Human Retrovirus Genomic RNA Packaging
Abstract
1. Introduction
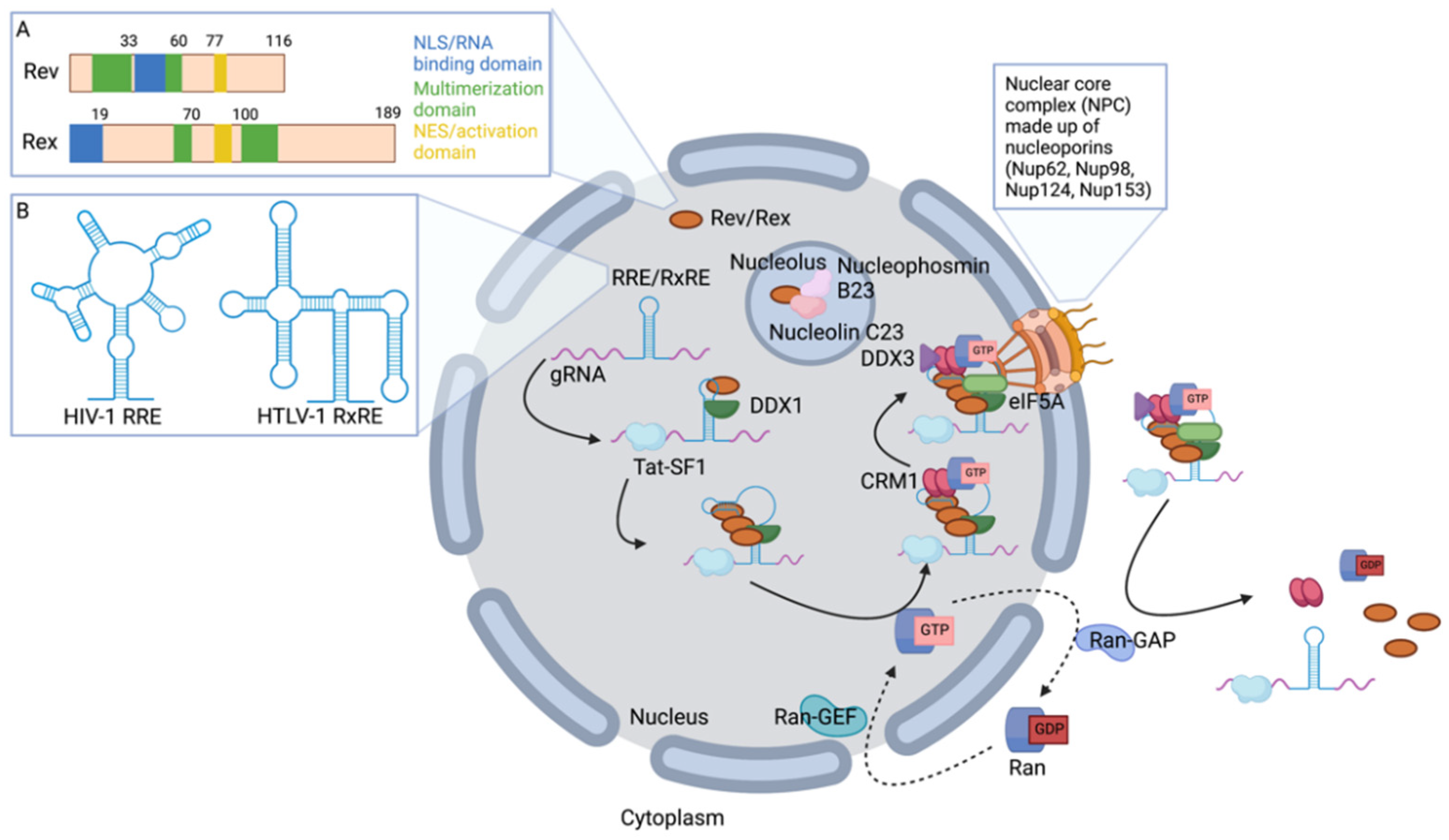
2. Genomic RNA (gRNA) Nuclear Export
2.1. Retroviral Rev and Rex Accessory Proteins Shuttle between the Nucleus and Cytoplasm
2.2. Rev and Rex Interact Directly with gRNA
2.3. Host Proteins Involved in gRNA Nuclear Export
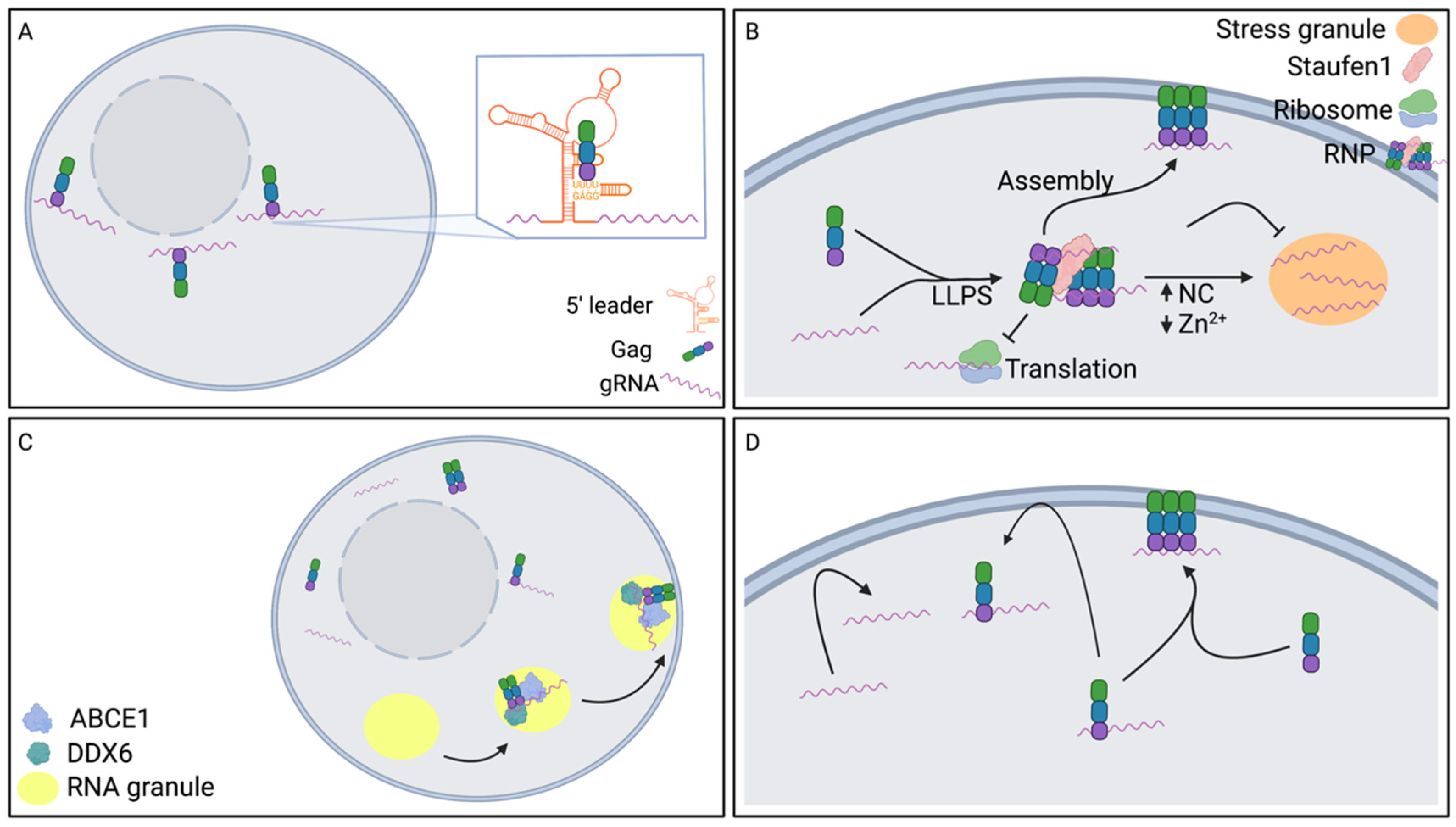
3. Translocation of gRNA to the Plasma Membrane
3.1. Influence of Nuclear Export Pathway on gRNA Translocation to the Plasma Membrane
3.2. Influence of Gag on gRNA Translocation to the Plasma Membrane
3.3. Role of Microtubule Organizing Center (MTOC) on gRNA Translocation to the Plasma Membrane
3.4. Role of Endosomal Vesicles on gRNA Translocation to the Plasma Membrane
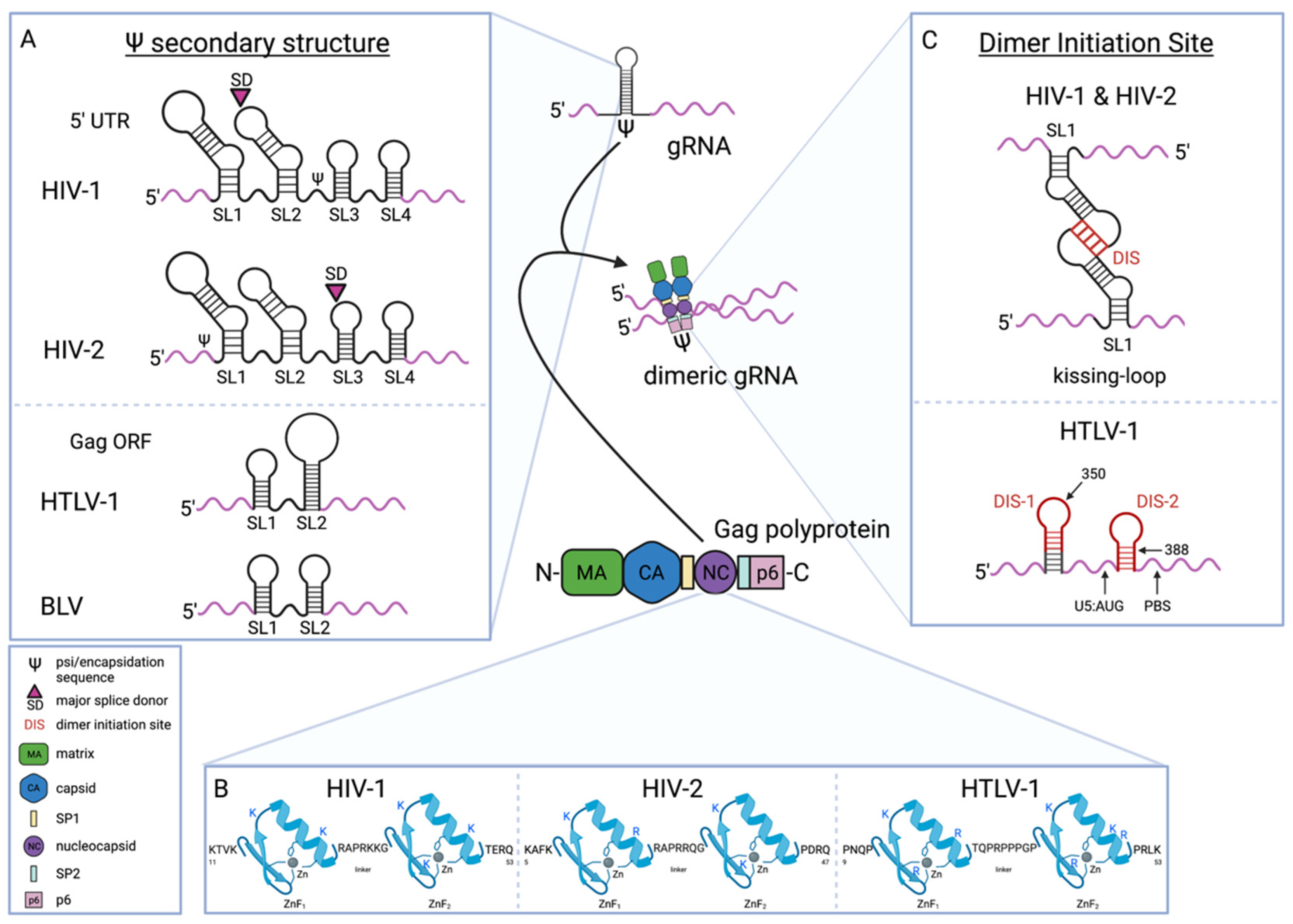
4. Packaging of Human Retroviral gRNA
4.1. gRNA Sequences Involved in the Gag-gRNA Interaction
4.2. Gag NC Determinants of Gag-gRNA Interaction
4.3. Role of Other Gag Domains in Gag-gRNA Interaction
4.4. Dimerization Initiation Site/Dimer Initiation Signal (DIS) Mediates gRNA-gRNA Interaction
4.5. Regulation of gRNA Dimerization by RNA Conformation
4.6. Role of Gag in Promoting gRNA Dimerization
4.7. Other RNA Sequences Contribute to gRNA Packaging: Secondary Packaging Signals
4.8. RNA Selection for gRNA Packaging Is Regulated on a Transcriptional Level
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Burdick, R.C.; Li, C.; Munshi, M.; Rawson, J.M.O.; Nagashima, K.; Hu, W.S.; Pathak, V.K. HIV-1 uncoats in the nucleus near sites of integration. Proc. Natl. Acad. Sci. USA 2020, 117, 5486–5493. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Burdick, R.C.; Nagashima, K.; Hu, W.S.; Pathak, V.K. HIV-1 cores retain their integrity until minutes before uncoating in the nucleus. Proc. Natl. Acad. Sci. USA 2021, 118, e2019467118. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, K.; Weiler, B.E.; Ugarkovic, D.; Bachman, M.; Schroder, H.C.; Muller, W.E.G. Evidence for specific nucleocytoplasmic transport pathways used by leucine-rich nuclear export signals. Eur. J. Biochem. 1991, 199, 53–64. [Google Scholar] [CrossRef]
- Bogerd, H.; Greene, W.C. Dominant Negative Mutants of Human T-Cell Leukemia Virus Type I Rex and Human Immunodeficiency Virus Type 1 Rev Fail To Multimerize In Vivo. J. Virol. 1993, 67, 2496–2502. [Google Scholar] [CrossRef] [PubMed]
- Madore, S.J.; Tiley, L.S.; Malim, M.H.; Cullen, B.R. Sequence Requirements for Rev Multimerization in Vivo. Virology 1994, 202, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Fritz, C.C.; Green, M.R. HIV Rev uses a conserved cellular protein export pathway for the nucleocytoplasmic transport of viral RNAs. Curr. Biol. 1996, 6, 848–854. [Google Scholar] [CrossRef][Green Version]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 Is an Export Receptor for Leucine-Rich Nuclear Export Signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef]
- Elfgang, C.; Rosorius, O.; Hofer, L.; Jaksche, H.; Hauber, J.; Bevec, D. Evidence for specific nucleocytoplasmic transport pathways used by leucine-rich nuclear export signals. Proc. Natl. Acad. Sci. USA 1999, 96, 6229–6234. [Google Scholar] [CrossRef]
- Yi, R.; Bogerd, H.P.; Cullen, B.R. Recruitment of the Crm1 nuclear export factor is sufficient to induce cytoplasmic expression of incompletely spliced human immunodeficiency virus mRNAs. J. Virol. 2002, 76, 2036–2042. [Google Scholar] [CrossRef][Green Version]
- Stade, K.; Ford, C.S.; Guthrie, C.; Weis, K. Exportin 1 (Crm1p) Is an Essential Nuclear Export Factor. Cell 1997, 90, 1041–1050. [Google Scholar] [CrossRef]
- Weichselbraun, I.; Farrington, G.K.; Rusche, J.R.; Bohnlein, E.; Hauber, J. Definition of the Human Immunodeficiency Virus Type 1 Rev and Human T-Cell Leukemia Virus Type I Rex Protein Activation Domain by Functional Exchange. J. Virol. 1992, 66, 2583–2587. [Google Scholar] [CrossRef] [PubMed]
- Hakata, Y.; Yamada, M.; Shida, H. A multifunctional domain in human CRM1 (exportin 1) mediates RanBP3 binding and multimerization of human T-cell leukemia virus type 1 Rex protein. Mol. Cell. Biol. 2003, 23, 8751–8761. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sodrowski, J.; Goh, W.C.; Rosen, C.; Dayton, A.; Terwilliger, E.; Haseltine, W. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature 1986, 321, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, E.; Burghoff, R.; Sia, R.; Sodroski, J.; Haseltine, W.; Rosen, G. The art Gene Product of Human Immunodeficiency Virus Is Required for Replication. J. Virol. 1988, 62, 655–658. [Google Scholar] [CrossRef] [PubMed]
- Emerman, M.; Vazeux, R.; Peden, K. The rev Gene Product of the Human Immunodeficiency Virus Affects Envelope-Specific RNA Localization. Cell 1989, 57, 1155–1165. [Google Scholar] [CrossRef]
- Hidaka, M.; Inoue, J.; Yoshida, M.; Seiki, M. Post-transcriptional regulator (rex) of HTLV-1 initiates expression of viral structural proteins but supresses expression of regulatory proteins. EMBO J. 1988, 7, 519–523. [Google Scholar] [CrossRef]
- Malim, M.H.; Hauber, J.; Le, S.-Y.; Maizel, J.V.; Cullen, B.R. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 1989, 338, 254–257. [Google Scholar] [CrossRef]
- Kalland, K.-H.; Szilvay, A.M.; Brokstad, K.A.; Saetrevik, W.; Haukenes, G. The Human Immunodeficiency Virus Type 1 Rev Protein Shuttles between the Cytoplasm and Nuclear Compartments. Mol. Cell. Biol. 1994, 14, 7436–7444. [Google Scholar]
- Fischer, U.; Meyer, S.; Teufel, M.; Heckel, C.; Luhrmann, R.; Rautmann, G. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 1994, 13, 4105–4112. [Google Scholar] [CrossRef]
- Palmeri, D.; Malim, M.H. The Human T-Cell Leukemia Virus Type 1 Posttranscriptional trans-Activator Rex Contains a Nuclear Export Signal. J. Virol. 1996, 70, 6442–6445. [Google Scholar] [CrossRef]
- Meyer, B.E.; Malim, M.H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994, 8, 1538–1547. [Google Scholar] [CrossRef] [PubMed]
- Richard, N.; Iacampo, S.; Cochrane, A. HIV-1 Rev Is Capable fo Shuttling between the Nucleus and Cytoplasm. Virology 1994, 204, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Szilvay, A.M.; Brokstad, K.A.; Kopperud, R.; Haukenes, G.; Kalland, K.-H. Nuclear Export of the Human Immunodeficiency Virus Type 1 Nucleocytoplasmic Shuttle Protein Rev Is Mediated by Its Activation Domain and Is Blocked by Transdominant Negative Mutants. J. Virol. 1995, 69, 3315–3323. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.; Aepinus, C.; Dobrovnik, M.; Fleckenstein, B.; Hauber, J.; Bohnlein, E. Mutational Analysis of Functional Domains in the HIV-1 Rev trans-Regulatory Protein. Virology 1991, 183, 630–635. [Google Scholar] [CrossRef]
- Dillon, P.J.; Nelbock, P.; Perkins, A.; Rosen, C.A. Structural and Functional ANalysis of the Human Immunodeficiency Virus Type 2 Rev Protein. J. Virol. 1991, 65, 445–449. [Google Scholar] [CrossRef]
- Nosaka, T.; Siomi, H.; Adachi, Y.; Ishibashi, M.; Kubota, S.; Maki, M.; Hatanaka, M. Nucleolar targeting signal of human T-cell leukemia virus type I rex-encoded protein is essential for cytoplasmic accumulation of unspliced viral mRNA. Proc. Natl. Acad. Sci. USA 1989, 86, 9798–9802. [Google Scholar] [CrossRef]
- Malim, M.H.; Bohnlein, S.; Hauber, J.; Cullen, B.R. Functional Dessiction of the HIV-1 Rev Trans-Activator- Derivation of a Trans-Dominant Repressor of Rev Function. Cell 1989, 58, 205–214. [Google Scholar] [CrossRef]
- Siomi, H.; Shida, H.; Nam, S.H.; Nosaka, T.; Maki, M.; Hatanaka, M. Sequence Requirements for Nucleolar Localization of Human T Cell Leukemia Virus Type I pX Protein Which Regulates Viral RNA Processing. Cell 1988, 55, 197–209. [Google Scholar] [CrossRef]
- Arizala, J.A.C.; Chomchan, P.; Li, H.; Moore, R.; Ge, H.; Ouellet, D.L.; Rossi, J.J. Identification of Nucleolar Factors During HIV-1 Replication through Rev Immunoprecipitation and Mass Spectrometry. J. Vis. Exp. 2019, 148, e59329. [Google Scholar] [CrossRef]
- Fischer, U.; Huber, J.; Boelens, W.C.; Mattaj, I.W.; Luhrmann, R. The HIV-1 Rev Activation Domain Is a Nuclear Export Signal That Accesses an Export Pathway Used by Specific Cellular RNAs. Cell 1995, 82, 475–483. [Google Scholar] [CrossRef]
- Bogerd, H.P.; Fridell, R.A.; Benson, R.E.; Hua, J.; Cullen, B.R. Protein Sequence Requirements for Function of the Human T-Cell Leukemia Virus Type 1 Rex Nuclear Export Signal Delineated by a Novel In Vivo Randomization-Selection Assay. Mol. Cell. Biol. 1996, 16, 4207–4214. [Google Scholar] [CrossRef]
- Hofer, L.; Weichselbraun, I.; Quick, S.; Farrington, G.K.; Bohnlein, E.; Hauber, J. Mutational Analysis of the Human T-Cell Leukemia Virus Type I trans-Acting rex Gene Product. J. Virol. 1991, 65, 3379–3383. [Google Scholar] [CrossRef] [PubMed]
- Hope, T.J.; Bond, B.L.; McDonald, C.; Klein, N.P.; Parslow, T.G. Effector DOmains of Human Immunodeficiency Virus Type 1 Rev and Human T-Cell Leukemia Virus Type I Rex Are Functionally Interchangeable and Share an Essential Peptide Motif. J. Virol. 1991, 65, 6001–6007. [Google Scholar] [CrossRef] [PubMed]
- Kesic, M.; Doueiri, R.; Ward, M.; Semmes, O.J.; Green, P.L. Phosphorylation regulates human T-cell leukemia virus type 1 Rex function. Retrovirology 2009, 6, 105. [Google Scholar] [CrossRef] [PubMed]
- Kim, F.J.; Beeche, A.A.; Hunter, J.J.; Chin, D.J.; Hope, T.J. Characterization of the Nuclear Export Signal of Human T-Cell Lymphotropic Virus Type 1 Rex Reveals that Nuclear Export Is Mediated by Position-Variable Hydrophobic Interactions. Mol. Cell. Biol. 1996, 16, 5147–5155. [Google Scholar] [CrossRef]
- Kubota, S.; Nosaka, T.; Cullen, B.R.; Maki, M.; Hatanaka, M. Effects of Chimeric Mutants of Human Immunodeficiency Virus Type 1 Rev and Human T-Cell Leukemia Virus Type I Rex on Nucleolar Targeting Signals. J. Virol. 1991, 65, 2452–2456. [Google Scholar] [CrossRef]
- Nawroth, I.; Mueller, F.; Basyuk, E.; Beerens, N.; Rahbek, U.L.; Darzacq, X.; Bertrand, E.; Kjems, J.; Schmidt, U. Stable assembly of HIV-1 export complexes occurs cotranscriptionally. RNA 2014, 20, 1–8. [Google Scholar] [CrossRef]
- Rosen, C.A.; Terwilliger, E.; Dayton, A.; Sodroski, J.G.; Haseltine, W.A. Intragenic cis-acting art gene-responsive sequences of the human immunodeficiency virus. Proc. Natl. Acad. Sci. USA 1988, 85, 2071–2075. [Google Scholar] [CrossRef]
- Felber, B.K.; Hadzopoulou-Cladaras, M.; Cladaras, C.; Copeland, T.; Pavlakis, G.N. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 1989, 86, 1495–1499. [Google Scholar] [CrossRef]
- Daly, T.J.; Cook, K.S.; Gray, G.S.; Maione, T.E.; Rusche, J.R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 1989, 342, 816–819. [Google Scholar] [CrossRef]
- Hadzopoulou-Cladaras, M.; Felber, B.K.; CLadaras, C.; Athanassopoulos, A.; Tse, A.; Pavlakis, G.N. The rev (trs/art) Protein of Human Immunodeficiency Virus Type 1 Affects Viral mRNA and Protein Expression via a cis-Acting Sequence in the env Region. J. Virol. 1989, 63, 1265–1274. [Google Scholar] [CrossRef] [PubMed]
- Le, S.-Y.; Malim, M.H.; Cullen, B.R.; Maizel, J.V. A highly conserved RNA folding region coincident with the Rev response element of primate immunodeficiency viruses. Nucleic Acids Res. 1990, 18, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, S.; Finch, J.T.; Gait, M.J.; Karn, J.; Singh, M. Human immunodeficiency virus type 1 regulator of virion expression, rev, forms nucleoprotein filaments after binding to a purine-rich “bubble” located within the rev-responsive region of viral mRNAs. Proc. Natl. Acad. Sci. USA 1991, 88, 7366–7370. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P.; Zapp, M.L.; Green, M.R.; Szostak, J.W. HIV-1 rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell 1991, 67, 529–536. [Google Scholar] [CrossRef]
- Tiley, L.S.; Malim, M.H.; Tewary, H.K.; Stockley, P.G.; Cullen, B.R. Identification of a high-affinity RNA-binding site for the human immunodeficiency virus type 1 Rev protein. Proc. Natl. Acad. Sci. USA 1992, 89, 758–762. [Google Scholar] [CrossRef]
- Jayaraman, B.; Crosby, D.C.; Homer, C.; Ribeiro, I.; Mavor, D.; Frankel, A.D. RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex. Elife 2014, 3, e04120. [Google Scholar] [CrossRef]
- Olsen, H.S.; Cochrane, A.W.; Dillon, P.J.; Nalin, C.M.; Rosen, C.A. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured reigon in env mRNA is dependent on multimer formation mediated through a basic stretch of amino acids. Genes Dev. 1990, 4, 1357–1364. [Google Scholar] [CrossRef]
- Bohnlein, E.; Berger, J.; Hauber, J. Functional Mapping of the Human Immunodeficiency Virus Type 1 Rev RNA Binding Domain:New Insights into the Domain Structure of Rev and Rex. J. Virol. 1991, 65, 7051–7055. [Google Scholar] [CrossRef]
- Jensen, T.H.; Leffers, H.; Kjems, J. Intermolecular Binding Sites of Human Immunodeficiency Virus Type 1 Rev Protein Determined by Protein Footprinting. J. Biol. Chem. 1995, 270, 13777–13784. [Google Scholar] [CrossRef]
- Jain, C.; Belasco, J.G. A Structural Model for the HIV-1 Rev-RRE Complex Deduced from Altered-Specificity Rev Variants Isolated by a Rapid Genetic Strategy. Cell 1996, 87, 115–125. [Google Scholar] [CrossRef]
- Keller, R.; Montagnier, L.; Cordonnier, A. Characterization of a Nuclear Retention Sequence within the 3′ Region of the HIV-2 Envelope Gene. Virology 1992, 192, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Paulous, S.; Emerman, M.; Keller, R.; Montagnier, L.; Cordonnier, A. Functional mapping of the rev-responsive element of human immunodeficiency virus type 2 (HIV02): Influence of HIV-2 envelope encoding sequences on HIV-1 gp120 expression in the presence or absence of Rev. J. Gen. Virol. 1992, 73, 1773–1780. [Google Scholar] [CrossRef] [PubMed]
- Lusvarghi, S.; Sztuba-Solinska, J.; Purzycka, K.J.; Pauly, G.T.; Rausch, J.W.; Grice, S.F. The HIV-2 Rev-response element: Determining secondary structure and defining folding intermediates. Nucleic Acids Res. 2013, 41, 6637–6649. [Google Scholar] [CrossRef] [PubMed]
- Seiki, M.; Inoue, J.-I.; Hidaka, M.; Yoshida, M. Two cis-acting elements responsible for posttranscriptional trans-regulation of gene expression of human T-cell leukemia virus type I. Proc. Natl. Acad. Sci. USA 1988, 85, 7124–7128. [Google Scholar] [CrossRef] [PubMed]
- Ballaun, C.; Farrington, G.K.; Dobrovnik, M.; Rusche, J.; Hauber, J.; Bohnlein, E. Function analysis of Human T-cell Leukemia Virus Type I rex-Response Element: Direct RNA Binding of Rex Protein Correlates with In Vivo Activity. J. Virol. 1991, 65, 4408–4413. [Google Scholar] [CrossRef] [PubMed]
- Bogerd, H.P.; Huckaby, G.L.; Ahmed, Y.F.; Hanly, S.M.; Greene, W.C. The type I human T-cell leukemia virus (HTLV-I) Rex trans-activator binds directly to the HTLV-I Rex and the type 1 human immunodeficiency virus Rev RNA response elements. Proc. Natl. Acad. Sci. USA 1991, 88, 5704–5708. [Google Scholar] [CrossRef]
- Grassmann, R.; Berchtold, S.; Aepinus, C.; Callaun, C.; Boehnlein, E.; Fleckenstein, B. In Vitro Binding of Human T-Cell Leukemia Virus rex Proteins to the rex-Response Elelment of Viral Transcripts. J. Virol. 1991, 65, 3721–3727. [Google Scholar] [CrossRef]
- Toyoshima, H.; Itoh, M.; Inoue, J.-I.; Seiki, M.; Takaku, F.; Yoshida, M. Secondary Structure of the Human T-Cell Leukemia Virus Type 1 rex-Responsive Element Is Essential for rex Regulation of RNA Processing and Transport of Unspliced RNAs. J. Virol. 1990, 64, 2825–2832. [Google Scholar] [CrossRef]
- Baskerville, S.; Zapp, M.; Ellington, A. High-Resolution Mapping of the Human T-Cell Leukemia Virus Type 1 Rex-Binding Element by In Vitro Selection. J. Virol. 1995, 69, 7559–7569. [Google Scholar] [CrossRef]
- Grone, M.; Hoffman, E.; Berchtold, S.; Cullen, B.R.; Grassmann, R. A Single Stem-Loop Structure within the HTLV-1 Rex Response Element Is Sufficient to Mediate Rex Activity in Vivo. Virology 1994, 204, 144–152. [Google Scholar] [CrossRef]
- Weichselbraun, I.; Berger, J.; Dobrovnik, M.; Bogerd, H.; Grassmann, R.; Greene, W.C.; Hauber, J.; Bohnlein, E. Dominant-Negative Mutants Are Clustered in a Domain of the Human T-Cell Leukemia Virus Type I Rex Protein: Implications for trans Dominance. J. Virol. 1992, 66, 4540–4545. [Google Scholar] [CrossRef] [PubMed]
- Rimsky, L.; Hauber, J.; Dukovich, M.; Malim, M.H.; Langlois, A.; Cullen, B.R.; Greene, W.C. Functional replacement of the HIV-1 rev protein by the HTLV-1 rex protein. Nature 1988, 335, 738–740. [Google Scholar] [CrossRef] [PubMed]
- Hanly, S.M.; Rimsky, L.T.; Malim, M.H.; Kim, J.H.; Hauber, J.; Dodon, M.D.; Le, S.-Y.; Maizel, J.N.; Cullen, B.R.; Greene, W.C. Comparative analysis of the HTLV-I Rex and HIV-1 Rev trans-regulatory proteins and their RNA response elements. Genes Dev. 1989, 3, 1534–1544. [Google Scholar] [CrossRef]
- Sakai, H.; Siomi, H.; Shida, H.; Kiyomasu, T.; Adachi, A. Functional Comparison of Transactivation by Human Retrovirus rec and rex Genes. J. Virol. 1990, 64, 5833–5839. [Google Scholar] [CrossRef] [PubMed]
- Lewis, N.; Williams, J.; Rekosh, D.; Hammarskjold, M.-L. Identification of a cis-Acting Element in Human Immunodeficiency Virus Type 2 (HIV-2) That is Responsive to the HIV-1 rev and Human T-Cell Leukemia Virus Types I and II rex Proteins. J. Virol. 1990, 64, 1690–1697. [Google Scholar] [CrossRef]
- Garrett, E.D.; Cullen, B.R. Comparative Analysis of Rev Function in Human Immunodeficiency Virus Types 1 and 2. J. Virol. 1992, 66, 4288–4294. [Google Scholar] [CrossRef]
- Dillon, P.J.; Nelbock, P.; Perkins, A.; Rosen, C.A. Function of the Human Immunodeficiency Virus Types 1 and 2 Rev Proteins Is Dependent on Their Ability To Interact with a Structured Region Present in env Gene mRNA. J. Virol. 1990, 64, 4428–4437. [Google Scholar] [CrossRef]
- Bai, Y.; Tambe, A.; Zhou, K.; Doudna, J.A. RNA-guided assembly of Rev-RRE nuclear export complexes. Elife 2014, 3, e03656. [Google Scholar] [CrossRef]
- Malim, M.H.; Cullen, B.R. HIV-1 Structural Gene Expression Requires the Binding of Multiple Rev Monomers to the Viral RRE: Implications for HIV-1 Latency. Cell 1991, 65, 241–248. [Google Scholar] [CrossRef]
- Mann, D.A.; Mikaelian, I.; Zemmel, R.W.; Green, S.M.; Lowe, A.D.; Kimura, T.; Singh, M.; Butler, P.J.G.; Gait, M.H.; Karn, J. Co-operative Rev Binding to Stem I of the Rev-response Element Modulates Human Immunodeficiency Virus Type-I Late Gene Expression. J. Mol. Biol. 1994, 241, 193–207. [Google Scholar] [CrossRef]
- Jain, C.; Belasco, J.G. Structural Model for the Cooperative Assembly of HIV-1 Rev Multimers on the RRE as Deduced from Analysis of Assembly-Defective Mutants. Mol. Cell 2001, 7, 602–614. [Google Scholar] [CrossRef]
- Pond, S.J.; Ridgeway, W.K.; Robertson, R.; Wang, J.; Millar, D.P. HIV-1 Rev protein assembles on viral RNA one molecule at a time. Proc. Natl. Acad. Sci. USA 2009, 106, 1404–1408. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, M.D.; Booth, D.S.; Jayaraman, B.; Cheng, Y.; Frankel, A.D. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc. Natl. Acad. Sci. USA 2010, 107, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Zapp, M.L.; Hope, T.J.; Parslow, T.G.; Green, M.R. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: A dual function for an arginie-rich binding motif. Proc. Natl. Acad. Sci. USA 1991, 88, 7734–7738. [Google Scholar] [CrossRef]
- Thomas, S.L.; Oft, M.; Jaksche, H.; Casari, G.; Heger, P.; Dobrovnik, M.; Bevec, D.; Hauber, J. Functional Analysis of the Human Immunodeficiency Virus Type 1 Rev Protein Oligomerization Interface. J. Virol. 1998, 72, 2935–2944. [Google Scholar] [CrossRef] [PubMed]
- Trikha, R.; Brighty, D.W. Phenotypic analysis of human immunodeficiency virus type 1 Rev trimerization-interface mutants in human cells. J. Gen. Virol. 2005, 86, 1509–1513. [Google Scholar] [CrossRef] [PubMed]
- Daly, T.J.; Rennert, P.; Lynch, P.; Barry, J.K.; Dundas, M.; Rusche, J.R.; Doten, R.C.; Auer, M.; Farrington, G.K. Perturbation of the carboxy terminus of HIV-1 Rev affects multimerization on the Rev responsive element. Biochemistry 1993, 32, 8945–8954. [Google Scholar] [CrossRef]
- Furuta, R.A.; Sakai, H.; Kawamura, M.; Tokunaga, K.; Satanaka, M.; Adachi, A. Functionality of Chimeric Rev Proteins of HIV/SIV. Virus Genes 1995, 11, 11–14. [Google Scholar] [CrossRef]
- Daugherty, M.D.; Liu, B.; Frankel, A.D. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct Mol. Biol. 2010, 17, 1337–1342. [Google Scholar] [CrossRef]
- Heger, P.; Rosorius, O.; Koch, C.; Casari, G.; Grassmann, R.; Hauber, J. Multimer Formation Is Not Essential for Nuclear Export of Human T-Cell Leukemia Virus Type 1 Rex trans-Activator Protein. J. Virol. 1998, 72, 8659–8668. [Google Scholar]
- Booth, D.S.; Cheng, Y.; Frankel, A.D. The export receptor Crm1 forms a dimer to promote nuclear export of HIV RNA. Elife 2014, 3, e04121. [Google Scholar] [CrossRef] [PubMed]
- Askjaer, P.; Jensen, T.H.; Nilsson, J.; Englmeier, L.; Kjems, J. The Specificty of the CRM1-Rev Nuclear Export Signal Interaction Is Mediated by RanGTP. J. Biol. Chem. 1998, 273, 33414–33422. [Google Scholar] [CrossRef] [PubMed]
- Edgcomb, S.P.; Carmel, A.B.; Naji, S.; Ambrus-Aikelin, G.; Reyes, J.R.; Saphire, A.C.; Gerace, L.; Williamson, J.R. DDX1 is an RNA-dependent ATPase involved in HIV-1 Rev function and virus replication. J. Mol. Biol. 2012, 415, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, R.; Hammond, J.A.; Pauszek, R.F., 3rd; Anderson, R.M.; Pedron, I.; van der Schans, E.; Williamson, J.R.; Millar, D.P. A DEAD-box protein acts through RNA to promote HIV-1 Rev-RRE assembly. Nucleic Acids Res. 2017, 45, 4632–4641. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hammond, J.A.; Lamichhane, R.; Millar, D.P.; Williamson, J.R. A DEAD-Box Helicase Mediates an RNA Structural Transition in the HIV-1 Rev Response Element. J. Mol. Biol. 2017, 429, 697–714. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yedavalli, V.S.; Neuveut, C.; Chi, Y.H.; Kleiman, L.; Jeang, K.T. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 2004, 119, 381–392. [Google Scholar] [CrossRef]
- Mahboobi, S.H.; Javanpour, A.A.; Mofrad, M.R. The interaction of RNA helicase DDX3 with HIV-1 Rev-CRM1-RanGTP complex during the HIV replication cycle. PLoS ONE 2015, 10, e0112969. [Google Scholar] [CrossRef]
- Liu, H.; Hu, P.W.; Budhiraja, S.; Misra, A.; Couturier, J.; Lloyd, R.E.; Lewis, D.E.; Kimata, J.T.; Rice, A.P. PACS1 is an HIV-1 cofactor that functions in Rev-mediated nuclear export of viral RNA. Virology 2020, 540, 88–96. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, H.; Na, L.; Du, C.; Zhang, Z.; Zheng, Y.H.; Wang, X. ANP32A and ANP32B are key factors in the Rev-dependent CRM1 pathway for nuclear export of HIV-1 unspliced mRNA. J. Biol. Chem. 2019, 294, 15346–15357. [Google Scholar] [CrossRef]
- Kiss, A.; Li, L.; Gettemeier, T.; Venkatesh, L.K. Functional analysis of the interaction of the human immunodeficiency virus type 1 Rev nuclear export signal with its cofactors. Virology 2003, 314, 591–600. [Google Scholar] [CrossRef][Green Version]
- Hofmann, W.; Reichart, B.; Ewald, A.; Muller, E.; Schmitt, I.; Stauber, R.H.; Lottspeich, F.; Jockusch, B.M.; Scheer, U.; Hauber, J.; et al. Cofactor Requirements for Nuclear Export of Rev Response Element (RRE)– and Constitutive Transport Element (CTE)–containing Retroviral RNAs- An Unexpected Role for Actin. J. Cell Biol. 2001, 152, 896–910. [Google Scholar] [CrossRef] [PubMed]
- Ruhl, M.; Himmelspach, M.; Bahr, G.M.; Hammerschmid, F.; Jaksche, H.; Wolff, B.; Sschauer, H.; Farrington, G.K.; Probst, H.; Bevec, D.; et al. Eukaryotic Initiation Factor 5A Is a Cellular Target of the Human Immunodeficiency Virus Type 1 Rev Activation Domain Mediating Trans-Activation. J. Cell Biol. 1993, 123, 1309–1320. [Google Scholar] [CrossRef] [PubMed]
- Fritz, C.C.; Zapp, M.L.; Green, M.R. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature 1995, 376, 530–533. [Google Scholar] [CrossRef] [PubMed]
- Stutz, F.; Neville, M.; Rosbash, M. Identification of a Novel Nuclear Pore-Associated Protein as a Functional Target of the HIV-1 Rev Protein in Yeast. Cell 1995, 82, 495–506. [Google Scholar] [CrossRef][Green Version]
- Bogerd, H.P.; Fridell, R.A.; Madore, S.; Cullen, B.R. Identification of a Novel Cellular Cofactor for the Rev/Rex Class of Retroviral Regulatory Proteins. Cell 1995, 82, 485–494. [Google Scholar] [CrossRef]
- Ajamian, L.; Abel, K.; Rao, S.; Vyboh, K.; Garcia-de-Gracia, F.; Soto-Rifo, R.; Kulozik, A.E.; Gehring, N.H.; Mouland, A.J. HIV-1 Recruits UPF1 but Excludes UPF2 to Promote Nucleocytoplasmic Export of the Genomic RNA. Biomolecules 2015, 5, 2808–2839. [Google Scholar] [CrossRef]
- Huang, F.; Zhang, J.; Zhang, Y.; Geng, G.; Liang, J.; Li, Y.; Chen, J.; Liu, C.; Zhang, H. RNA helicase MOV10 functions as a co-factor of HIV-1 Rev to facilitate Rev/RRE-dependent nuclear export of viral mRNAs. Virology 2015, 486, 15–26. [Google Scholar] [CrossRef]
- Banerjee, A.; Benjamin, R.; Balakrishnan, K.; Ghosh, P.; Banerjee, S. Human protein Staufen-2 promotes HIV-1 proliferation by positively regulating RNA export activity of viral protein Rev. Retrovirology 2014, 11, 18. [Google Scholar] [CrossRef]
- Hulver, M.J.; Trautman, J.P.; Goodwin, A.P.; Roszczenko, S.K.; Fogarty, K.H.; Miller, H.B. Human Tat-specific factor 1 binds the HIV-1 genome and selectively transports HIV-1 RNAs. Mol. Biol. Rep. 2020, 47, 1759–1772. [Google Scholar] [CrossRef]
- Gudleski, N.; Flanagan, J.M.; Ryan, E.P.; Bewley, M.C.; Parent, L.J. Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 9358–9363. [Google Scholar] [CrossRef]
- Tuffy, K.M.; Maldonado, R.J.K.; Chang, J.; Rosenfeld, P.; Cochrane, A.; Parent, L.J. HIV-1 Gag Forms Ribonucleoprotein Complexes with Unspliced Viral RNA at Transcription Sites. Viruses 2020, 12, 1281. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Grunwald, D.; Sardo, L.; Galli, A.; Plisov, S.; Nikolaitchik, O.A.; Chen, D.; Lockett, S.; Larson, D.R.; Pathak, V.K.; et al. Cytoplasmic HIV-1 RNA is mainly transported by diffusion in the presence or absence of Gag protein. Proc. Natl. Acad. Sci. USA 2014, 111, E5205–E5213. [Google Scholar] [CrossRef] [PubMed]
- Swanson, C.M.; Puffer, B.A.; Ahmad, K.M.; Doms, R.W.; Malim, M.H. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 2004, 23, 2632–2640. [Google Scholar] [CrossRef]
- Moore, M.D.; Nikolaitchik, O.A.; Chen, J.; Hammarskjold, M.L.; Rekosh, D.; Hu, W.S. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009, 5, e1000627. [Google Scholar] [CrossRef]
- Chen, J.; Umunnakwe, C.; Sun, D.Q.; Nikolaitchik, O.A.; Pathak, V.K.; Berkhout, B.; Das, A.T.; Hu, W.S. Impact of Nuclear Export Pathway on Cytoplasmic HIV-1 RNA Transport Mechanism and Distribution. mBio 2020, 11, e01578-20. [Google Scholar] [CrossRef] [PubMed]
- Pocock, G.M.; Becker, J.T.; Swanson, C.M.; Ahlquist, P.; Sherer, N.M. HIV-1 and M-PMV RNA Nuclear Export Elements Program Viral Genomes for Distinct Cytoplasmic Trafficking Behaviors. PLoS Pathog. 2016, 12, e1005565. [Google Scholar] [CrossRef]
- Jouvenet, N.; Simon, S.M.; Bieniasz, P.D. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. USA 2009, 106, 19114–19119. [Google Scholar] [CrossRef] [PubMed]
- Bernacchi, S.; Abd El-Wahab, E.W.; Dubois, N.; Hijnen, M.; Smyth, R.P.; Mak, J.; Marquet, R.; Paillart, J.C. HIV-1 Pr55(Gag) binds genomic and spliced RNAs with different affinity and stoichiometry. RNA Biol. 2017, 14, 90–103. [Google Scholar] [CrossRef]
- Poole, E.; Strappe, P.; Mok, H.P.; Hicks, R.; Lever, A.M. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic 2005, 6, 741–755. [Google Scholar] [CrossRef]
- Kemler, I.; Meehan, A.; Poeschla, E.M. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J. Virol. 2010, 84, 6352–6366. [Google Scholar] [CrossRef]
- Ding, P.; Kharytonchyk, S.; Waller, A.; Mbaekwe, U.; Basappa, S.; Kuo, N.; Frank, H.M.; Quasney, C.; Kidane, A.; Swanson, C.; et al. Identification of the initial nucleocapsid recognition element in the HIV-1 RNA packaging signal. Proc. Natl. Acad. Sci. USA 2020, 117, 17737–17746. [Google Scholar] [CrossRef] [PubMed]
- Durand, S.; Seigneuret, F.; Burlaud-Gaillard, J.; Lemoine, R.; Tassi, M.F.; Moreau, A.; Mougel, M.; Roingeard, P.; Tauber, C.; de Rocquigny, H. Quantitative analysis of the formation of nucleoprotein complexes between HIV-1 Gag protein and genomic RNA using transmission electron microscopy. J. Biol. Chem. 2021, 298, 101500. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Clerte, C.; Chamontin, C.; Basyuk, E.; Laine, S.; Hottin, J.; Bertrand, E.; Margeat, E.; Mougel, M. Imaging HIV-1 RNA dimerization in cells by multicolor super-resolution and fluctuation microscopies. Nucleic Acids Res. 2016, 44, 7922–7934. [Google Scholar] [CrossRef] [PubMed]
- Boutant, E.; Bonzi, J.; Anton, H.; Nasim, M.B.; Cathagne, R.; Real, E.; Dujardin, D.; Carl, P.; Didier, P.; Paillart, J.C.; et al. Zinc Fingers in HIV-1 Gag Precursor Are Not Equivalent for gRNA Recruitment at the Plasma Membrane. Biophys. J. 2020, 119, 419–433. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, J.; Baumgartel, V.; Schrimpf, W.; Ivanchenko, S.; Digman, M.A.; Gratton, E.; Krausslich, H.G.; Muller, B.; Lamb, D.C. Live-cell observation of cytosolic HIV-1 assembly onset reveals RNA-interacting Gag oligomers. J. Cell Biol. 2015, 210, 629–646. [Google Scholar] [CrossRef]
- Monette, A.; Niu, M.; Chen, L.; Rao, S.; Gorelick, R.J.; Mouland, A.J. Pan-retroviral Nucleocapsid-Mediated Phase Separation Regulates Genomic RNA Positioning and Trafficking. Cell Rep. 2020, 31, 107520. [Google Scholar] [CrossRef]
- Mouland, A.J.; Mercier, J.; Luo, M.; Bernier, L.; DesGroseillers, L.; Cohen, E.A. The Double-Stranded RNA-Binding Protein Staufen Is Incorporated in Human Immunodeficiency Virus Type 1: Evidence for a ROle in Genomic RNA Encapsidation. J. Virol. 2000, 74, 5441–5451. [Google Scholar] [CrossRef]
- Abrahamyan, L.G.; Chatel-Chaix, L.; Ajamian, L.; Milev, M.P.; Monette, A.; Clement, J.F.; Song, R.; Lehmann, M.; DesGroseillers, L.; Laughrea, M.; et al. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci 2010, 123, 369–383. [Google Scholar] [CrossRef]
- Rao, S.; Hassine, S.; Monette, A.; Amorim, R.; Desgroseillers, L.; Mouland, A.J. HIV-1 requires Staufen1 to dissociate stress granules and to produce infectious viral particles. RNA 2019, 25, 727–736. [Google Scholar] [CrossRef]
- Soto-Rifo, R.; Valiente-Echeverria, F.; Rubilar, P.S.; Garcia-de-Gracia, F.; Ricci, E.P.; Limousin, T.; Decimo, D.; Mouland, A.J.; Ohlmann, T. HIV-2 genomic RNA accumulates in stress granules in the absence of active translation. Nucleic Acids Res. 2014, 42, 12861–12875. [Google Scholar] [CrossRef]
- Barajas, B.C.; Tanaka, M.; Robinson, B.A.; Phuong, D.J.; Chutiraka, K.; Reed, J.C.; Lingappa, J.R. Identifying the assembly intermediate in which Gag first associates with unspliced HIV-1 RNA suggests a novel model for HIV-1 RNA packaging. PLoS Pathog. 2018, 14, e1006977. [Google Scholar] [CrossRef] [PubMed]
- Sardo, L.; Hatch, S.C.; Chen, J.; Nikolaitchik, O.; Burdick, R.C.; Chen, D.; Westlake, C.J.; Lockett, S.; Pathak, V.K.; Hu, W.S. Dynamics of HIV-1 RNA Near the Plasma Membrane during Virus Assembly. J. Virol. 2015, 89, 10832–10840. [Google Scholar] [CrossRef] [PubMed]
- Crist, R.M.; Datta, S.A.; Stephen, A.G.; Soheilian, F.; Mirro, J.; Fisher, R.J.; Nagashima, K.; Rein, A. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: Implications for retrovirus assembly. J. Virol. 2009, 83, 2216–2225. [Google Scholar] [CrossRef]
- Kutluay, S.B.; Bieniasz, P.D. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010, 6, e1001200. [Google Scholar] [CrossRef] [PubMed]
- Dilley, K.A.; Nikolaitchik, O.A.; Galli, A.; Burdick, R.C.; Levine, L.; Li, K.; Rein, A.; Pathak, V.K.; Hu, W.-S. Interactions between HIV-1 Gag and Viral RNA Genome Enhance Virion Assembly. J. Virol. 2017, 91, e02319-16. [Google Scholar] [CrossRef] [PubMed]
- Clavel, F.; Orenstein, J.M. A Mutant of Human Immunodeficiency Virus with Reduced RNA Packaging and Abnormal Particle Morphology. J. Virol. 1990, 64, 5230–5234. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Aldovini, A. RNA incorporation is critical for retroviral particle integrity after cell membrane assembly of Gag complexes. J. Virol. 2002, 76, 11853–11865. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Noonan, K.; Aldovini, A. Nucleocapsid-RNA interactions are essential to structural stability but not to assembly of retroviruses. J. Virol. 2004, 78, 716–723. [Google Scholar] [CrossRef]
- Comas-Garcia, M.; Kroupa, T.; Datta, S.A.; Harvin, D.P.; Hu, W.S.; Rein, A. Efficient support of virus-like particle assembly by the HIV-1 packaging signal. Elife 2018, 7, e38438. [Google Scholar] [CrossRef]
- Mouland, A.J.; Xu, H.; Cui, H.; Krueger, W.; Munro, T.P.; Prasol, M.; Mercier, J.; Rekosh, D.; Smith, R.; Barbarese, E.; et al. RNA trafficking signals in human immunodeficiency virus type 1. Mol. Cell. Biol. 2001, 21, 2133–2143. [Google Scholar] [CrossRef]
- Levesque, K.; Halvorsen, M.; Abrahamyan, L.; Chatel-Chaix, L.; Poupon, V.; Gordon, H.; DesGroseillers, L.; Gatignol, A.; Mouland, A.J. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic 2006, 7, 1177–1193. [Google Scholar] [CrossRef] [PubMed]
- Molle, D.; Segura-Morales, C.; Camus, G.; Berlioz-Torrent, C.; Kjems, J.; Basyuk, E.; Bertrand, E. Endosomal trafficking of HIV-1 gag and genomic RNAs regulates viral egress. J. Biol. Chem. 2009, 284, 19727–19743. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, M.; Milev, M.P.; Abrahamyan, L.; Yao, X.J.; Pante, N.; Mouland, A.J. Intracellular transport of human immunodeficiency virus type 1 genomic RNA and viral production are dependent on dynein motor function and late endosome positioning. J. Biol. Chem. 2009, 284, 14572–14585. [Google Scholar] [CrossRef] [PubMed]
- Luban, J.; Goff, S.P. Mutationsal Analysis of cis-Acting Packaging Signals in Human Immunodeficiency Virus Type 1 RNA. J. Virol. 1994, 68, 3784–3793. [Google Scholar] [CrossRef]
- Chen, J.; Nikolaitchik, O.; Singh, J.; Wright, A.; Bencsics, C.E.; Coffin, J.M.; Ni, N.; Lockett, S.; Pathak, V.K.; Hu, W.S. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. USA 2009, 106, 13535–13540. [Google Scholar] [CrossRef]
- Nikolaitchik, O.A.; Dilley, K.A.; Fu, W.; Gorelick, R.J.; Tai, S.H.; Soheilian, F.; Ptak, R.G.; Nagashima, K.; Pathak, V.K.; Hu, W.S. Dimeric RNA recognition regulates HIV-1 genome packaging. PLoS Pathog. 2013, 9, e1003249. [Google Scholar] [CrossRef]
- Dilley, K.A.; Ni, N.; Nikolaitchik, O.A.; Chen, J.; Galli, A.; Hu, W.S. Determining the frequency and mechanisms of HIV-1 and HIV-2 RNA copackaging by single-virion analysis. J. Virol. 2011, 85, 10499–10508. [Google Scholar] [CrossRef]
- Berkhout, B.; Wamel, J.L.B.V. Role of the DIS Hairpin in Replication of Human Immunodeficiency Virus Type 1. J. Virol. 1996, 70, 6723–6732. [Google Scholar] [CrossRef]
- Laughrea, M.; Jette, L.; Mak, J.; Kleiman, L.; Liang, C.; Wainberg, M.A. Mutations in the Kissing-Loop Hairpin of Human Immunodeficiency Virus Type 1 Reduce Viral Infectivity as well as Genomic RNA Packaging and Dimerization. J. Virol. 1997, 71, 3397–3406. [Google Scholar] [CrossRef]
- Sakuragi, J.; Iwamoto, A.; Shioda, T. Dissociation of genome dimerization from packaging functions and virion maturation of human immunodeficiency virus type 1. J. Virol. 2002, 76, 959–967. [Google Scholar] [CrossRef][Green Version]
- CLever, J.L.; Parslow, T.G. Mutant Human Immunodeficiency Virus Type 1 Genomes with Defects in RNA Dimerization or Encapsidation. J. Virol. 1997, 71, 3407–3414. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Hatterschide, J.; Syu, Y.C.; Cantara, W.A.; Blower, R.J.; Hanson, H.M.; Mansky, L.M.; Musier-Forsyth, K. Human T-cell leukemia virus type 1 Gag domains have distinct RNA-binding specificities with implications for RNA packaging and dimerization. J. Biol. Chem. 2018, 293, 16261–16276. [Google Scholar] [CrossRef] [PubMed]
- Lever, A.; Gottlinger, H.; Haseltine, W.; Sodroski, J. Identification of a Sequence Required for Efficient Packaging of Human Immunodeficiency Virus Type 1 RNA into Virions. J. Virol. 1989, 63, 4085–4087. [Google Scholar] [CrossRef] [PubMed]
- Aldovini, A.; Young, R.A. Mutations of RNA and Protein Sequences Involved in Human Immunodeficiency Virus Type 1 Packaging Result in Production of Noninfectious Virus. J. Virol. 1990, 64, 1920–1926. [Google Scholar] [CrossRef]
- Garzino-Demo, A.; Gallo, R.C.; Arya, S.K. Human Immunodeficiency Virus Type 2 (HIV-2)—Packaging Signal and Associated Negative Regulatory Element. Hum. Gene Ther. 1995, 6, 177–184. [Google Scholar] [CrossRef]
- Damgaard, C.K.; Andersen, E.S.; Knudsen, B.; Gorodkin, J.; Kjems, J. RNA interactions in the 5′ region of the HIV-1 genome. J. Mol. Biol. 2004, 336, 369–379. [Google Scholar] [CrossRef]
- Luban, J.; Goff, S.P. Binding of Human Immunodeficiency Virus Type 1 (HIV-1) RNA to Recombinant HIV-1 gag Polyprotein. J. Virol. 1991, 65, 3203–3212. [Google Scholar] [CrossRef]
- Berkowitz, R.D.; Luban, J.; Goff, S.P. Specific Binding of Human Immunodeficiency Virus Type 1 gag Polyprotein and Nucleocapsid Protein to Viral RNAs Detected by RNA Mobility Shift Assays. J. Virol. 1993, 67, 7190–7200. [Google Scholar] [CrossRef]
- McBride, M.S.; Panganiban, A.R. The Human Immunodeficiency Virus Type 1 Encapsidation Site Is a Multipartite RNA Element Composed of Functional Hairpin Structures. J. Virol. 1996, 70, 2963–2973. [Google Scholar] [CrossRef]
- Heng, X.; Kharytonchyk, S.; Garcia, E.L.; Lu, K.; Divakaruni, S.S.; LaCotti, C.; Edme, K.; Telesnitsky, A.; Summers, M.F. Identification of a minimal region of the HIV-1 5′-leader required for RNA dimerization, NC binding, and packaging. J. Mol. Biol. 2012, 417, 224–239. [Google Scholar] [CrossRef]
- Liu, Y.; Nikolaitchik, O.A.; Rahman, S.A.; Chen, J.; Pathak, V.K.; Hu, W.S. HIV-1 Sequence Necessary and Sufficient to Package Non-viral RNAs into HIV-1 Particles. J. Mol. Biol. 2017, 429, 2542–2555. [Google Scholar] [CrossRef] [PubMed]
- McCann, E.M.; Lever, A.M.L. Location of cis-Acting Signals Important for RNA Encapsidation in the Leader Sequence of Human Immunodeficiency Virus Type 2. J. Virol. 1997, 71, 4133–4137. [Google Scholar] [CrossRef] [PubMed]
- Griffin, S.D.; Allen, J.F.; Lever, A.M. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: Cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 2001, 75, 12058–12069. [Google Scholar] [CrossRef] [PubMed]
- Poeschla, E.; Gilbert, J.; Li, X.; Huang, S.; Ho, A.; Wong-Staal, F. Identification of a Human Immunodeficiency Virus Type 2 (HIV-2) Encapsidation Determinant and Transduction of Nondividing Human Cells by HIV-2-Based Lentivirus Vectors. J. Virol. 1998, 72, 6527–6536. [Google Scholar] [CrossRef]
- Tsukahara, T.; Komatsu, H.; Kubo, M.; Obata, F.; Tozawa, H. Binding Properties of Human Immunodeficiency Virus Type-2 (HIV-2) RNA Corresponding to the Packaging Signal to its Nucleocapsid Protein. Biochem. Mol. Biol. Int. 1996, 40, 33–42. [Google Scholar] [CrossRef]
- Clever, J.; Sassetti, C.; Parslow, T.G. RNA Secondary Structure and Binding Sites for gag Gene Products in the 59 Packaging Signal of Human Immunodeficiency Virus Type 1. J. Virol. 1995, 69, 2101–2109. [Google Scholar] [CrossRef]
- Bacharach, E.; GOff, S.P. Binding of the Human Immunodeficiency Virus Type 1 Gag Protein to the Viral RNA Encapsidation Signal in the Yeast Three-Hybrid System. J. Virol. 1998, 72, 6944–6949. [Google Scholar] [CrossRef]
- Berkowitz, R.D.; Goff, S.P. Analysis of Binding Elements in the Human Immunodeficiency Virus Type 1 Genomic RNA and Nucleocapsid Protein. Virology 1994, 202, 233–246. [Google Scholar] [CrossRef]
- McBride, M.S.; Panganiban, A.T. Position Dependence of Functional Hairpins Important for Human Immunodeficiency Virus Type 1 RNA Encapsidation In Vivo. J. Virol. 1997, 71, 2050–2058. [Google Scholar] [CrossRef]
- Rong, L.; Russell, R.S.; Hu, J.; Laughrea, M.; Wainberg, M.A.; Liang, C. Deletion of stem-loop 3 is compensated by second-site mutations within the Gag protein of human immunodeficiency virus type 1. Virology 2003, 314, 221–228. [Google Scholar] [CrossRef][Green Version]
- Wilkinson, K.A.; Gorelick, R.J.; Vasa, S.M.; Guex, N.; Rein, A.; Mathews, D.H.; Giddings, M.C.; Weeks, K.M. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008, 6, e96. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Jun, H.J.; Jang, S.I.; You, J.C. The determination of importance of sequences neighboring the Psi sequence in lentiviral vector transduction and packaging efficiency. PLoS ONE 2012, 7, e50148. [Google Scholar] [CrossRef] [PubMed]
- Nikolaitchik, O.A.; Somoulay, X.; Rawson, J.M.O.; Yoo, J.A.; Pathak, V.K.; Hu, W.S. Unpaired Guanosines in the 5’ Untranslated Region of HIV-1 RNA Act Synergistically To Mediate Genome Packaging. J. Virol. 2020, 94, e00439-20. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, K.; Zambrano, N.; Baldwin, E.T.; Shapiro, B.A.; Erickson, J.W.; Omichinski, J.G.; CLore, M.; Gronenborn, A.M.; Appella, E. Identification of a binding site for the human immunodeficiency virus type 1 nucleocapsid protein. Proc. Natl. Acad. Sci. USA 1993, 90, 5219–5223. [Google Scholar] [CrossRef] [PubMed]
- Houzet, L.; Paillart, J.C.; Smagulova, F.; Maurel, S.; Morichaud, Z.; Marquet, R.; Mougel, M. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 2007, 35, 2695–2704. [Google Scholar] [CrossRef]
- Lu, K.; Heng, X.; Summers, M.F. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011, 410, 609–633. [Google Scholar] [CrossRef]
- Abd El-Wahab, E.W.; Smyth, R.P.; Mailler, E.; Bernacchi, S.; Vivet-Boudou, V.; Hijnen, M.; Jossinet, F.; Mak, J.; Paillart, J.C.; Marquet, R. Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat. Commun. 2014, 5, 4304. [Google Scholar] [CrossRef]
- Smyth, R.P.; Despons, L.; Huili, G.; Bernacchi, S.; Hijnen, M.; Mak, J.; Jossinet, F.; Weixi, L.; Paillart, J.C.; von Kleist, M.; et al. Mutational interference mapping experiment (MIME) for studying RNA structure and function. Nat. Methods 2015, 12, 866–872. [Google Scholar] [CrossRef]
- Webb, J.A.; Jones, C.P.; Parent, L.J.; Rouzina, I.; Musier-Forsyth, K. Distinct binding interactions of HIV-1 Gag to Psi and non-Psi RNAs: Implications for viral genomic RNA packaging. RNA 2013, 19, 1078–1088. [Google Scholar] [CrossRef]
- Umunnakwe, C.N.; Duchon, A.; Nikolaitchik, O.A.; Rahman, S.A.; Liu, Y.; Chen, J.; Tai, S.; Pathak, V.K.; Hu, W.S. Specific Guanosines in the HIV-2 Leader RNA are Essential for Efficient Viral Genome Packaging. J. Mol. Biol. 2021, 433, 166718. [Google Scholar] [CrossRef]
- Sakuragi, S.; Kotani, O.; Yokoyama, M.; Shioda, T.; Sato, H.; Sakuragi, J.I. Identification of a Novel Cis-Acting Regulator of HIV-1 Genome Packaging. Int. J. Mol. Sci. 2021, 22, 3435. [Google Scholar] [CrossRef] [PubMed]
- Mansky, L.M.; Gajary, L.C. The primary nucleotide sequence of the bovine leukemia virus RNA packaging signal can influence efficient RNA packaging and virus replication. Virology 2002, 301, 272–280. [Google Scholar] [CrossRef] [PubMed]
- Gorelick, R.J.; Jr, S.M.N.; Jr, J.W.B.; Arthur, L.O.; Henderson, L.E.; Rein, A. Noninfectious Human Immunodeficiency Virus Type 1 Mutants Deficient in Genomic RNA. J. Virol. 1990, 64, 3207–3211. [Google Scholar] [CrossRef] [PubMed]
- Jowett, H.B.M.; Hockley, D.J.; Nermut, M.V.; Jones, I.M. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 1992, 73, 3079–3086. [Google Scholar] [CrossRef] [PubMed]
- Gorelick, R.J.; Gargliardi, T.D.; Bosche, W.J.; Wiltrout, T.A.; Coren, L.V.; Chabot, D.J.; Lifson, J.D.; Henderson, L.E.; Aurthur, L.O. Strict Conservation of the Retroviral Nucleocapsid Protein Zinc Finger Is Strongly Influenced by Its Role in Viral Infection Processes: Characterization of HIV-1 Particles Containing Mutant Nucleocapsid Zinc-Coordinating Sequences. Virology 1999, 256, 92–104. [Google Scholar] [CrossRef]
- Mark-Danieli, M.; Laham, N.; Kenan-Eichler, M.; Castiel, A.; Melamed, D.; Landau, M.; Bouvier, N.M.; Evans, M.J.; Bacharach, E. Single point mutations in the zinc finger motifs of the human immunodeficiency virus type 1 nucleocapsid alter RNA binding specificities of the gag protein and enhance packaging and infectivity. J. Virol. 2005, 79, 7756–7767. [Google Scholar] [CrossRef][Green Version]
- Gorelick, R.J.; Chabot, D.J.; Rein, A.; Henderson, L.E.; Aurthur, L.O. The Two Zinc FIngers in the Human Immunodeficiency VIrus Type 1 Nucleocapsid Protein Are Not FUnctionally Equivalent. J. Virol. 1993, 67, 4027–4036. [Google Scholar] [CrossRef]
- Dorfman, T.; Luban, J.; Goff, S.P.; Haseltine, W.A.; Gottlinger, H.G. Mapping of Functionally Important Residues of a Cysteine-Histidine Box in the Human Immunodeficiency Virus Type 1 Nucleocapsid Protein. J. Virol. 1993, 67, 6159–6169. [Google Scholar] [CrossRef]
- Dannull, J.; Surovoy, A.; Jung, G.; Moelling, K. Specific binding of HIV-1 nucleocapsid proteins to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994, 13, 1525–1533. [Google Scholar] [CrossRef]
- Schwartz, M.D.; Fiore, D.; Panganiban, A.T. Distinct Functions and Requirements for the Cys-His Boxes of the Human Immunodeficiency Virus Type 1 Nucleocapsid Protein during RNA Encapsidation and Replication. J. Virol. 1997, 71, 9295–9305. [Google Scholar] [CrossRef]
- Guo, C.; Yao, X.; Wang, K.; Wang, J.; Wang, Y. Comparison of HIV-1 Gag and NCp7 in their selectivity for package signal, affinity for stem-loop 3, and Zn2+ content. Biochimie 2020, 179, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Rocquigny, H.D.; Gabus, C.; Vincent, A.; Fournie-Zaluski, M.-C.; Roques, B.; Darlix, J.-L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl. Acad. Sci. USA 1992, 89, 6472–6476. [Google Scholar] [CrossRef] [PubMed]
- Poon, D.T.K.; Wu, J.; Aldovini, A. Charged Amino Acid Residues of Human Immunodeficiency Virus Type 1 Nucleocapsid p7 Protein Involved in RNA Packaging and Infectivity. J. Virol. 1996, 70, 6607–6616. [Google Scholar] [CrossRef] [PubMed]
- Schmalzbauer, E.; Strack, B.; Dannull, J.; Guehmann, S.; Moelling, K. Mutations of Basic Amino Acids of MCp7 of Human Immunodeficiency Virus Type 1 Affect RNA Binding In Vitro. J. Virol. 1996, 70, 771–777. [Google Scholar] [CrossRef]
- Zhang, Y.; Barklis, E. Nucleocapsid Protein Effects on the Specificity of Retrovirus RNA Encapsidation. J. Virol. 1995, 69, 5716–5722. [Google Scholar] [CrossRef]
- Guzman, R.N.D.; Wu, Z.R.; Stalling, C.C.; Pappalardo, L.; Borer, P.N.; Summers, M.F. Structure of the HIV-1 Nucleocapsid Protein Bound to the SL3 Ψ-RNA Recognition Element. Science 1998, 279, 384–388. [Google Scholar] [CrossRef]
- Zhang, Y.; Barklis, E. Effects of Nucleocapsid Mutations on Human Immunodeficiency Virus Assembly and RNA Encapsidation. J. Virol. 1997, 71, 6765–6776. [Google Scholar] [CrossRef]
- Russell, R.S.; Roldan, A.; Detorio, M.; Hu, J.; Wainberg, M.A.; Liang, C. Effects of a single amino acid substitution within the p2 region of human immunodeficiency virus type 1 on packaging of spliced viral RNA. J. Virol. 2003, 77, 12986–12995. [Google Scholar] [CrossRef]
- Dubois, N.; Khoo, K.K.; Ghossein, S.; Seissler, T.; Wolff, P.; McKinstry, W.J.; Mak, J.; Paillart, J.C.; Marquet, R.; Bernacchi, S. The C-terminal p6 domain of the HIV-1 Pr55(Gag) precursor is required for specific binding to the genomic RNA. RNA Biol. 2018, 15, 923–936. [Google Scholar] [CrossRef]
- Sun, M.; Grigsby, I.F.; Gorelick, R.J.; Mansky, L.M.; Musier-Forsyth, K. Retrovirus-specific differences in matrix and nucleocapsid protein-nucleic acid interactions: Implications for genomic RNA packaging. J. Virol. 2014, 88, 1271–1280. [Google Scholar] [CrossRef]
- Pachulska-Wieczorek, K.; Blaszczyk, L.; Biesiada, M.; Adamiak, R.W.; Purzycka, K.J. The matrix domain contributes to the nucleic acid chaperone activity of HIV-2 Gag. Retrovirology 2016, 13, 18. [Google Scholar] [CrossRef] [PubMed]
- Duchon, A.; Santos, S.; Chen, J.; Brown, M.; Nikolaitchik, O.A.; Tai, S.; Chao, J.A.; Freed, E.O.; Pathak, V.K.; Hu, W.-S. Plasma Membran Anchoring and Gag:Gag Multimerization on Viral RNA are Critical Properties of HIV-1 Gag Required to Mediate Efficient Genome Packaging. mBio 2021, 12, e0325421. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Fu, W.; Nikolaitchik, O.; Chen, J.; Ptak, R.G.; Hu, W.S. Dimer initiation signal of human immunodeficiency virus type 1: Its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 2007, 81, 4002–4011. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Rahman, S.A.; Nikolaitchik, O.A.; Grunwald, D.; Sardo, L.; Burdick, R.C.; Plisov, S.; Liang, E.; Tai, S.; Pathak, V.K.; et al. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc. Natl. Acad. Sci. USA 2016, 113, E201–E208. [Google Scholar] [CrossRef]
- Darlix, J.-L.; Gabus, C.; Nugeyre, M.-T.; Clavel, F.; Barre-Sinoussi, F. Cis Elements and Trans-acting Factors Involved in the RNA Dimerization of the Human Immunodeficiency Virus HIV-1. J. Mol. Biol. 1990, 216, 689–699. [Google Scholar] [CrossRef]
- Skripkin, E.; Paillart, J.-C.; Marquet, R.; Ehresmann, B.; Ehresmann, C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA 1994, 91, 4945–4949. [Google Scholar] [CrossRef]
- Paillart, J.-C.; Berthoux, L.; Ottmann, M.; Darlix, J.-L.; Marquet, R.; Ehresmann, B.; Ehresmann, C. A Dual Role of the Putative RNA Dimerization Initiation Site of Human Immunodeficiency Virus Type 1 in Genomic RNA Packaging and Proviral DNA Synthesis. J. Virol. 1996, 70, 8348–8354. [Google Scholar] [CrossRef]
- Lanchy, J.M.; Ivanovitch, J.D.; Lodmell, J.S. A structural linkage between the dimerization and encapsidation signals in HIV-2 leader RNA. RNA 2003, 9, 1007–1018. [Google Scholar] [CrossRef]
- Sakuragi, J.; Ueda, S.; Iwamoto, A.; Shioda, T. Possible role of dimerization in human immunodeficiency virus type 1 genome RNA packaging. J. Virol. 2003, 77, 4060–4069. [Google Scholar] [CrossRef][Green Version]
- Awang, G.; Sen, D. Mode of Dimerization of HIV-1 Genomic RNA. Biochemistry 1993, 32, 11453–11457. [Google Scholar] [CrossRef]
- Clever, J.L.; Wong, M.L.; Parslow, T.G. Requirements for Kissing-Loop-Mediated DImerization of Human Immunodeficiency Virus RNA. J. Virol. 1996, 70, 5902–5908. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.-C.; Skripkin, E.; Ehresmann, B.; Ehresmann, C.; Marquet, R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 1996, 93, 5572–5577. [Google Scholar] [CrossRef] [PubMed]
- Blakemore, R.J.; Burnett, C.; Swanson, C.; Kharytonchyk, S.; Telesnitsky, A.; Munro, J.B. Stability and conformation of the dimeric HIV-1 genomic RNA 5’UTR. Biophys. J. 2021, 120, 4874–4890. [Google Scholar] [CrossRef] [PubMed]
- Dirac, A.M.G.; Huthoff, H.; Kjems, J.; Berkhout, B. The Dimer Initiation Site Hairpin Mediates Dimerization of the Human Immunodeficiency Virus, Type 2 RNA Genome. J. Biol. Chem. 2001, 276, 32345–32352. [Google Scholar] [CrossRef]
- Lanchy, J.-M.; Stephen Lodmell, J. Alternate Usage of Two Dimerization Initiation Sites in HIV-2 Viral RNA In Vitro. J. Mol. Biol. 2002, 319, 637–648. [Google Scholar] [CrossRef]
- Purzycka, K.J.; Pachulska-Wieczorek, K.; Adamiak, R.W. The in vitro loose dimer structure and rearrangements of the HIV-2 leader RNA. Nucleic Acids Res. 2011, 39, 7234–7248. [Google Scholar] [CrossRef]
- Berkhout, B.; Essink, B.B.; Schoneveld, I. In vitro dimerization of HIV-2 leader RNA in the absence of PuGGAPuA motifs. FASEB J. 1993, 7, 181–187. [Google Scholar] [CrossRef]
- Jossinet, F.; Lodmell, J.S.; Ehresmann, C.; Ehresmann, B.; Marquet, R. Identification of the in vitro HIV-2/SIV RNA dimerization site reveals striking differences with HIV-1. J. Biol. Chem. 2001, 276, 5598–5604. [Google Scholar] [CrossRef]
- Deer, E.L.; Douk, B.; Lanchy, J.-M.; Lodmell, J.S. Elucidation and characterization of oligonucleotide-accessible sites on HIV-2 leader region RNA. Antisense Nucleic Acid Drug Dev. 2003, 13, 45–55. [Google Scholar] [CrossRef]
- Seif, E.; Niu, M.; Kleiman, L. Annealing to sequences within the primer binding site loop promotes an HIV-1 RNA conformation favoring RNA dimerization and packaging. RNA 2013, 19, 1384–1393. [Google Scholar] [CrossRef]
- Dirac, A.M.G.; Huthoff, H.; Kjems, J.; Berkhout, B. Requirements for RNA heterodimerization of the human immunodeficiency virus type 1 (HIV-1) and HIV-2 genomes. J. Gen. Virol. 2002, 83, 2533–2542. [Google Scholar] [CrossRef] [PubMed]
- Greatorex, J.S.; Laisse, V.; Dokhelar, M.-C.; Lever, A.M.L. Sequences involved in the dimerisation of human T cell leukaemia virus type-1 RNA. Nucleic Acids Res. 1996, 24, 2919–2923. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Blanc, I.L.; Greatorex, J.; Dokhelar, M.-C.; Lever, A.M. A 37 base sequence in the leader region of human T-cell leukemia virus type I is a high affinity dimerization site but is not essential for virus replication. J. Gen. Virol. 2000, 81, 105–108. [Google Scholar]
- Monie, T.; Greatorex, J.; Lever, A.M.L. Oligonucleotide mapping of the core genomic RNA dimer linkage in human T-cell leukaemia virus type-1. Virus Res. 2001, 78, 45–56. [Google Scholar] [CrossRef]
- Monie, T.P.; Greatorex, J.S.; Zacharias, M.; Lever, A.M.L. The Human T-Cell Lymphotropic Virus Type-I Dimerization Initiation Site Forms a Hairpin Loop, Unlike Previously Characterized Retroviral Dimerization Motifs. Biochemistry 2004, 43, 6085–6090. [Google Scholar] [CrossRef] [PubMed]
- Huthoff, H.; Berkhout, B. Two alternating structures of the HIV-1 leader RNA. RNA 2001, 7, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Dirac, A.M.G.; Huthoff, H.; Kjems, J.; Berkhout, B. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic Acids Res. 2002, 30, 2647–2655. [Google Scholar] [CrossRef] [PubMed]
- Lanchy, J.-M.; Rentz, C.A.; Ivanovitch, J.D.; Lodmell, J.S. Elements located upstream and downstream of the major splice donor site incluence the ability of HIV-2 leader RNA to dimerize in vitro. Biochemistry 2003, 42, 2634–2642. [Google Scholar] [CrossRef]
- Huthoff, H.; Berkhout, B. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res. 2001, 29, 2594–2600. [Google Scholar] [CrossRef]
- Abbink, T.E.; Berkhout, B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003, 278, 11601–11611. [Google Scholar] [CrossRef]
- Ooms, M.; Huthoff, H.; Russell, R.; Liang, C.; Berkhout, B. A riboswitch regulates RNA dimerization and packaging in human immunodeficiency virus type 1 virions. J. Virol. 2004, 78, 10814–10819. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Heng, X.; Garyu, L.; Monti, S.; Garcia, E.L.; Kharytonchyk, S.; Dorjsuren, B.; Kulandaivel, G.; Jones, S.; Hiremath, A.; et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 2011, 334, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.; Liu, Y.; Marchant, J.; Monti, S.; Seu, M.; Zaki, J.; Yang, A.L.; Bohn, J.; Ramakrishnan, V.; Singh, R.; et al. Conserved determinants of lentiviral genome dimerization. Retrovirology 2015, 12, 83. [Google Scholar] [CrossRef]
- Strong, C.L.; Lanchy, J.M.; Lodmell, J.S. Viral SELEX reveals individual and cooperative roles of the C-box and G-box in HIV-2 replication. RNA 2011, 17, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Deforges, J.; Chamond, N.; Sargueil, B. Structural investigation of HIV-1 genomic RNA dimerization process reveals a role for the Major Splice-site Donor stem loop. Biochimie 2012, 94, 1481–1489. [Google Scholar] [CrossRef]
- L’Hernault, A.; Greatorex, J.S.; Crowther, R.A.; Lever, A.M. Dimerisation of HIV-2 genomic RNA is linked to efficient RNA packaging, normal particle maturation and viral infectivity. Retrovirology 2007, 4, 90. [Google Scholar] [CrossRef] [PubMed]
- Lanchy, J.M.; Lodmell, J.S. An extended stem-loop 1 is necessary for human immunodeficiency virus type 2 replication and affects genomic RNA encapsidation. J. Virol. 2007, 81, 3285–3292. [Google Scholar] [CrossRef]
- Baig, T.T.; Lanchy, J.M.; Lodmell, J.S. Randomization and in vivo selection reveal a GGRG motif essential for packaging human immunodeficiency virus type 2 RNA. J. Virol. 2009, 83, 802–810. [Google Scholar] [CrossRef]
- Baig, T.T.; Lanchy, J.M.; Lodmell, J.S. HIV-2 RNA dimerization is regulated by intramolecular interactions in vitro. RNA 2007, 13, 1341–1354. [Google Scholar] [CrossRef]
- McBride, M.S.; Schwartz, M.D.; Panganiban, A.T. Efficient Encapsidation of Human Immunodeficiency VIrus Type 1 Vectors and Further Characterization of cis Elements Required for Encapsidation. J. Virol. 1997, 71, 4544–4554. [Google Scholar] [CrossRef]
- Kharytonchyk, S.; Brown, J.D.; Stilger, K.; Yasin, S.; Iyer, A.S.; Collins, J.; Summers, M.F.; Telesnitsky, A. Influence of gag and RRE Sequences on HIV-1 RNA Packaging Signal Structure and Function. J. Mol. Biol. 2018, 430, 2066–2079. [Google Scholar] [CrossRef] [PubMed]
- Keane, S.C.; Van, V.; Frank, H.M.; Sciandra, C.A.; McCowin, S.; Santos, J.; Heng, X.; Summers, M.F. NMR detection of intermolecular interaction sites in the dimeric 5′-leader of the HIV-1 genome. Proc. Natl. Acad. Sci. USA 2016, 113, 13033–13038. [Google Scholar] [CrossRef] [PubMed]
- Kaye, J.F.; Richardson, J.H.; Lever, A.M.L. cis-Acting Sequences Involved in Human Immunodeficiency Virus Type 1 RNA Packaging. J. Virol. 1995, 69, 6588–6592. [Google Scholar] [CrossRef] [PubMed]
- Blissenbach, M.; Grewe, B.; Hoffmann, B.; Brandt, S.; Uberla, K. Nuclear RNA export and packaging functions of HIV-1 Rev revisited. J. Virol. 2010, 84, 6598–6604. [Google Scholar] [CrossRef] [PubMed]
- Cockrell, A.S.; van Praag, H.; Santistevan, N.; Ma, H.; Kafri, T. The HIV-1 Rev/RRE system is required for HIV-1 5′ UTR cis elements to augment encapsidation of heterologous RNA into HIV-1 viral particles. Retrovirology 2011, 8, 51. [Google Scholar] [CrossRef]
- Grewe, B.; Ehrhardt, K.; Hoffmann, B.; Blissenbach, M.; Brandt, S.; Uberla, K. The HIV-1 Rev protein enhances encapsidation of unspliced and spliced, RRE-containing lentiviral vector RNA. PLoS ONE 2012, 7, e48688. [Google Scholar] [CrossRef]
- D’Costa, J.; Brown, H.M.; Kundra, P.; Davis-Warren, A.; Arya, S.K. Human immunodeficiency virus type 2 lentiviral vectors: Packaging signal and splice donor in expression and encapsidation. J. Gen. Virol. 2001, 82, 425–434. [Google Scholar] [CrossRef]
- Chamanian, M.; Purzycka, K.J.; Wille, P.T.; Ha, J.S.; McDonald, D.; Gao, Y.; Le Grice, S.F.; Arts, E.J. A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation. Cell Host Microbe 2013, 13, 181–192. [Google Scholar] [CrossRef]
- Nikolaitchik, O.A.; Hu, W.S. Deciphering the role of the Gag-Pol ribosomal frameshift signal in HIV-1 RNA genome packaging. J. Virol. 2014, 88, 4040–4046. [Google Scholar] [CrossRef]
- Kaye, J.F.; Lever, A.M.L. Nonreciprocal Packaging of Human Immunodeficiency Virus Type 1 and Type 2 RNA- a Possible Role for the p2 Domain of Gag in RNA Encapsidation. J. Virol. 1998, 72, 5877–5885. [Google Scholar] [CrossRef]
- Strappe, P.M.; Hampton, D.W.; Brown, D.; Cachon-Gonzalez, B.; Caldwell, M.; Fawcett, J.W.; Lever, A.M. Identification of unique reciprocal and non reciprocal cross packaging relationships between HIV-1, HIV-2 and SIV reveals an efficient SIV/HIV-2 lentiviral vector system with highly favourable features for in vivo testing and clinical usage. Retrovirology 2005, 2, 55. [Google Scholar] [CrossRef]
- Butsch, M.; Boris-Lawrie, K. Translation Is Not Required To Generate Virion Precursor RNA in Human Immunodeficiency Virus Type 1-Infected T Cells. J. Virol. 2000, 74, 11531–11537. [Google Scholar] [CrossRef] [PubMed]
- Dorman, N.; Lever, A. Comparison of Viral Genomic RNA Sorting Mechanisms in Human Immunodeficiency Virus Type 1 (HIV-1), HIV-2, and Moloney Murine Leukemia Virus. J. Virol. 2000, 74, 11413–11417. [Google Scholar] [CrossRef] [PubMed]
- Nikolaitchik, O.; Rhodes, T.D.; Ott, D.; Hu, W.S. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J. Virol. 2006, 80, 4691–4697. [Google Scholar] [CrossRef] [PubMed]
- Kharytonchyk, S.; Monti, S.; Smaldino, P.J.; Van, V.; Bolden, N.C.; Brown, J.D.; Russo, E.; Swanson, C.; Shuey, A.; Telesnitsky, A.; et al. Transcriptional start site heterogeneity modulates the structure and function of the HIV-1 genome. Proc. Natl. Acad. Sci. USA 2016, 113, 13378–13383. [Google Scholar] [CrossRef]
- Masuda, T.; Sato, Y.; Huang, Y.L.; Koi, S.; Takahata, T.; Hasegawa, A.; Kawai, G.; Kannagi, M. Fate of HIV-1 cDNA intermediates during reverse transcription is dictated by transcription initiation site of virus genomic RNA. Sci Rep. 2015, 5, 17680. [Google Scholar] [CrossRef]
- Brown, J.D.; Kharytonchyk, S.; Chaudry, I.; Iyer, A.S.; Carter, H.; Becker, G.; Desai, Y.; Glang, L.; Choi, S.H.; Singh, K.; et al. Structural basis for transcriptional start site control of HIV-1 RNA fate. Science 2020, 368, 413–417. [Google Scholar] [CrossRef]
- Ding, P.; Kharytonchyk, S.; Kuo, N.; Cannistraci, E.; Flores, H.; Chaudhary, R.; Sarkar, M.; Dong, X.; Telesnitsky, A.; Summers, M.F. 5′-Cap sequestration is an essential determinant of HIV-1 genome packaging. Proc. Natl. Acad. Sci. USA 2021, 118, e2112475118. [Google Scholar] [CrossRef]
- Sakuragi, S.; Shioda, T.; Sakuragi, J.I. Relationship between genome packaging and Gag translation/AUG of primate lentiviruses. Microbes Infect. 2019, 21, 119–123. [Google Scholar] [CrossRef]
- Chen, J.; Liu, Y.; Wu, B.; Nikolaitchik, O.A.; Mohan, P.R.; Chen, J.; Pathak, V.K.; Hu, W.S. Visualizing the translation and packaging of HIV-1 full-length RNA. Proc. Natl. Acad. Sci. USA 2020, 117, 6145–6155. [Google Scholar] [CrossRef]
- Ni, N.; Nikolaitchik, O.A.; Dilley, K.A.; Chen, J.; Galli, A.; Fu, W.; Prasad, V.V.; Ptak, R.G.; Pathak, V.K.; Hu, W.S. Mechanisms of human immunodeficiency virus type 2 RNA packaging: Efficient trans packaging and selection of RNA copackaging partners. J. Virol. 2011, 85, 7603–7612. [Google Scholar] [CrossRef] [PubMed]
- Kaye, J.F.; Lever, A.M.L. Human Immunodeficiency Virus Types 1 and 2 Differ in the Predominant Mechanism Used for Selection of Genomic RNA for Encapsidation. J. Virol. 1999, 73, 3023–3031. [Google Scholar] [CrossRef] [PubMed]
- Poon, D.T.; Chertova, E.N.; Ott, D.E. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55(Gag): Functional linkage between translation and RNA packaging. Virology 2002, 293, 368–378. [Google Scholar] [CrossRef] [PubMed]
| Protein | Function in gRNA Nuclear Export |
|---|---|
| CRM1 | Major nuclear export receptor |
| Ran | G protein, Ran GTP hydrolysis cycle drives export of CRM1 complex |
| DDX1 | Nucleates Rev oligomerization on RRE |
| DDX3 | Restructures gRNA for translocation through NPC |
| Nup62, Nup98, Nup124, Nup153 | Nucleoporins that have been identified as part of the NPC involved in gRNA nuclear export |
| PACS1 | Nucleocytoplasmic shuttle protein that interacts with Rev-RRE-CRM1 complex |
| ANP32A/B | Mediate export of viral RNAs via interaction with Rev-RRE-CRM1 complex |
| eIF5a | Mediates Rev-RRE-CRM1 interaction with NPC |
| Rab/hRIP | Interacts with CRM1 |
| UPF1 | Nucleocytoplasmic shuttle protein that interacts with Rev-RRE-CRM1 complex |
| MOV10 | Rev cofactor |
| Staufen2 | Regulates Rev nuclear export |
| Tat-SF1 | Interacts with gRNA and promotes nuclear export |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanson, H.M.; Willkomm, N.A.; Yang, H.; Mansky, L.M. Human Retrovirus Genomic RNA Packaging. Viruses 2022, 14, 1094. https://doi.org/10.3390/v14051094
Hanson HM, Willkomm NA, Yang H, Mansky LM. Human Retrovirus Genomic RNA Packaging. Viruses. 2022; 14(5):1094. https://doi.org/10.3390/v14051094
Chicago/Turabian StyleHanson, Heather M., Nora A. Willkomm, Huixin Yang, and Louis M. Mansky. 2022. "Human Retrovirus Genomic RNA Packaging" Viruses 14, no. 5: 1094. https://doi.org/10.3390/v14051094
APA StyleHanson, H. M., Willkomm, N. A., Yang, H., & Mansky, L. M. (2022). Human Retrovirus Genomic RNA Packaging. Viruses, 14(5), 1094. https://doi.org/10.3390/v14051094








