | |
| Replicate 3 | 1.312 ± 0.049 | 24.60 ± 0.12 | |
| Replicate 4 | 1.471 ± 0.175 | 24.20 ± 0.44 | |
| Mean ± SD | 1.400 ± 0.079 | 25.95 ± 0.20 |
| T12 | T24 | |||||
|---|---|---|---|---|---|---|
| Repetition 1 | Repetition 2 | Repetition 3 | Repetition 1 | Repetition 2 | Repetition 3 | |
| No. positive wells/Total 1 | 11/88 | 8/88 | 8/88 | 7/88 | 11/88 | 13/88 |
| Probability (0) | 0.875 | 0.909 | 0.909 | 0.920 | 0.875 | 0.852 |
| Probability (1) | 0.117 | 0.0866 | 0.0866 | 0.0763 | 0.117 | 0.136 |
| Probability (2) | 0.00780 | 0.00413 | 0.00413 | 0.00316 | 0.00780 | 0.0109 |
| Probability (3) | 0.000347 | 0.000131 | 0.000131 | 0.0000874 | 0.000347 | 0.000580 |
| LC50 | Relative | 95% Fiducial Limits | Expected | |||
|---|---|---|---|---|---|---|
| Host | Virus Treatment 1 | (OBs/mL) | Potency | Low | High | LC50m |
| H. armigera | HearMNPV | 1.0 × 105 | 1 | - | - | - |
| HearSNPV | 1.5 × 104 | 6.6 | 2.5 | 17.2 | - | |
| T12 mixed-virus | 2.2 × 104 | 4.5 | 1.9 | 10.8 | 3.1 × 104 | |
| T24 mixed-virus | 3.5 × 104 | 2.9 | 1.5 | 5.9 | 2.4 × 104 | |
| 50% SNPV + 50% MNPV | 3.0 × 104 | 3.4 | 1.6 | 7.1 | 2.7 × 104 | |
| S. frugiperda | HearMNPV | 2.0 × 107 | 1 | - | - | - |
| HearSNPV | - | - | - | - | - | |
| T12 mixed-virus | 3.8 × 107 | 0.5 | 0.3 | 0.9 | 3.4 × 107 | |
| T24 mixed-virus | 3.6 × 107 | 0.6 | 0.3 | 0.9 | 4.7 × 107 | |
| 50% SNPV + 50% MNPV | 4.2 × 107 | 0.5 | 0.3 | 0.8 | 4.0 × 107 | |
| M. brassicae | HearMNPV | 2.0 × 105 | 1 | - | - | - |
| HearSNPV | - | - | - | - | - | |
| T12 mixed-virus | 3.9 × 105 | 0.5 | 0.3 | 0.9 | 3.3 × 105 | |
| T24 mixed-virus | 4.1 × 105 | 0.5 | 0.2 | 0.9 | 4.6 × 105 | |
| 50% SNPV + 50% MNPV | 5.5 × 105 | 0.4 | 0.2 | 0.7 | 3.9 × 105 | |
| Host Species | Virus Sample | P0 1 | P1 | P2 | P3 | P4 | P5 |
|---|---|---|---|---|---|---|---|
| H. armigera | T0 | 96.76 | 98.92147 | - | - | - | - |
| T12 | 41.01 | 99.91097 | 100.00 | 100.00 | 100.00 | 100.00 | |
| T24 | 57.34 | 96.46719 | 100.00 | 100.00 | 100.00 | 100.00 | |
| T48 | 3.20 | 63.90424 | - | - | - | - | |
| T72 | 4.41 | 88.43874 | - | - | - | - | |
| 50% SNPV + 50% MNPV | 50.00 | 97.88138 | 100.00 | 100.00 | 100.00 | 100.00 | |
| S. frugiperda | T0 | 96.76 | 0.00008 | - | - | - | - |
| T12 | 41.01 | 0.00129 | 0.00003 | 0.00004 | 0.00003 | 0.00006 | |
| T24 | 57.34 | 0.00106 | 0.00001 | 0.00002 | 0.00001 | 0.00001 | |
| T48 | 3.20 | 0.00002 | - | - | - | - | |
| T72 | 4.41 | 0.00017 | - | - | - | - | |
| 50% SNPV + 50% MNPV | 50.00 | 0.00013 | 0.00002 | 0.00001 | 0.00002 | 0.00002 | |
| M. brassicae | T0 | 96.76 | 0.00084 | - | - | - | - |
| T12 | 41.01 | 0.01113 | 0.01987 | 0.00166 | 0.00348 | 0.00569 | |
| T24 | 57.34 | 0.00109 | 0.00311 | 0.00071 | 0.00026 | 0.00018 | |
| T48 | 3.20 | 0.02164 | - | - | - | - | |
| T72 | 4.41 | 0.02384 | - | - | - | - | |
| 50% SNPV + 50% MNPV | 50.00 | 0.02933 | 0.00348 | 0.00087 | 0.00108 | 0.00064 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arrizubieta, M.; Simón, O.; Ricarte-Bermejo, A.; López-Ferber, M.; Williams, T.; Caballero, P. Coocclusion of Helicoverpa armigera Single Nucleopolyhedrovirus (HearSNPV) and Helicoverpa armigera Multiple Nucleopolyhedrovirus (HearMNPV): Pathogenicity and Stability in Homologous and Heterologous Hosts. Viruses 2022, 14, 687. https://doi.org/10.3390/v14040687
Arrizubieta M, Simón O, Ricarte-Bermejo A, López-Ferber M, Williams T, Caballero P. Coocclusion of Helicoverpa armigera Single Nucleopolyhedrovirus (HearSNPV) and Helicoverpa armigera Multiple Nucleopolyhedrovirus (HearMNPV): Pathogenicity and Stability in Homologous and Heterologous Hosts. Viruses. 2022; 14(4):687. https://doi.org/10.3390/v14040687
Chicago/Turabian StyleArrizubieta, Maite, Oihane Simón, Adriana Ricarte-Bermejo, Miguel López-Ferber, Trevor Williams, and Primitivo Caballero. 2022. "Coocclusion of Helicoverpa armigera Single Nucleopolyhedrovirus (HearSNPV) and Helicoverpa armigera Multiple Nucleopolyhedrovirus (HearMNPV): Pathogenicity and Stability in Homologous and Heterologous Hosts" Viruses 14, no. 4: 687. https://doi.org/10.3390/v14040687
APA StyleArrizubieta, M., Simón, O., Ricarte-Bermejo, A., López-Ferber, M., Williams, T., & Caballero, P. (2022). Coocclusion of Helicoverpa armigera Single Nucleopolyhedrovirus (HearSNPV) and Helicoverpa armigera Multiple Nucleopolyhedrovirus (HearMNPV): Pathogenicity and Stability in Homologous and Heterologous Hosts. Viruses, 14(4), 687. https://doi.org/10.3390/v14040687