Genetic Variations among Different Variants of G1-like Avian Influenza H9N2 Viruses and Their Pathogenicity in Chickens
Abstract
:1. Introduction
2. Material and Methods
2.1. Viruses and Virus Isolation
2.2. Genetic and Phylogenetic Analysis
2.3. Ethical Approval
2.4. Pathogenicity and Transmission
2.5. Antigenic Characterization
3. Results
3.1. Genetic and Antigenic Characterization
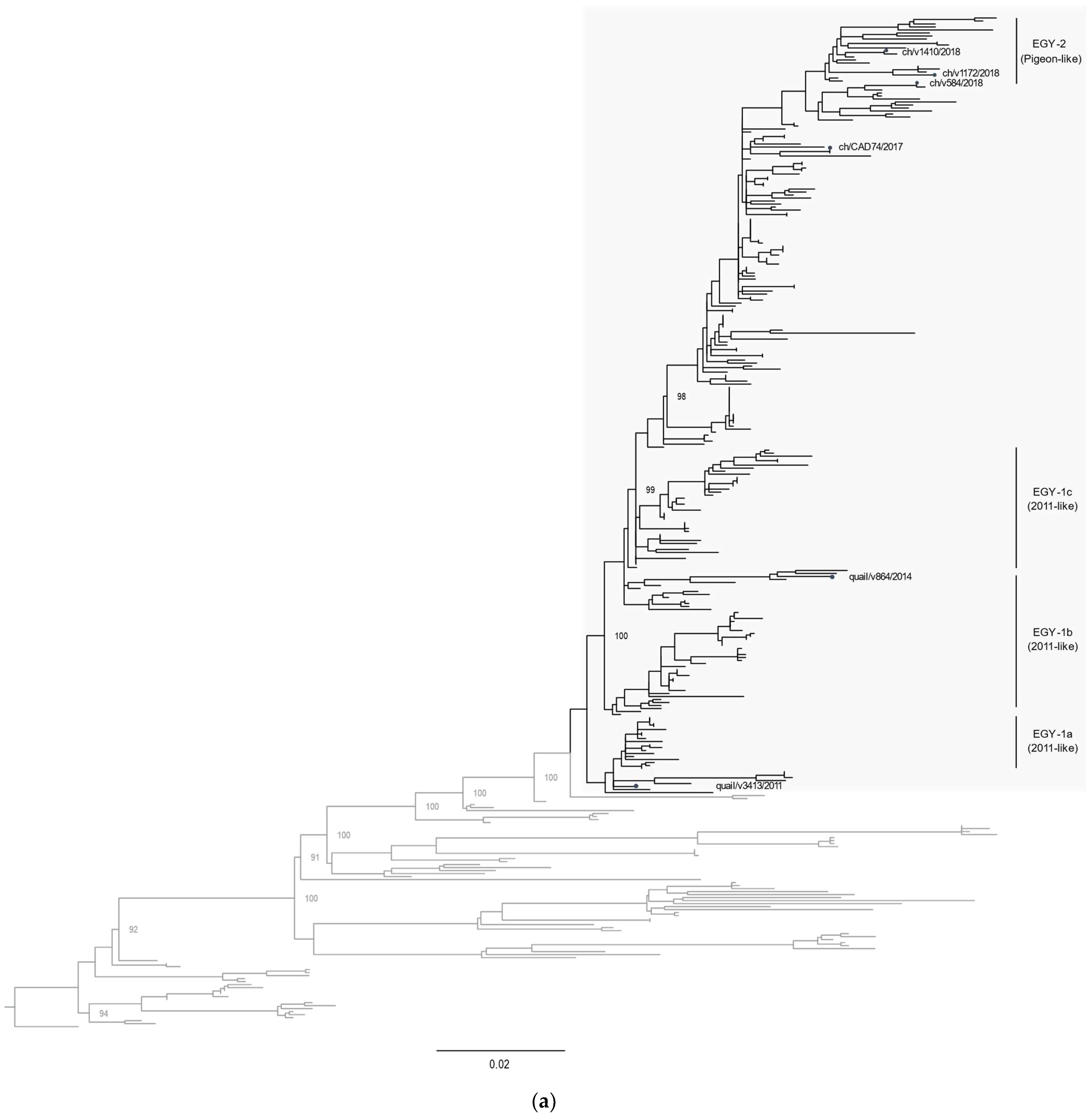
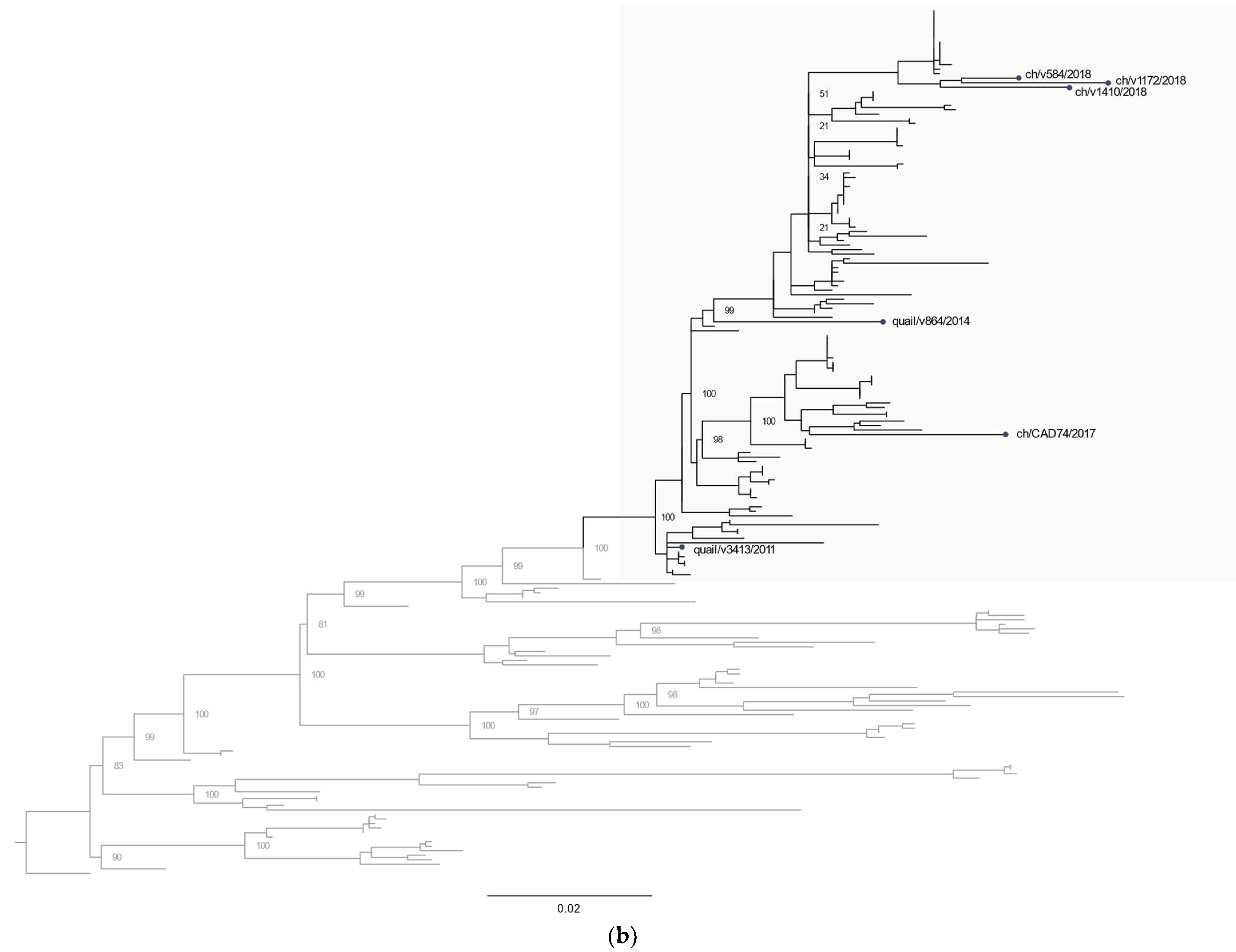
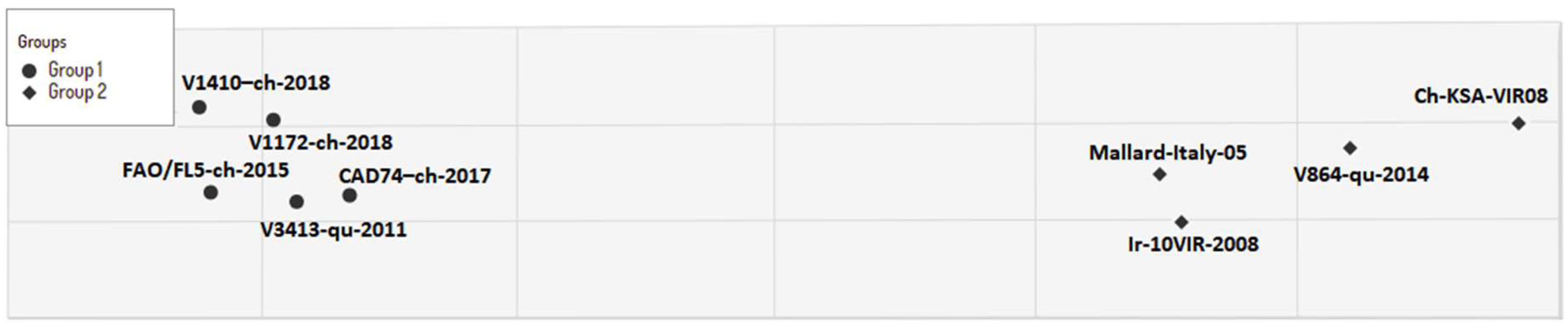
3.2. Phylogenetic Characterization
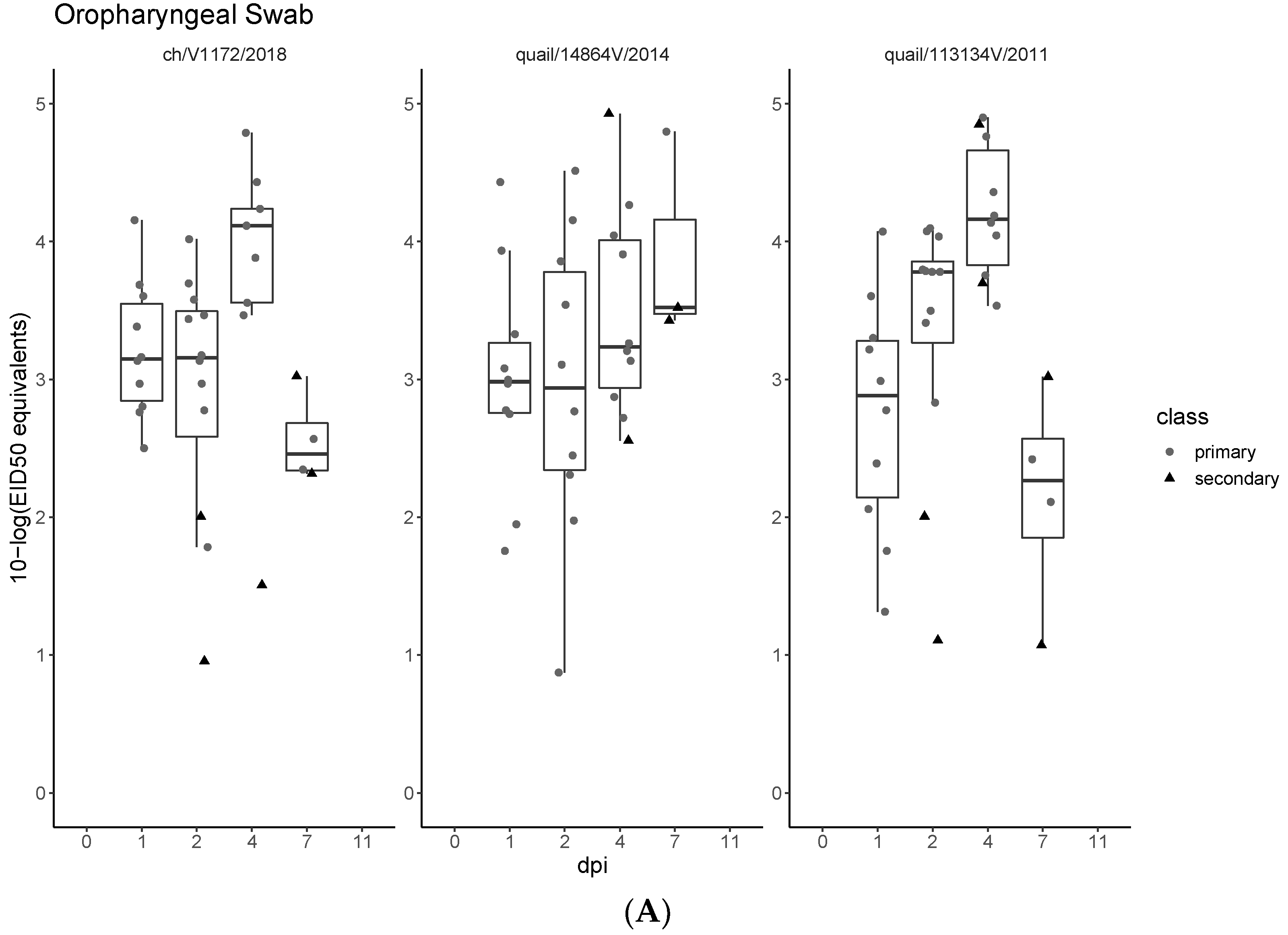
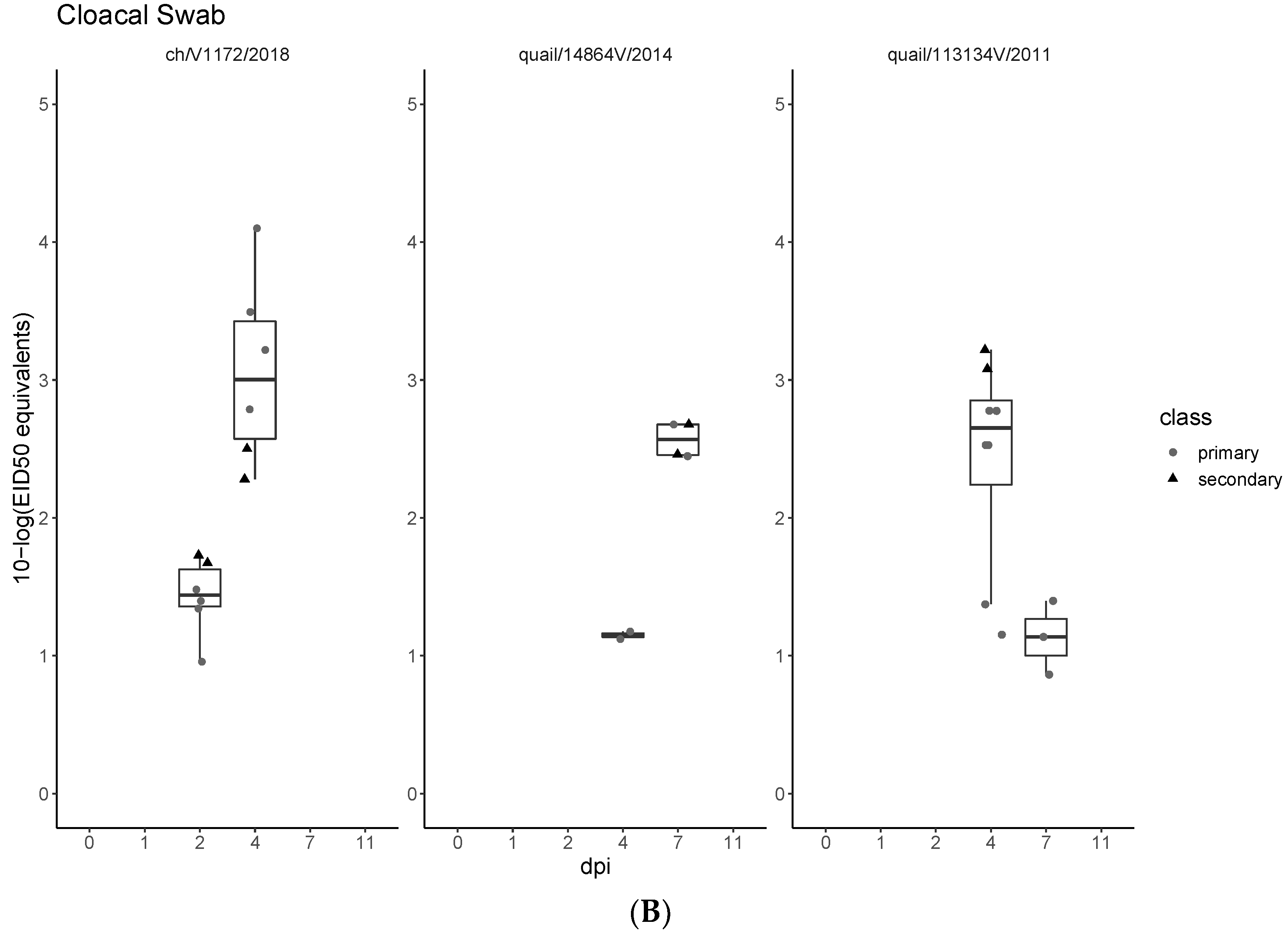
3.3. Pathogenicity and Transmission in Chickens
3.4. Cross Antigenicity
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Peacock, T.H.P.; James, J.; Sealy, J.E.; Iqbal, M. A Global Perspective on H9N2 Avian Influenza Virus. Viruses 2019, 11, 620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Y.; Liu, J. H9N2 influenza virus in China: A cause of concern. Protein Cell 2015, 6, 18–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalonda, A.; Saasa, N.; Nkhoma, P.; Kajihara, M.; Sawa, H.; Takada, A.; Simulundu, E. Avian Influenza Viruses Detected in Birds in Sub-Saharan Africa: A Systematic Review. Viruses 2020, 12, 993. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Qin, K. Human-infecting influenza A (H9N2) virus: A forgotten potential pandemic strain? Zoonoses Public Health 2020, 67, 203–212. [Google Scholar] [CrossRef]
- Li, R.; Adel, A.; Bohlin, J.; Lundkvist, Å.; Olsen, B.; Pettersson, J.H.; Naguib, M.M. Phylogeographic Dynamics of Influenza A(H9N2) Virus Crossing Egypt. Front. Microbiol. 2020, 11, 392. [Google Scholar] [CrossRef] [Green Version]
- Steel, J.; Lowen, A.C. Influenza A virus reassortment. Curr. Top. Microbiol. Immunol. 2014, 385, 377–401. [Google Scholar] [CrossRef]
- Guan, Y.; Shortridge, K.F.; Krauss, S.; Webster, R.G. Molecular characterization of H9N2 influenza viruses: Were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 1999, 96, 9363–9367. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.P.; Peng, L.; Chen, L.; Jiang, L.F.; Zhang, Z.J.; Xiong, C.L.; Zhao, G.M.; Chen, Y.; Jiang, Q.W. Avian influenza viruses (AIVs) H9N2 are in the course of reassorting into novel AIVs. J. Zhejiang Univ. Sci. B 2018, 19, 409–414. [Google Scholar] [CrossRef]
- El-Zoghby, E.F.; Arafa, A.S.; Hassan, M.K.; Aly, M.M.; Selim, A.; Kilany, W.H.; Selim, U.; Nasef, S.; Aggor, M.G.; Abdelwhab, E.M.; et al. Isolation of H9N2 avian influenza virus from bobwhite quail (Colinus virginianus) in Egypt. Arch. Virol. 2012, 157, 1167–1172. [Google Scholar] [CrossRef]
- Adel, A.; Arafa, A.; Hussein, H.A.; El-Sanousi, A.A. Molecular and antigenic traits on hemagglutinin gene of avian influenza H9N2 viruses: Evidence of a new escape mutant in Egypt adapted in quails. Res. Vet. Sci. 2017, 112, 132–140. [Google Scholar] [CrossRef]
- Kandeil, A.; El-Shesheny, R.; Maatouq, A.; Moatasim, Y.; Cai, Z.; McKenzie, P.; Webby, R.; Kayali, G.; Ali, M.A. Novel reassortant H9N2 viruses in pigeons and evidence for antigenic diversity of H9N2 viruses isolated from quails in Egypt. J. Gen. Virol. 2017, 98, 548–562. [Google Scholar] [CrossRef] [PubMed]
- Hassan, K.E.; Saad, N.; Abozeid, H.H.; Shany, S.; El-Kady, M.F.; Arafa, A.; El-Sawah, A.A.A.; Pfaff, F.; Hafez, H.M.; Beer, M.; et al. Genotyping and reassortment analysis of highly pathogenic avian influenza viruses H5N8 and H5N2 from Egypt reveals successive annual replacement of genotypes. Infect. Genet. Evol. 2020, 84, 104375. [Google Scholar] [CrossRef] [PubMed]
- Naguib, M.M.; El-Kady, M.F.; Lüschow, D.; Hassan, K.E.; Arafa, A.S.; El-Zanaty, A.; Hassan, M.K.; Hafez, H.M.; Grund, C.; Harder, T.C. New real time and conventional RT-PCRs for updated molecular diagnosis of infectious bronchitis virus infection (IBV) in chickens in Egypt associated with frequent co-infections with avian influenza and Newcastle Disease viruses. J. Virol. Methods 2017, 245, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Hagag, N.M.; Erfan, A.M.; El-Husseiny, M.; Shalaby, A.G.; Saif, M.A.; Tawakol, M.M.; Nour, A.A.; Selim, A.A.; Arafa, A.S.; Hassan, M.K.; et al. Isolation of a Novel Reassortant Highly Pathogenic Avian Influenza (H5N2) Virus in Egypt. Viruses 2019, 11, 565. [Google Scholar] [CrossRef] [Green Version]
- Adel, A.; Mosaad, Z.; Shalaby, A.G.; Selim, K.; Samy, M.; Abdelmagid, M.A.; Hagag, N.M.; Arafa, A.S.; Hassan, W.M.; Shahien, M.A. Molecular evolution of the hemagglutinin gene and epidemiological insight into low-pathogenic avian influenza H9N2 viruses in Egypt. Res. Vet. Sci. 2021, 136, 540–549. [Google Scholar] [CrossRef]
- OIE. Avian Influenza (Including Infection with Highly Pathogenic Avian Influenza Viruses); OIE: Paris, France, 2021. [Google Scholar]
- Meir, R.; Maharat, O.; Farnushi, Y.; Simanov, L. Development of a real-time TaqMan RT-PCR assay for the detection of infectious bronchitis virus in chickens, and comparison of RT-PCR and virus isolation. J. Virol. Methods 2010, 163, 190–194. [Google Scholar] [CrossRef]
- Spackman, E.; Senne, D.A.; Myers, T.J.; Bulaga, L.L.; Garber, L.P.; Perdue, M.L.; Lohman, K.; Daum, L.T.; Suarez, D.L. Development of a real-time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J. Clin. Microbiol. 2002, 40, 3256–3260. [Google Scholar] [CrossRef] [Green Version]
- Wise, M.G.; Suarez, D.L.; Seal, B.S.; Pedersen, J.C.; Senne, D.A.; King, D.J.; Kapczynski, D.R.; Spackman, E. Development of a real-time reverse-transcription PCR for detection of newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 2004, 42, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Naguib, M.M.; Arafa, A.S.; El-Kady, M.F.; Selim, A.A.; Gunalan, V.; Maurer-Stroh, S.; Goller, K.V.; Hassan, M.K.; Beer, M.; Abdelwhab, E.M.; et al. Evolutionary trajectories and diagnostic challenges of potentially zoonotic avian influenza viruses H5N1 and H9N2 co-circulating in Egypt. Infect. Genet. Evol. 2015, 34, 278–291. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 1st ed.; Springer: New York, NY, USA, 2016. [Google Scholar]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model, 1st ed.; Springer: New York, NY, USA, 2000; ISBN 0-387-98784-3. [Google Scholar]
- Rhodes, A. Fixation of tissues. In Bancroft’s Theory and Practice of Histological Techniques, 7th ed.; Suvarna, S.K., Layton, C., Bancroft, J.D., Eds.; Churchill Livingstone: Oxford, UK, 2013; p. 6993. [Google Scholar]
- Landmann, M.; Scheibner, D.; Graaf, A.; Gischke, M.; Koethe, S.; Fatola–, O.I.; Raddatz, B.; Mettenleiter, T.C.; Beer, M.; Grund, C.; et al. A Semiquantitative Scoring System for Histopathological and Immunohistochemical Assessment of Lesions and Tissue Tropism in Avian Influenza. Viruses 2021, 13, 868. [Google Scholar] [CrossRef] [PubMed]
- Barnett, J.L.; Yang, J.; Cai, Z.; Zhang, T.; Wan, X.-F. AntigenMap 3D: An online antigenic cartography resource. Bioinformatics 2012, 28, 1292–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorrell, E.M.; Wan, H.; Araya, Y.; Song, H.; Perez, D.R. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc. Natl. Acad. Sci. USA 2009, 106, 7565–7570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adel, A.; Mady, W.; Mosad, Z.; Amer, F.; Shaaban, A.; Said, D.; Maged, M.; Arafa, A.; Morsi, M.; Hassan, M. Molecular and epidemiological overview on low pathogenic avian influenza H9N2 in Egypt between 2015 and 2016. Hosts Viruses 2019, 6, 30–41. [Google Scholar] [CrossRef]
- Paterson, D.; te Velthuis, A.J.; Vreede, F.T.; Fodor, E. Host restriction of influenza virus polymerase activity by PB2 627E is diminished on short viral templates in a nucleoprotein-independent manner. J. Virol. 2014, 88, 339–344. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Song, Y.J.; Park, J.H.; Lee, J.H.; Baek, Y.H.; Song, M.S.; Oh, T.K.; Han, H.S.; Pascua, P.N.; Choi, Y.K. Emergence of amantadine-resistant H3N2 avian influenza A virus in South Korea. J. Clin. Microbiol. 2008, 46, 3788–3790. [Google Scholar] [CrossRef] [Green Version]
- Samy, A.; Naguib, M.M. Avian Respiratory Coinfection and Impact on Avian Influenza Pathogenicity in Domestic Poultry: Field and Experimental Findings. Vet. Sci. 2018, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Hassan, K.E.; King, J.; El-Kady, M.; Afifi, M.; Abozeid, H.H.; Pohlmann, A.; Beer, M.; Harder, T. Novel Reassortant Highly Pathogenic Avian Influenza A(H5N2) Virus in Broiler Chickens, Egypt. Emerg. Infect. Dis. 2020, 26, 129–133. [Google Scholar] [CrossRef]
- Naguib, M.M.; Arafa, A.S.; Parvin, R.; Beer, M.; Vahlenkamp, T.; Harder, T.C. Insights into genetic diversity and biological propensities of potentially zoonotic avian influenza H9N2 viruses circulating in Egypt. Virology 2017, 511, 165–174. [Google Scholar] [CrossRef]
- Chrzastek, K.; Leng, J.; Zakaria, M.K.; Bialy, D.; La Ragione, R.; Shelton, H. Low pathogenic avian influenza virus infection retards colon microbiota diversification in two different chicken lines. Anim. Microbiome 2021, 3, 64. [Google Scholar] [CrossRef]
- Spickler, A.R.; Trampel, D.W.; Roth, J.A. The onset of virus shedding and clinical signs in chickens infected with high-pathogenicity and low-pathogenicity avian influenza viruses. Avian Pathol. 2008, 37, 555–577. [Google Scholar] [CrossRef] [PubMed]
- van der Goot, J.A.; de Jong, M.C.; Koch, G.; Van Boven, M. Comparison of the transmission characteristics of low and high pathogenicity avian influenza A virus (H5N2). Epidemiol. Infect. 2003, 131, 1003–1013. [Google Scholar] [CrossRef] [PubMed]
- Kye, S.J.; Park, M.J.; Kim, N.Y.; Lee, Y.N.; Heo, G.B.; Baek, Y.K.; Shin, J.I.; Lee, M.H.; Lee, Y.J. Pathogenicity of H9N2 low pathogenic avian influenza viruses of different lineages isolated from live bird markets tested in three animal models: SPF chickens, Korean native chickens, and ducks. Poult. Sci. 2021, 100, 101318. [Google Scholar] [CrossRef] [PubMed]
- Bano, S.; Naeem, K.; Malik, S.A. Evaluation of pathogenic potential of avian influenza virus serotype H9N2 in chickens. Avian Dis. 2003, 47, 817–822. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, Y.; Chen, L.; Zhang, B.; Zhang, M.; Wang, J.; Jiang, Y.; Yang, C.; Jiang, T. Genetic Characteristics and Pathogenicity Analysis in Chickens and Mice of Three H9N2 Avian Influenza Viruses. Viruses 2019, 11, 1127. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adel, A.; Abdelmagid, M.A.; Mohamed, A.A.-E.; Wasberg, A.; Mosaad, Z.; Selim, K.; Shaaban, A.; Tarek, M.; Hagag, N.M.; Lundkvist, Å.; et al. Genetic Variations among Different Variants of G1-like Avian Influenza H9N2 Viruses and Their Pathogenicity in Chickens. Viruses 2022, 14, 1030. https://doi.org/10.3390/v14051030
Adel A, Abdelmagid MA, Mohamed AA-E, Wasberg A, Mosaad Z, Selim K, Shaaban A, Tarek M, Hagag NM, Lundkvist Å, et al. Genetic Variations among Different Variants of G1-like Avian Influenza H9N2 Viruses and Their Pathogenicity in Chickens. Viruses. 2022; 14(5):1030. https://doi.org/10.3390/v14051030
Chicago/Turabian StyleAdel, Amany, Marwa A. Abdelmagid, Ahmed Abd-Elhalem Mohamed, Anishia Wasberg, Zienab Mosaad, Karim Selim, Asmaa Shaaban, Mohamed Tarek, Naglaa M. Hagag, Åke Lundkvist, and et al. 2022. "Genetic Variations among Different Variants of G1-like Avian Influenza H9N2 Viruses and Their Pathogenicity in Chickens" Viruses 14, no. 5: 1030. https://doi.org/10.3390/v14051030
APA StyleAdel, A., Abdelmagid, M. A., Mohamed, A. A.-E., Wasberg, A., Mosaad, Z., Selim, K., Shaaban, A., Tarek, M., Hagag, N. M., Lundkvist, Å., Ellström, P., & Naguib, M. M. (2022). Genetic Variations among Different Variants of G1-like Avian Influenza H9N2 Viruses and Their Pathogenicity in Chickens. Viruses, 14(5), 1030. https://doi.org/10.3390/v14051030






