Preparation of a Single-Chain Antibody against Nucleocapsid Protein of Porcine Deltacoronavirus by Phage Display Technology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria Strains, Plasmids, Cell Lines and Viruses
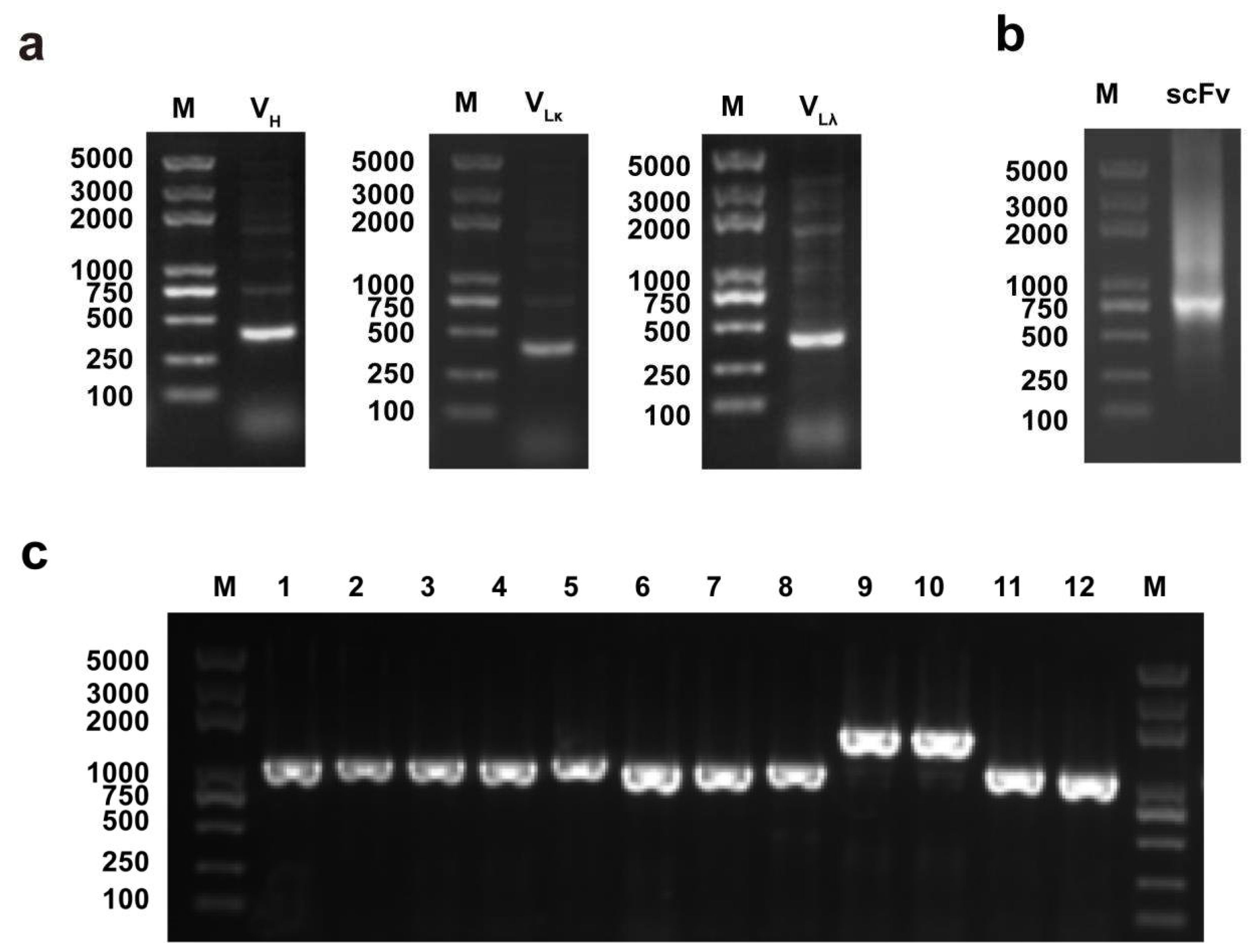
2.2. Antibody Variable Region Amplification and Full-Length scFv Assembly
2.3. Construction of the Phage Display scFv Library
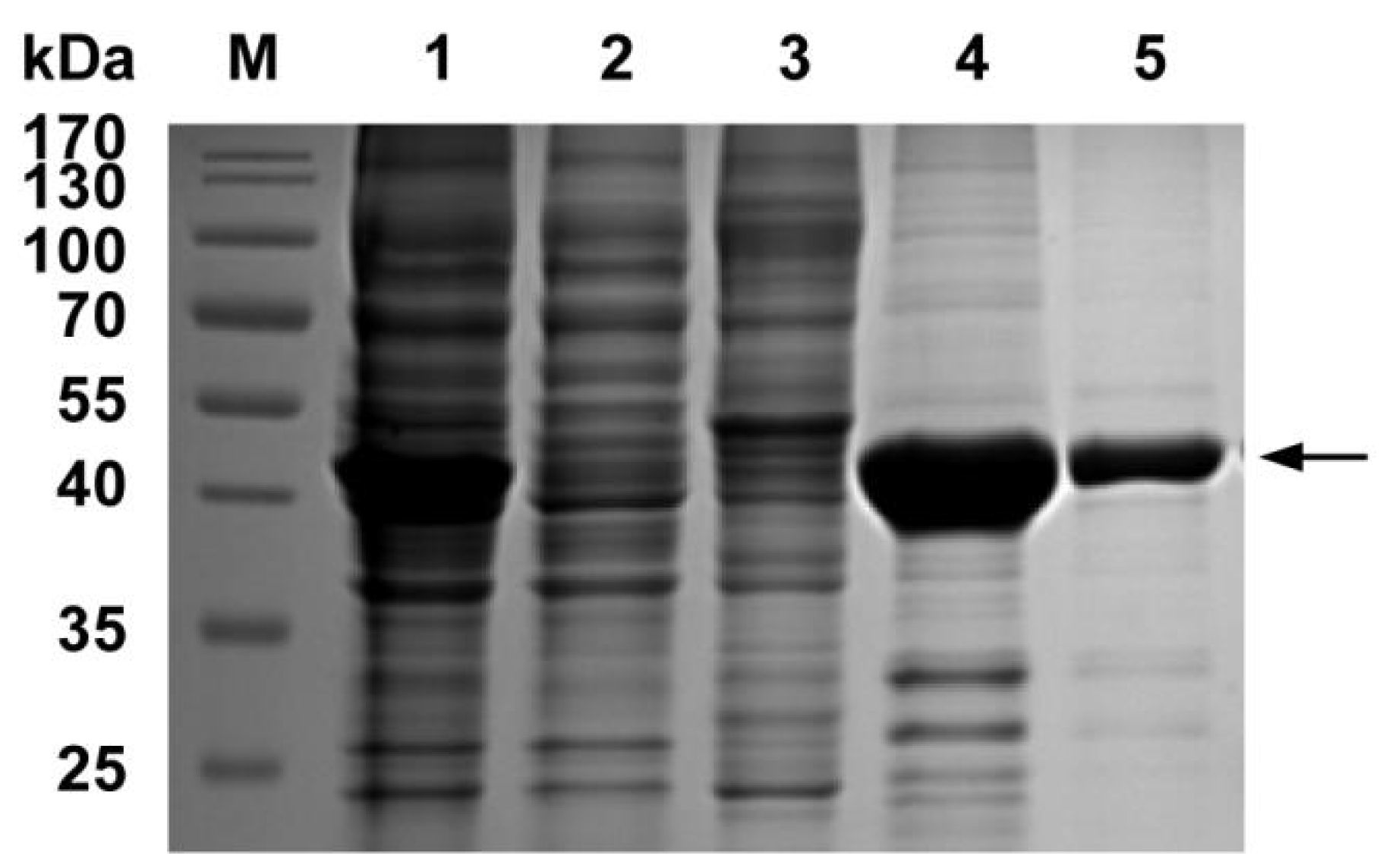
2.4. Expression of the Complete and Truncated PDCoV N Protein
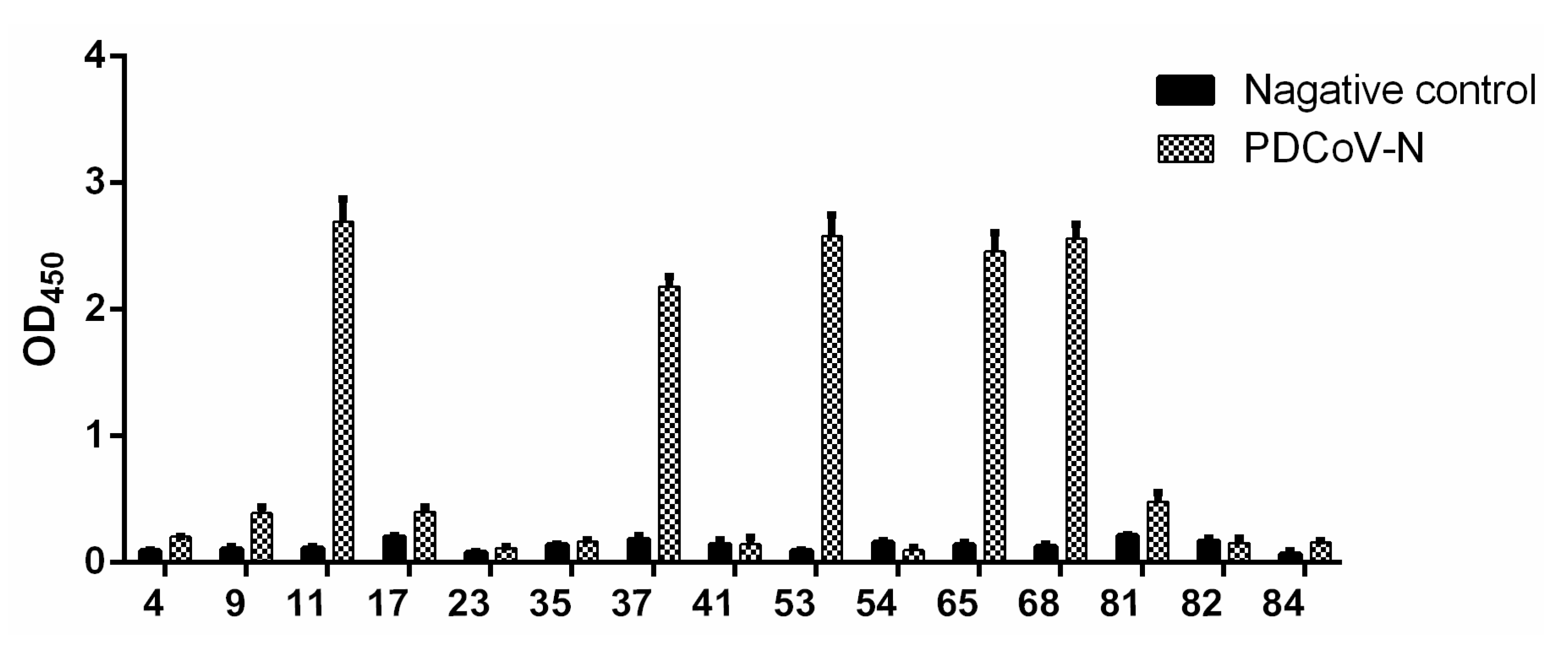
2.5. Enrichment and Screening of scFv
2.6. Phage–Indirect Enzyme-Linked Immunosorbent Assay (ELISA)
2.7. Expression and Purification of scFv Antibodies
2.8. Indirect ELISA
2.9. Western Blot
2.10. Indirect Immunofluorescence Assay
2.11. Dot Blot
2.12. Protein Modeling and Molecular Docking
3. Results
3.1. Construction of Phage Display scFv Library
3.2. Expression and Purification of PDCoV N Protein
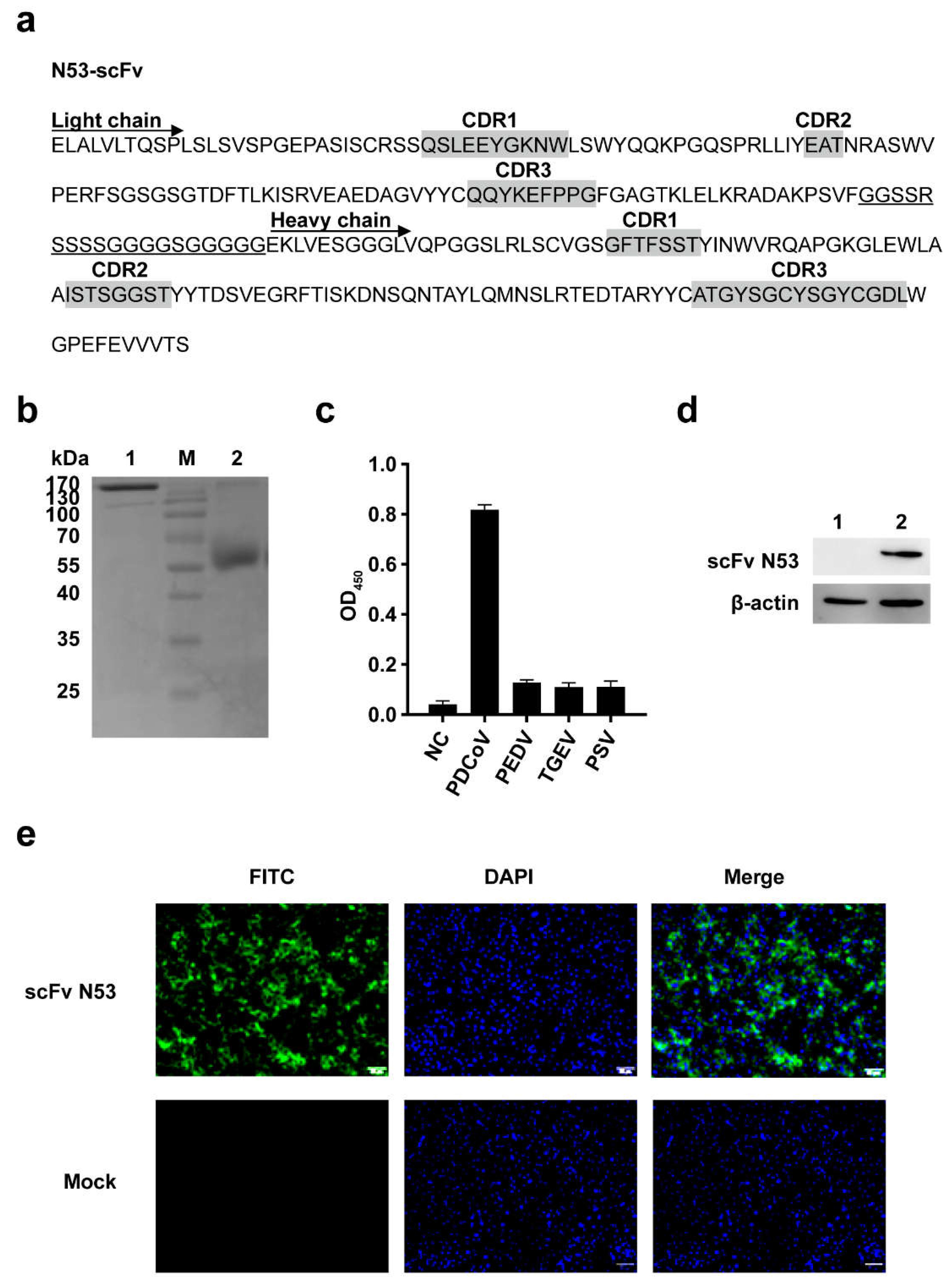
3.3. Screening and Identification of scFv against PDCoV N Protein
3.4. Expression and Purification of scFv-Fc
3.5. Analysis of the Specificity and Affinity of the Purified scFv N53
3.6. Identification of Epitope Recognized by the scFv N53
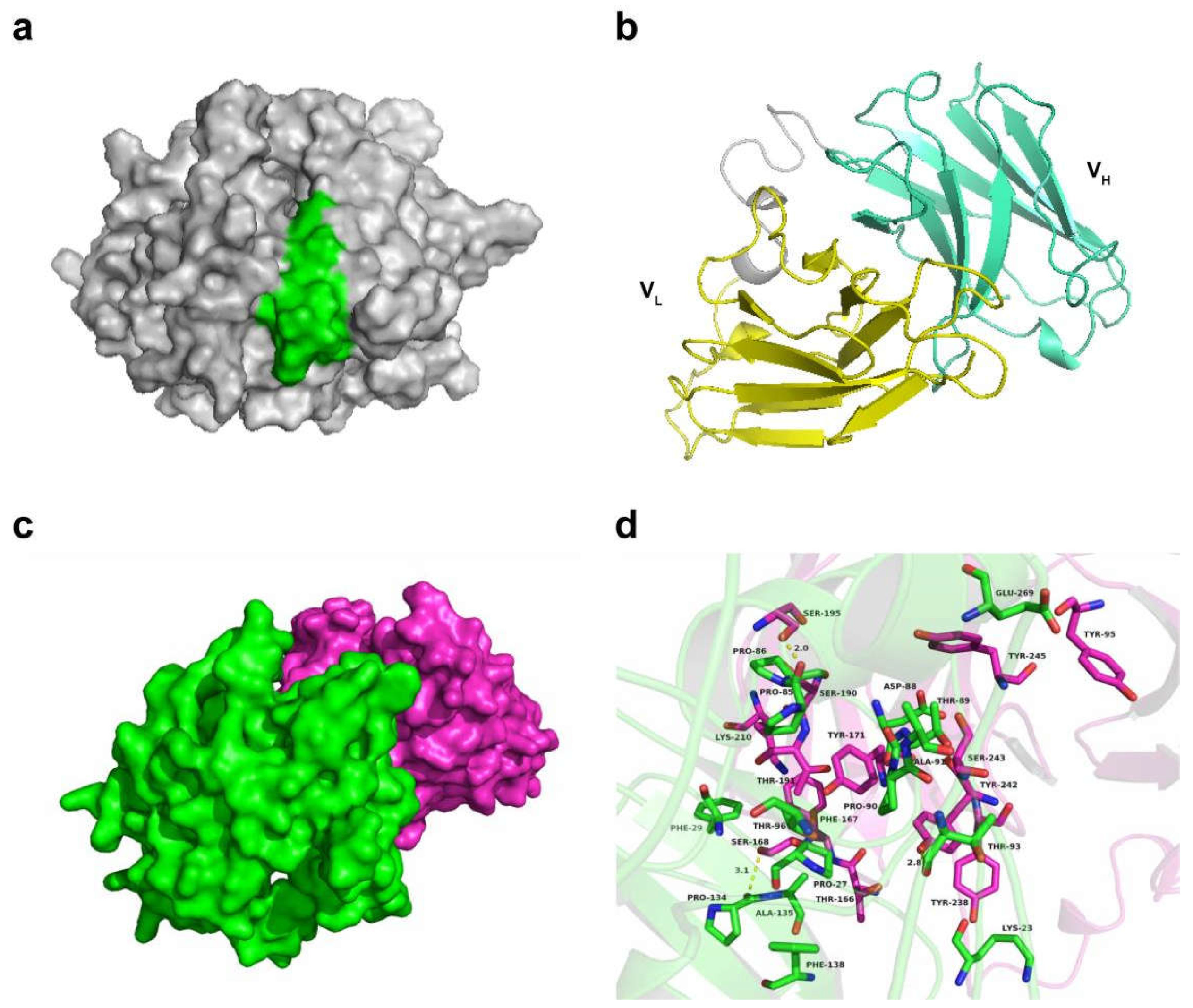
3.7. Docking Analysis of scFv and N Protein Interactions
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chen, Q.; Gauger, P.; Stafne, M.; Thomas, J.; Arruda, P.; Burrough, E.; Madson, D.; Brodie, J.; Magstadt, D.; Derscheid, R.; et al. Pathogenicity and pathogenesis of a United States porcine deltacoronavirus cell culture isolate in 5-day-old neonatal piglets. Virology 2015, 482, 51–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J. Porcine deltacoronavirus: Overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016, 226, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Jung, K.; Vlasova, A.N.; Chepngeno, J.; Lu, Z.; Wang, Q.; Saif, L.J. Isolation and characterization of porcine deltacoronavirus from pigs with diarrhea in the United States. J. Clin. Microbiol. 2015, 53, 1537–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Byrum, B.; Zhang, Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014, 20, 1227–1230. [Google Scholar] [CrossRef]
- Marthaler, D.; Raymond, L.; Jiang, Y.; Collins, J.; Rossow, K.; Rovira, A. Rapid detection, complete genome sequencing, and phylogenetic analysis of porcine deltacoronavirus. Emerg. Infect. Dis. 2014, 20, 1347–1350. [Google Scholar] [CrossRef] [Green Version]
- Woo, P.C.; Lau, S.K.; Lam, C.S.; Lau, C.C.; Tsang, A.K.; Lau, J.H.; Bai, R.; Teng, J.L.; Tsang, C.C.; Wang, M.; et al. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Hulswit, R.J.G.; Kenney, S.P.; Widjaja, I.; Jung, K.; Alhamo, M.A.; van Dieren, B.; van Kuppeveld, F.J.M.; Saif, L.J.; Bosch, B.J. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc. Natl. Acad. Sci. USA 2018, 115, E5135–E5143. [Google Scholar] [CrossRef] [Green Version]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature 2021, 600, 133–137. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, Y.; Liang, X.; Lou, F.; Oglesbee, M.; Krakowka, S.; Li, J. Origin, evolution, and virulence of porcine deltacoronaviruses in the United States. mBio 2015, 6, e00064. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Chen, Q.; Harmon, K.M.; Yoon, K.J.; Schwartz, K.J.; Hoogland, M.J.; Gauger, P.C.; Main, R.G.; Zhang, J. Full-Length Genome Sequence of Porcine Deltacoronavirus Strain USA/IA/2014/8734. Genome Announc. 2014, 2, e00278-14. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Lee, C. Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res. 2015, 208, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Duan, C. An Updated Review of Porcine Deltacoronavirus in Terms of Prevalence, Pathogenicity, Pathogenesis and Antiviral Strategy. Front. Vet. Sci. 2021, 8, 811187. [Google Scholar] [CrossRef] [PubMed]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar] [CrossRef] [PubMed]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef] [PubMed]
- Furuta, M.; Uchikawa, M.; Ueda, Y.; Yabe, T.; Taima, T.; Tsumoto, K.; Kojima, S.; Juji, T.; Kumagai, I. Construction of mono- and bivalent human single-chain Fv fragments against the D antigen in the Rh blood group: Multimerization effect on cell agglutination and application to blood typing. Protein Eng. 1998, 11, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Park, H.; Kim, M.; Seo, Y.; Lee, Y.; Byun, S.J.; Lee, S.; Kwon, M.H. Functional stability of 3D8 scFv, a nucleic acid-hydrolyzing single chain antibody, under different biochemical and physical conditions. Int. J. Pharm. 2015, 496, 561–570. [Google Scholar] [CrossRef] [Green Version]
- Weisser, N.E.; Hall, J.C. Applications of single-chain variable fragment antibodies in therapeutics and diagnostics. Biotechnol. Adv. 2009, 27, 502–520. [Google Scholar] [CrossRef]
- Hagemeyer, C.E.; von Zur Muhlen, C.; von Elverfeldt, D.; Peter, K. Single-chain antibodies as diagnostic tools and therapeutic agents. Thromb. Haemost. 2009, 101, 1012–1019. [Google Scholar]
- Hu, H.; Jung, K.; Kenney, S.P.; Saif, L.J. Isolation and Tissue Culture Adaptation of Porcine Deltacoronavirus: A Case Study. Methods Mol. Biol. 2020, 2203, 77–88. [Google Scholar] [CrossRef]
- Zhang, Y.; Xia, L.; Yuan, Y.; Li, Q.; Han, L.; Yang, G.; Hu, H. Rhodanine derivative LJ001 inhibits TGEV and PDCoV replication in vitro. Virus Res. 2020, 289, 198167. [Google Scholar] [CrossRef]
- Zhang, H.; Liang, Q.; Li, B.; Cui, X.; Wei, X.; Ding, Q.; Wang, Y.; Hu, H. Prevalence, phylogenetic and evolutionary analysis of porcine deltacoronavirus in Henan province, China. Prev. Vet. Med. 2019, 166, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; Zhang, H.; Li, B.; Ding, Q.; Wang, Y.; Gao, W.; Guo, D.; Wei, Z.; Hu, H. Susceptibility of Chickens to Porcine Deltacoronavirus Infection. Viruses 2019, 11, 573. [Google Scholar] [CrossRef] [Green Version]
- Jung, K.; Hu, H.; Saif, L.J. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch. Virol. 2017, 162, 2357–2362. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, F.; Yuan, Y.; Zeng, X.; Wei, Z.; Zhu, L.; Sun, B.; Xie, Q.; Cao, Y.; Xue, C.; et al. Sequence and phylogenetic analysis of nucleocapsid genes of porcine epidemic diarrhea virus (PEDV) strains in China. Arch. Virol. 2013, 158, 1267–1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Z.; Luo, S.; Gong, W.; Wang, L.; Ding, N.; Chen, J.; Chen, J.; Wang, T.; Ye, Y.; Song, D.; et al. Subcellular localization of the porcine deltacoronavirus nucleocapsid protein. Virus Genes 2020, 56, 687–695. [Google Scholar] [CrossRef]
- Gu, X.; Jia, X.; Feng, J.; Shen, B.; Huang, Y.; Geng, S.; Sun, Y.; Wang, Y.; Li, Y.; Long, M. Molecular modeling and affinity determination of scFv antibody: Proper linker peptide enhances its activity. Ann. Biomed. Eng. 2010, 38, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Skerra, A.; Plückthun, A. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science 1988, 240, 1038–1041. [Google Scholar] [CrossRef]
- Jørgensen, M.L.; Friis, N.A.; Just, J.; Madsen, P.; Petersen, S.V.; Kristensen, P. Expression of single-chain variable fragments fused with the Fc-region of rabbit IgG in Leishmania tarentolae. Microb. Cell Factories 2014, 13, 9. [Google Scholar] [CrossRef] [Green Version]
- Jäger, V.; Büssow, K.; Wagner, A.; Weber, S.; Hust, M.; Frenzel, A.; Schirrmann, T. High level transient production of recombinant antibodies and antibody fusion proteins in HEK293 cells. BMC Biotechnol. 2013, 13, 52. [Google Scholar] [CrossRef] [Green Version]

| Primer | Sequence(5’−3’) |
|---|---|
| VH-1F | GGTGGTTCCTCTAGATCTTCCTCCTCTGGTGGCGGTGGCTCGGGCGGTGGTGGGGAGGWGAAGCTGGTGGAGTCYGG |
| VH-2F | GGTGGTTCCTCTAGATCTTCCTCCTCTGGTGGCGGTGGCTCGGGCGGTGGTGGGSAGGTSCAGCTGGTRCAGTCTGG |
| VH-3F | GGTGGTTCCTCTAGATCTTCCTCCTCTGGTGGCGGTGGCTCGGGCGGTGGTGGGSAGGTGCAGCTGKTGGAG |
| VH-1R | CCTGGCCGGCCTGGCCACTAGTCACGACGACTTCAACGCCTGG |
| VH-2R | CCTGGCCGGCCTGGCCACTAGTCACGACGACTTCRAYGCCTGG |
| VH-3R | CCTGGCCGGCCTGGCCACTAGTCACGACGACTTCRACKCCTGG |
| VLκ-1F | GGGCCCAGGCGGCCGAGCTCGCCMTYGTGCTGACCCAGTCTCCA |
| VLκ-2F | GGGCCCAGGCGGCCGAGCTCGAGMTCGTSATGACCCAGTCTCCA |
| VLκ-3F | GGGCCCAGGCGGCCGAGCTCGMCATCCRGWTGACCCAGTCTCCA |
| VLκ-1R | GGAAGATCTAGAGGAACCACCTTTGAKYTCCAGCTTGGTCCC |
| VLκ-2R | GGAAGATCTAGAGGAACCACCTTTGATATCCACTTTGGTCCC |
| VLλ-1F | GGGCCCAGGCGGCCGAGCTCTCTTCTAAGCTGACTCAGCCCCCGGGGGTGT |
| VLλ-1R | GGAAGATCTAGAGGAACCACCCCGTGGGAGYGGCCTTGGGCTGACC |
| scFv-F | GAGGAGGAGGAGGAGGAGGCGGGGCCCAGGCGGCCGAGCTC |
| scFv-R | GAGGAGGAGGAGGAGGAGCCTGGCCGGCCTGGCCACTAGT |
| Primers | Sequences(5′–3′) | Amino Acid Position |
|---|---|---|
| PDCoV N | (F) CGGAATTCATGGCCGCACCAGTAGTCC | 1–342 |
| (R) CCCTCGAGCGCTGCTGATTCCTGCTTTA | ||
| NP-1 | (F) CGGAATTCATGGCCGCACCAGTAGTCC (R) CCGCTCGAGATAATAAAAGGCATAGGATGGAGGA | 1–70 |
| NP-2 | (F) CGGAATTCACTCCGATTCCTCCATCCTATGCCT (R) CCGCTCGAGTTTAGGATTGTTGGGGTTGCGTTTG | 60–120 |
| NP-3 | (F) CGGAATTCCATGTTGCCAAACGCAACCC (R) CCGCTCGAGGGGCTGATTGCCTGTGCCTCT | 110–170 |
| NP-4 | (F) CGGAATTCTCTGTTAACTCCAGAGGCACAGG (R) CCGCTCGAGCTCAGTGTCTGCAGAGCCGACAT | 160–220 |
| NP-5 | (F) CGGAATTCACTGGTGCCAATGTCGGCT (R) CCGCTCGAGCGCATCCTTAAGTCTCTCATAGTCA | 210–280 |
| NP-6 | (F) CGGAATTCGGTTCTCCTGACTATGAGAGACTTA (R) CCGCTCGAGCGCTGGTGATTCCTGCTTTATCTCA | 270–342 |
| NP-2-1 | (F) CGGAATTCTATACTGGCACAGGTCCCAGAGGAA (R) CCGCTCGAGTTTAGGATTGTTGGGGTTGCGTTTG | 70–120 |
| NP-2-2 | (F)CGGAATTCAAGTATGGTGAACTCCCTCCTAATG (R) CCGCTCGAGTTTAGGATTGTTGGGGTTGCGTTTG | 80–120 |
| NP-2-3 | (F) CGGAATTCCCAGCAACCACTCGTGTTACTTGG (R) CCGCTCGAGTTTAGGATTGTTGGGGTTGCGTTTG | 90–120 |
| NP-2-4 | (F) CGGAATTCACTCCGATTCCTCCATCCTATGCCT (R) CCGCTCGAGATGAGGTTTAATAGAAGTGTCAGCT | 60–110 |
| NP-2-5 | (F) CGGAATTCACTCCGATTCCTCCATCCTATGCCT (R) CCGCTCGAGACCCTTAACCCAAGTAACACGAGTG | 60–100 |
| NP-2-6 | (F) CGGAATTCACTCCGATTCCTCCATCCTATGCCT (R) CCGCTCGAGTGGGGTATCATTAGGAGGGAGTTCA | 60–90 |
| NP-2-7 | (F) CGGAATTCTATGGTGAACTCCCTCCTAATGATA (R) CCGCTCGAGTTTAGGATTGTTGGGGTTGCGTTTG | 81–120 |
| NP-2-8 | (F) CGGAATTCGGTGAACTCCCTCCTAATGATACCC (R) CCGCTCGAGTTTAGGATTGTTGGGGTTGCGTTTG | 82–120 |
| NP-2-9 | (F) CGGAATTCGAACTCCCTCCTAATGATACCCCAG (R) CCGCTCGAGTTTAGGATTGTTGGGGTTGCGTTTG | 83–120 |
| NP-2-10 | (F) CGGAATTCACTCCGATTCCTCCATCCTATGCCT (R) CCGCTCGAGAACACGAGTGGTTGCTGGGGTAT | 60–95 |
| NP-2-11 | (F) CGGAATTCACTCCGATTCCTCCATCCTATGCCT (R) CCGCTCGAGAGTAACACGAGTGGTTGCTGGGGTA | 60–96 |
| NP-2-12 | (F) CGGAATTCACTCCGATTCCTCCATCCTATGCCT (R) CCGCTCGAGCCAAGTAACACGAGTGGTTGCTG | 60–97 |
| Round of Screening | Input Number of Phage (pfu) | Output Number of Phage (pfu) | Output/Input |
|---|---|---|---|
| 1 | 4.5 × 109 | 3.8 × 104 | 8.4 × 10−6 |
| 2 | 1.2 × 1010 | 3 × 106 | 2.5 × 10−4 |
| 3 | 3.9 × 109 | 8.7 × 106 | 2.2 × 10−3 |
| 4 | 1.7 × 1010 | 4.5 × 107 | 2.6 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Song, Y.; Ren, H.; Zeng, Q.; Yuan, Y.; Xia, L.; Wei, Z. Preparation of a Single-Chain Antibody against Nucleocapsid Protein of Porcine Deltacoronavirus by Phage Display Technology. Viruses 2022, 14, 772. https://doi.org/10.3390/v14040772
Zhang Y, Song Y, Ren H, Zeng Q, Yuan Y, Xia L, Wei Z. Preparation of a Single-Chain Antibody against Nucleocapsid Protein of Porcine Deltacoronavirus by Phage Display Technology. Viruses. 2022; 14(4):772. https://doi.org/10.3390/v14040772
Chicago/Turabian StyleZhang, Yixuan, Yue Song, Haojie Ren, Quan Zeng, Yixin Yuan, Lu Xia, and Zhanyong Wei. 2022. "Preparation of a Single-Chain Antibody against Nucleocapsid Protein of Porcine Deltacoronavirus by Phage Display Technology" Viruses 14, no. 4: 772. https://doi.org/10.3390/v14040772
APA StyleZhang, Y., Song, Y., Ren, H., Zeng, Q., Yuan, Y., Xia, L., & Wei, Z. (2022). Preparation of a Single-Chain Antibody against Nucleocapsid Protein of Porcine Deltacoronavirus by Phage Display Technology. Viruses, 14(4), 772. https://doi.org/10.3390/v14040772






