NK Cells in Protection from HIV Infection
Abstract
:1. Introduction
2. Basis of NK Cell Education
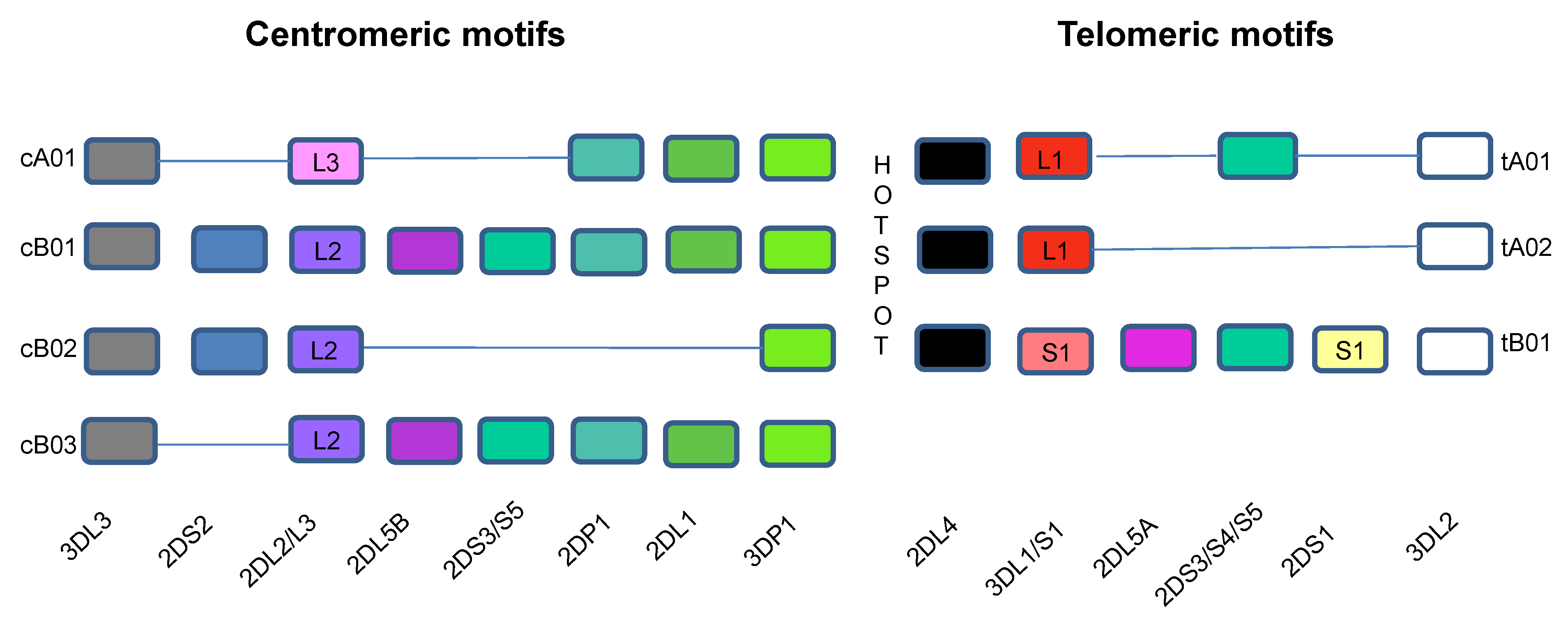
3. KIR NK Receptors
4. Epidemiological Studies Supporting a Role for NK Cells in Protection from HIV Infection
5. The Contribution of NK Cell Education to Function
6. Alloreactive NK Cells in Protection from Sexual HIV Transmission
7. Mechanisms of Protection from HIV Infection
8. Adaptive NK Cells in Protection from HIV Infection
9. Can Vaccines Induce NK Cell Responses That Prevent HIV Infection?
10. Concluding Remarks and Future Directions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Meyers, A.F.; Fowke, K.R. International symposium on natural immunity to HIV: A gathering of the HIV-exposed seronegative clan. J. Infect. Dis. 2010, 202 (Suppl. S3), S327–S328. [Google Scholar] [CrossRef]
- Beretta, A.; Furci, L.; Burastero, S.; Cosma, A.; Dinelli, M.E.; Lopalco, L.; DeSantis, C.; Tambussi, G.; Carrow, E.; Sabbatani, S.; et al. HIV-1-specific immunity in persistently seronegative individuals at high risk for HIV infection. Immunol. Lett. 1996, 51, 39–43. [Google Scholar] [CrossRef]
- Clerici, M.; Giorgi, J.V.; Chou, C.C.; Gudeman, V.K.; Zack, J.A.; Gupta, P.; Ho, H.N.; Nishanian, P.G.; Berzofsky, J.A.; Shearer, G.M. Cell-mediated immune response to human immunodeficiency virus (HIV) type 1 in seronegative homosexual men with recent sexual exposure to HIV-1. J. Infect. Dis. 1992, 165, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Clerici, M.; Levin, J.M.; Kessler, H.A.; Harris, A.; Berzofsky, J.A.; Landay, A.L.; Shearer, G.M. HIV-specific T-helper activity in seronegative health care workers exposed to contaminated blood. JAMA 1994, 271, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Mazzoli, S.; Trabattoni, D.; Lo, C.S.; Piconi, S.; Ble, C.; Meacci, F.; Ruzzante, S.; Salvi, A.; Semplici, F.; Longhi, R.; et al. HIV-specific mucosal and cellular immunity in HIV-seronegative partners of HIV-seropositive individuals. Nat. Med. 1997, 3, 1250–1257. [Google Scholar] [CrossRef] [PubMed]
- Pinto, L.A.; Sullivan, J.; Berzofsky, J.A.; Clerici, M.; Kessler, H.A.; Landay, A.L.; Shearer, G.M. ENV-specific cytotoxic T lymphocyte responses in HIV seronegative health care workers occupationally exposed to HIV-contaminated body fluids. J. Clin. Investig. 1995, 96, 867–876. [Google Scholar] [CrossRef] [Green Version]
- Ranki, A.; Mattinen, S.; Yarchoan, R.; Broder, S.; Ghrayeb, J.; Lahdevirta, J.; Krohn, K. T-cell response towards HIV in infected individuals with and without zidovudine therapy, and in HIV-exposed sexual partners. AIDS 1989, 3, 63–69. [Google Scholar] [CrossRef]
- Kaul, R.; Rowland-Jones, S.L.; Kimani, J.; Dong, T.; Yang, H.B.; Kiama, P.; Rostron, T.; Njagi, E.; Bwayo, J.J.; MacDonald, K.S.; et al. Late seroconversion in HIV-resistant Nairobi prostitutes despite pre-existing HIV-specific CD8+ responses. J. Clin. Investig. 2001, 107, 341–349. [Google Scholar] [CrossRef] [Green Version]
- Kaul, R.; MacDonald, K.S.; Nagelkerke, N.J.; Kimani, J.; Fowke, K.; Ball, T.B.; Luo, M.; Kariri, A.; Jaoko, W.; Moses, S.; et al. HIV viral set point and host immune control in individuals with HIV-specific CD8+ T-cell responses prior to HIV acquisition. AIDS 2010, 24, 1449–1454. [Google Scholar] [CrossRef]
- Liu, Y.; Woodward, A.; Zhu, H.; Andrus, T.; McNevin, J.; Lee, J.; Mullins, J.I.; Corey, L.; McElrath, M.J.; Zhu, T. Preinfection human immunodeficiency virus (HIV)-specific cytotoxic T lymphocytes failed to prevent HIV type 1 infection from strains genetically unrelated to viruses in long-term exposed partners. J. Virol. 2009, 83, 10821–10829. [Google Scholar] [CrossRef] [Green Version]
- Alimonti, J.B.; Kimani, J.; Matu, L.; Wachihi, C.; Kaul, R.; Plummer, F.A.; Fowke, K.R. Characterization of CD8 T-cell responses in HIV-1-exposed seronegative commercial sex workers from Nairobi, Kenya. Immunol. Cell Biol. 2006, 84, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Kaul, R.; Rowland-Jones, S.L.; Kimani, J.; Fowke, K.; Dong, T.; Kiama, P.; Rutherford, J.; Njagi, E.; Mwangi, F.; Rostron, T.; et al. New insights into HIV-1 specific cytotoxic T-lymphocyte responses in exposed, persistently seronegative Kenyan sex workers. Immunol. Lett. 2001, 79, 3–13. [Google Scholar] [CrossRef]
- Buchbinder, S.P.; Mehrotra, D.V.; Duerr, A.; Fitzgerald, D.W.; Mogg, R.; Li, D.; Gilbert, P.B.; Lama, J.R.; Marmor, M.; del Rio, C.; et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): A double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 2008, 372, 1881–1893. [Google Scholar] [CrossRef] [Green Version]
- McElrath, M.J.; De Rosa, S.C.; Moodie, Z.; Dubey, S.; Kierstead, L.; Janes, H.; Defawe, O.D.; Carter, D.K.; Hural, J.; Akondy, R.; et al. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: A case-cohort analysis. Lancet 2008, 372, 1894–1905. [Google Scholar] [CrossRef] [Green Version]
- Whitney, J.B.; Hill, A.L.; Sanisetty, S.; Penaloza-MacMaster, P.; Liu, J.; Shetty, M.; Parenteau, L.; Cabral, C.; Shields, J.; Blackmore, S.; et al. Rapid seeding of the viral reservoir prior to SIV viraemia in rhesus monkeys. Nature 2014, 512, 74–77. [Google Scholar] [CrossRef]
- Haase, A.T. Early events in sexual transmission of HIV and SIV and opportunities for interventions. Annu. Rev. Med. 2011, 62, 127–139. [Google Scholar] [CrossRef]
- Tomescu, C.; Abdulhaqq, S.; Montaner, L.J. Evidence for the innate immune response as a correlate of protection in human immunodeficiency virus (HIV)-1 highly exposed seronegative subjects (HESN). Clin. Exp. Immunol 2011, 164, 158–169. [Google Scholar] [CrossRef]
- Alter, G.; Teigen, N.; Ahern, R.; Streeck, H.; Meier, A.; Rosenberg, E.S.; Altfeld, M. Evolution of innate and adaptive effector cell functions during acute HIV-1 infection. J. Infect. Dis 2007, 195, 1452–1460. [Google Scholar] [CrossRef]
- Trinchieri, G. Biology of natural killer cells. Adv. Immunol. 1989, 47, 187–376. [Google Scholar]
- Vivier, E.; Raulet, D.H.; Moretta, A.; Caligiuri, M.A.; Zitvogel, L.; Lanier, L.L.; Yokoyama, W.M.; Ugolini, S. Innate or adaptive immunity? The example of natural killer cells. Science 2011, 331, 44–49. [Google Scholar] [CrossRef] [Green Version]
- Tremblay-McLean, A.; Bruneau, J.; Lebouche, B.; Lisovsky, I.; Song, R.; Bernard, N.F. Expression Profiles of Ligands for Activating Natural Killer Cell Receptors on HIV Infected and Uninfected CD4(+) T Cells. Viruses 2017, 9, 295. [Google Scholar] [CrossRef] [PubMed]
- Apps, R.; Del Prete, G.Q.; Chatterjee, P.; Lara, A.; Brumme, Z.L.; Brockman, M.A.; Neil, S.; Pickering, S.; Schneider, D.K.; Piechocka-Trocha, A.; et al. HIV-1 Vpu Mediates HLA-C Downregulation. Cell Host. Microbe 2016, 19, 686–695. [Google Scholar] [CrossRef] [Green Version]
- Cohen, G.B.; Gandhi, R.T.; Davis, D.M.; Mandelboim, O.; Chen, B.K.; Strominger, J.L.; Baltimore, D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 1999, 10, 661–671. [Google Scholar] [CrossRef]
- Kiani, Z.; Bruneau, J.; Geraghty, D.E.; Bernard, N.F. HLA-F on Autologous HIV-Infected Cells Activates Primary NK Cells Expressing the Activating Killer Immunoglobulin-Like Receptor KIR3DS1. J. Virol. 2019, 93, e00933-19. [Google Scholar] [CrossRef] [Green Version]
- Bonaparte, M.I.; Barker, E. Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood 2004, 104, 2087–2094. [Google Scholar] [CrossRef] [PubMed]
- Biron, C.A.; Nguyen, K.B.; Pien, G.C.; Cousens, L.P.; Salazar-Mather, T.P. Natural killer cells in antiviral defense: Function and regulation by innate cytokines. Annu. Rev. Immunol. 1999, 17, 189–220. [Google Scholar] [CrossRef]
- Oliva, A.; Kinter, A.L.; Vaccarezza, M.; Rubbert, A.; Catanzaro, A.; Moir, S.; Monaco, J.; Ehler, L.; Mizell, S.; Jackson, R.; et al. Natural killer cells from human immunodeficiency virus (HIV)-infected individuals are an important source of CC-chemokines and suppress HIV-1 entry and replication in vitro. J. Clin. Investig. 1998, 102, 223–231. [Google Scholar] [CrossRef] [Green Version]
- Song, R.; Lisovsky, I.; Lebouche, B.; Routy, J.P.; Bruneau, J.; Bernard, N.F. HIV Protective KIR3DL1/S1-HLA-B Genotypes Influence NK Cell-Mediated Inhibition of HIV Replication in Autologous CD4 Targets. PLoS Pathog. 2014, 10, e1003867. [Google Scholar] [CrossRef]
- Long, E.O.; Kim, H.S.; Liu, D.; Peterson, M.E.; Rajagopalan, S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu. Rev. Immunol. 2013, 31, 227–258. [Google Scholar] [CrossRef] [Green Version]
- Long, E.O. Regulation of immune responses through inhibitory receptors. Annu. Rev. Immunol. 1999, 17, 875–904. [Google Scholar] [CrossRef]
- Watzl, C.; Stebbins, C.C.; Long, E.O. NK cell inhibitory receptors prevent tyrosine phosphorylation of the activation receptor 2B4 (CD244). J. Immunol. 2000, 165, 3545–3548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anfossi, N.; Andre, P.; Guia, S.; Falk, C.S.; Roetynck, S.; Stewart, C.A.; Breso, V.; Frassati, C.; Reviron, D.; Middleton, D.; et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity 2006, 25, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Joncker, N.T.; Fernandez, N.C.; Treiner, E.; Vivier, E.; Raulet, D.H. NK cell responsiveness is tuned commensurate with the number of inhibitory receptors for self-MHC class I: The rheostat model. J. Immunol. 2009, 182, 4572–4580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Poursine-Laurent, J.; Truscott, S.M.; Lybarger, L.; Song, Y.J.; Yang, L.; French, A.R.; Sunwoo, J.B.; Lemieux, S.; Hansen, T.H.; et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 2005, 436, 709–713. [Google Scholar] [CrossRef]
- Boudreau, J.E.; Mulrooney, T.J.; Le Luduec, J.B.; Barker, E.; Hsu, K.C. KIR3DL1 and HLA-B Density and Binding Calibrate NK Education and Response to HIV. J. Immunol. 2016, 196, 3398–3410. [Google Scholar] [CrossRef]
- Raulet, D.H.; Vance, R.E. Self-tolerance of natural killer cells. Nat. Rev. Immunol. 2006, 6, 520–531. [Google Scholar] [CrossRef]
- Joncker, N.T.; Raulet, D.H. Regulation of NK cell responsiveness to achieve self-tolerance and maximal responses to diseased target cells. Immunol. Rev. 2008, 224, 85–97. [Google Scholar] [CrossRef]
- Boudreau, J.E.; Hsu, K.C. Natural Killer Cell Education and the Response to Infection and Cancer Therapy: Stay Tuned. Trends Immunol. 2018, 39, 222–239. [Google Scholar] [CrossRef]
- Braud, V.M.; Allan, D.S.; O’Callaghan, C.A.; Soderstrom, K.; D’Andrea, A.; Ogg, G.S.; Lazetic, S.; Young, N.T.; Bell, J.I.; Phillips, J.H.; et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998, 391, 795–799. [Google Scholar] [CrossRef]
- Trowsdale, J.; Barten, R.; Haude, A.; Stewart, C.A.; Beck, S.; Wilson, M.J. The genomic context of natural killer receptor extended gene families. Immunol. Rev. 2001, 181, 20–38. [Google Scholar] [CrossRef]
- Raulet, D.H.; Vance, R.E.; McMahon, C.W. Regulation of the natural killer cell receptor repertoire. Annu. Rev. Immunol. 2001, 19, 291–330. [Google Scholar] [CrossRef]
- Jiang, W.; Johnson, C.; Jayaraman, J.; Simecek, N.; Noble, J.; Moffatt, M.F.; Cookson, W.O.; Trowsdale, J.; Traherne, J.A. Copy number variation leads to considerable diversity for B but not A haplotypes of the human KIR genes encoding NK cell receptors. Genome Res. 2012, 22, 1845–1854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Sunwoo, J.B.; Yang, L.; Choi, T.; Song, Y.J.; French, A.R.; Vlahiotis, A.; Piccirillo, J.F.; Cella, M.; Colonna, M.; et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc. Natl. Acad. Sci. USA 2008, 105, 3053–3058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moesta, A.K.; Norman, P.J.; Yawata, M.; Yawata, N.; Gleimer, M.; Parham, P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J. Immunol. 2008, 180, 3969–3979. [Google Scholar] [CrossRef] [PubMed]
- Carr, W.H.; Pando, M.J.; Parham, P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 2005, 175, 5222–5229. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Heller, G.; Chewning, J.; Kim, S.; Yokoyama, W.M.; Hsu, K.C. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J. Immunol. 2007, 179, 5977–5989. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, N.C.; Lozier, A.; Flament, C.; Ricciardi-Castagnoli, P.; Bellet, D.; Suter, M.; Perricaudet, M.; Tursz, T.; Maraskovsky, E.; Zitvogel, L. Dendritic cells directly trigger NK cell functions: Cross-talk relevant in innate anti-tumor immune responses in vivo. Nat. Med. 1999, 5, 405–411. [Google Scholar] [CrossRef]
- Roe, D.; Vierra-Green, C.; Pyo, C.W.; Geraghty, D.E.; Spellman, S.R.; Maiers, M.; Kuang, R. A Detailed View of KIR Haplotype Structures and Gene Families as Provided by a New Motif-Based Multiple Sequence Alignment. Front. Immunol. 2020, 11, 585731. [Google Scholar] [CrossRef]
- Marsh, S.G.; Parham, P.; Dupont, B.; Geraghty, D.E.; Trowsdale, J.; Middleton, D.; Vilches, C.; Carrington, M.; Witt, C.; Guethlein, L.A.; et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Hum. Immunol. 2003, 64, 648–654. [Google Scholar] [CrossRef]
- Carrington, M.; Norman, P. The KIR Gene Cluster; National Center for Biotechnology Information US: Bethesda MD, USA, 2003; pp. 1–165. [Google Scholar]
- Cella, M.; Longo, A.; Ferrara, G.B.; Strominger, J.L.; Colonna, M. NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J. Exp. Med. 1994, 180, 1235–1242. [Google Scholar] [CrossRef] [Green Version]
- Gumperz, J.E.; Litwin, V.; Phillips, J.H.; Lanier, L.L.; Parham, P. The Bw4 public epitope of HLA-B molecules confers reactivity with natural killer cell clones that express NKB1, a putative HLA receptor. J. Exp. Med. 1995, 181, 1133–1144. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, D.H.; Allen, T.M.; Vogel, T.U.; Jing, P.; DeSouza, I.P.; Dodds, E.; Dunphy, E.J.; Melsaether, C.; Mothe, B.; Yamamoto, H.; et al. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 2002, 8, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Trundley, A.; Frebel, H.; Jones, D.; Chang, C.; Trowsdale, J. Allelic expression patterns of KIR3DS1 and 3DL1 using the Z27 and DX9 antibodies. Eur. J. Immunol. 2007, 37, 780–787. [Google Scholar] [CrossRef]
- Colonna, M.; Borsellino, G.; Falco, M.; Ferrara, G.B.; Strominger, J.L. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc. Natl. Acad. Sci. USA 1993, 90, 12000–12004. [Google Scholar] [CrossRef] [Green Version]
- Winter, C.C.; Gumperz, J.E.; Parham, P.; Long, E.O.; Wagtmann, N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J. Immunol. 1998, 161, 571–577. [Google Scholar]
- Moesta, A.K.; Graef, T.; Abi-Rached, L.; Older Aguilar, A.M.; Guethlein, L.A.; Parham, P. Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J. Immunol. 2010, 185, 4233–4237. [Google Scholar] [CrossRef]
- Hsu, K.C.; Chida, S.; Geraghty, D.E.; Dupont, B. The killer cell immunoglobulin-like receptor (KIR) genomic region: Gene-order, haplotypes and allelic polymorphism. Immunol. Rev. 2002, 190, 40–52. [Google Scholar] [CrossRef]
- Pyo, C.W.; Guethlein, L.A.; Vu, Q.; Wang, R.; Abi-Rached, L.; Norman, P.J.; Marsh, S.G.; Miller, J.S.; Parham, P.; Geraghty, D.E. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE 2010, 5, e15115. [Google Scholar] [CrossRef] [Green Version]
- Uhrberg, M.; Valiante, N.M.; Shum, B.P.; Shilling, H.G.; Lienert-Weidenbach, K.; Corliss, B.; Tyan, D.; Lanier, L.L.; Parham, P. Human diversity in killer cell inhibitory receptor genes. Immunity 1997, 7, 753–763. [Google Scholar] [CrossRef] [Green Version]
- Pyo, C.W.; Wang, R.; Vu, Q.; Cereb, N.; Yang, S.Y.; Duh, F.M.; Wolinsky, S.; Martin, M.P.; Carrington, M.; Geraghty, D.E. Recombinant structures expand and contract inter and intragenic diversification at the KIR locus. BMC Genom. 2013, 14, 89. [Google Scholar] [CrossRef] [Green Version]
- Maxwell, L.D.; Wallace, A.; Middleton, D.; Curran, M.D. A common KIR2DS4 deletion variant in the human that predicts a soluble KIR molecule analogous to the KIR1D molecule observed in the rhesus monkey. Tissue Antigens 2002, 60, 254–258. [Google Scholar] [CrossRef]
- Middleton, D.; Gonzalez, A.; Gilmore, P.M. Studies on the expression of the deleted KIR2DS4*003 gene product and distribution of KIR2DS4 deleted and nondeleted versions in different populations. Hum. Immunol. 2007, 68, 128–134. [Google Scholar] [CrossRef]
- Wilson, M.J.; Torkar, M.; Haude, A.; Milne, S.; Jones, T.; Sheer, D.; Beck, S.; Trowsdale, J. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc. Natl. Acad. Sci. USA 2000, 97, 4778–4783. [Google Scholar] [CrossRef] [Green Version]
- Yawata, M.; Yawata, N.; McQueen, K.L.; Cheng, N.W.; Guethlein, L.A.; Rajalingam, R.; Shilling, H.G.; Parham, P. Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics 2002, 54, 543–550. [Google Scholar] [CrossRef]
- Truong, L.X.; Luong, T.T.; Scott-Algara, D.; Versmisse, P.; David, A.; Perez-Bercoff, D.; Nguyen, N.V.; Tran, H.K.; Cao, C.T.; Fontanet, A.; et al. CD4 cell and CD8 cell-mediated resistance to HIV-1 infection in exposed uninfected intravascular drug users in Vietnam. AIDS 2003, 17, 1425–1434. [Google Scholar] [CrossRef]
- Martin, M.P.; Qi, Y.; Gao, X.; Yamada, E.; Martin, J.N.; Pereyra, F.; Colombo, S.; Brown, E.E.; Shupert, W.L.; Phair, J.; et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007, 39, 733–740. [Google Scholar] [CrossRef]
- Martin, M.P.; Gao, X.; Lee, J.H.; Nelson, G.W.; Detels, R.; Goedert, J.J.; Buchbinder, S.; Hoots, K.; Vlahov, D.; Trowsdale, J.; et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002, 31, 429–434. [Google Scholar] [CrossRef]
- Kiani, Z.; Dupuy, F.P.; Bruneau, J.; Lebouche, B.; Retiere, C.; Geraghty, D.E.; Bernard, N.F. The Education of NK Cells Determines Their Responsiveness to Autologous HIV-Infected CD4 T Cells. J. Virol. 2019, 93, e01185-19. [Google Scholar] [CrossRef]
- Boulet, S.; Kleyman, M.; Kim, J.Y.; Kamya, P.; Sharafi, S.; Simic, N.; Bruneau, J.; Routy, J.P.; Tsoukas, C.M.; Bernard, N.F. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS 2008, 22, 1487–1491. [Google Scholar] [CrossRef]
- Boulet, S.; Sharafi, S.; Simic, N.; Bruneau, J.; Routy, J.P.; Tsoukas, C.M.; Bernard, N.F. Increased proportion of KIR3DS1 homozygotes in HIV-exposed uninfected individuals. AIDS 2008, 22, 595–599. [Google Scholar] [CrossRef]
- Bruneau, J.; Daniel, M.; Abrahamowicz, M.; Zang, G.; Lamothe, F.; Vincelette, J. Trends in human immunodeficiency virus incidence and risk behavior among injection drug users in montreal, Canada: A 16-year longitudinal study. Am. J. Epidemiol. 2011, 173, 1049–1058. [Google Scholar] [CrossRef] [Green Version]
- Mehraj, V.; Cox, J.; Lebouche, B.; Costiniuk, C.; Cao, W.; Li, T.; Ponte, R.; Thomas, R.; Szabo, J.; Baril, J.G.; et al. Socio-economic status and time trends associated with early ART initiation following primary HIV infection in Montreal, Canada: 1996 to 2015. J. Int. AIDS Soc. 2018, 21, e25034. [Google Scholar] [CrossRef] [Green Version]
- Tallon, B.J.; Bruneau, J.; Tsoukas, C.M.; Routy, J.P.; Kiani, Z.; Tan, X.; Bernard, N.F. Time to seroconversion in HIV-exposed subjects carrying protective versus non protective KIR3DS1/L1 and HLA-B genotypes. PLoS ONE 2014, 9, e110480. [Google Scholar] [CrossRef] [Green Version]
- Tremblay-McLean, A.; Coenraads, S.; Kiani, Z.; Dupuy, F.P.; Bernard, N.F. Expression of ligands for activating natural killer cell receptors on cell lines commonly used to assess natural killer cell function. BMC Immunol. 2019, 20, 8. [Google Scholar] [CrossRef] [Green Version]
- Boulet, S.; Song, R.; Kamya, P.; Bruneau, J.; Shoukry, N.H.; Tsoukas, C.M.; Bernard, N.F. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. J. Immunol. 2010, 184, 2057–2064. [Google Scholar] [CrossRef] [Green Version]
- Parsons, M.S.; Boulet, S.; Song, R.; Bruneau, J.; Shoukry, N.H.; Routy, J.P.; Tsoukas, C.M.; Bernard, N.F. Mind the gap: Lack of association between KIR3DL1*004/HLA-Bw4-induced natural killer cell function and protection from HIV infection. J. Infect. Dis. 2010, 202 (Suppl. S3), S356–S360. [Google Scholar] [CrossRef] [Green Version]
- Jackson, E.; Zhang, C.X.; Kiani, Z.; Lisovsky, I.; Tallon, B.; Del Corpo, A.; Gilbert, L.; Bruneau, J.; Thomas, R.; Cote, P.; et al. HIV exposed seronegative (HESN) compared to HIV infected individuals have higher frequencies of telomeric Killer Immunoglobulin-like Receptor (KIR) B motifs; Contribution of KIR B motif encoded genes to NK cell responsiveness. PLoS ONE 2017, 12, e0185160. [Google Scholar] [CrossRef] [Green Version]
- Ravet, S.; Scott-Algara, D.; Bonnet, E.; Tran, H.K.; Tran, T.; Nguyen, N.; Truong, L.X.; Theodorou, I.; Barre-Sinoussi, F.; Pancino, G.; et al. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood 2007, 109, 4296–4305. [Google Scholar] [CrossRef]
- Guerini, F.R.; Lo, C.S.; Gori, A.; Bandera, A.; Mazzotta, F.; Uglietti, A.; Zanzottera, M.; Maserati, R.; Clerici, M. Under Representation of the Inhibitory KIR3DL1 Molecule and the KIR3DL1+/BW4+ Complex in HIV Exposed Seronegative Individuals. J. Infect. Dis. 2011, 203, 1235–1239. [Google Scholar] [CrossRef] [Green Version]
- Habegger de, S.A.; Sinchi, J.L.; Marinic, K.; Lopez, R.; Iliovich, E. KIR-HLA-A and B alleles of the Bw4 epitope against HIV infection in discordant heterosexual couples in Chaco Argentina. Immunology 2013, 140, 273–279. [Google Scholar] [CrossRef]
- Garcia-Beltran, W.F.; Holzemer, A.; Martrus, G.; Chung, A.W.; Pacheco, Y.; Simoneau, C.R.; Rucevic, M.; Lamothe-Molina, P.A.; Pertel, T.; Kim, T.E.; et al. Open conformers of HLA-F are high-affinity ligands of the activating NK-cell receptor KIR3DS1. Nat. Immunol. 2016, 17, 1067–1074. [Google Scholar] [CrossRef]
- Gillespie, G.M.; Bashirova, A.; Dong, T.; McVicar, D.W.; Rowland-Jones, S.L.; Carrington, M. Lack of KIR3DS1 binding to MHC class I Bw4 tetramers in complex with CD8+ T cell epitopes. AIDS Res. Hum. Retrovir. 2007, 23, 451–455. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, G.M.; Vivian, J.P.; Gostick, E.; Pymm, P.; Lafont, B.A.; Price, D.A.; Rossjohn, J.; Brooks, A.G.; McVicar, D.W. Peptide-Dependent Recognition of HLA-B*57:01 by KIR3DS1. J. Virol. 2015, 89, 5213–5221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fauriat, C.; Ivarsson, M.A.; Ljunggren, H.G.; Malmberg, K.J.; Michaelsson, J. Education of human natural killer cells by activating killer cell immunoglobulin-like receptors. Blood 2010, 115, 1166–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, C.A.; Laugier-Anfossi, F.; Vely, F.; Saulquin, X.; Riedmuller, J.; Tisserant, A.; Gauthier, L.; Romagne, F.; Ferracci, G.; Arosa, F.A.; et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc. Natl. Acad. Sci. USA 2005, 102, 13224–13229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiani, Z.; Dupuy, F.P.; Bruneau, J.; Lebouche, B.; Zhang, C.X.; Jackson, E.; Lisovsky, I.; da Fonseca, S.; Geraghty, D.E.; Bernard, N.F. HLA-F on HLA-Null 721.221 Cells Activates Primary NK Cells Expressing the Activating Killer Ig-like Receptor KIR3DS1. J. Immunol. 2018, 201, 113–123. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Ishitani, A.; Geraghty, D.E. HLA-F is a surface marker on activated lymphocytes. Eur. J. Immunol. 2010, 40, 2308–2318. [Google Scholar] [CrossRef] [Green Version]
- Burian, A.; Wang, K.L.; Finton, K.A.; Lee, N.; Ishitani, A.; Strong, R.K.; Geraghty, D.E. HLA-F and MHC-I Open Conformers Bind Natural Killer Cell Ig-Like Receptor KIR3DS1. PLoS ONE 2016, 11, e0163297. [Google Scholar] [CrossRef]
- Alter, G.; Martin, M.P.; Teigen, N.; Carr, W.H.; Suscovich, T.J.; Schneidewind, A.; Streeck, H.; Waring, M.; Meier, A.; Brander, C.; et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J. Exp. Med. 2007, 204, 3027–3036. [Google Scholar] [CrossRef] [Green Version]
- Merino, A.; Malhotra, R.; Morton, M.; Mulenga, J.; Allen, S.; Hunter, E.; Tang, J.; Kaslow, R.A. Impact of a functional KIR2DS4 allele on heterosexual HIV-1 transmission among discordant Zambian couples. J. Infect. Dis. 2011, 203, 487–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merino, A.M.; Dugast, A.S.; Wilson, C.M.; Goepfert, P.A.; Alter, G.; Kaslow, R.A.; Tang, J. KIR2DS4 promotes HIV-1 pathogenesis: New evidence from analyses of immunogenetic data and natural killer cell function. PLoS ONE 2014, 9, e99353. [Google Scholar] [CrossRef] [Green Version]
- Olvera, A.; Perez-Alvarez, S.; Ibarrondo, J.; Ganoza, C.; Lama, J.R.; Lucchetti, A.; Cate, S.; Hildebrand, W.; Bernard, N.; Gomez, L.; et al. The HLA-C*04: 01/KIR2DS4 gene combination and human leukocyte antigen alleles with high population frequency drive rate of HIV disease progression. AIDS 2015, 29, 507–517. [Google Scholar] [CrossRef]
- Graef, T.; Moesta, A.K.; Norman, P.J.; Abi-Rached, L.; Vago, L.; Older Aguilar, A.M.; Gleimer, M.; Hammond, J.A.; Guethlein, L.A.; Bushnell, D.A.; et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J. Exp. Med. 2009, 206, 2557–2572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aversa, F.; Terenzi, A.; Tabilio, A.; Falzetti, F.; Carotti, A.; Ballanti, S.; Felicini, R.; Falcinelli, F.; Velardi, A.; Ruggeri, L.; et al. Full haplotype-mismatched hematopoietic stem-cell transplantation: A phase II study in patients with acute leukemia at high risk of relapse. J. Clin. Oncol. 2005, 23, 3447–3454. [Google Scholar] [CrossRef] [PubMed]
- Locatelli, F.; Pende, D.; Mingari, M.C.; Bertaina, A.; Falco, M.; Moretta, A.; Moretta, L. Cellular and molecular basis of haploidentical hematopoietic stem cell transplantation in the successful treatment of high-risk leukemias: Role of alloreactive NK cells. Front. Immunol. 2013, 4, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moretta, L.; Locatelli, F.; Pende, D.; Marcenaro, E.; Mingari, M.C.; Moretta, A. Killer Ig-like receptor-mediated control of natural killer cell alloreactivity in haploidentical hematopoietic stem cell transplantation. Blood 2011, 117, 764–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.; Hubner, W.; Spinelli, M.A.; Chen, B.K. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J. Virol. 2007, 81, 12582–12595. [Google Scholar] [CrossRef] [Green Version]
- Mazurov, D.; Ilinskaya, A.; Heidecker, G.; Lloyd, P.; Derse, D. Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog. 2010, 6, e1000788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard-Stoecklin, S.; Gommet, C.; Corneau, A.B.; Guenounou, S.; Torres, C.; Dejucq-Rainsford, N.; Cosma, A.; Dereuddre-Bosquet, N.; Le Grand, R. Semen CD4+ T cells and macrophages are productively infected at all stages of SIV infection in macaques. PLoS Pathog. 2013, 9, e1003810. [Google Scholar] [CrossRef]
- Anderson, D.J.; Politch, J.A.; Nadolski, A.M.; Blaskewicz, C.D.; Pudney, J.; Mayer, K.H. Targeting Trojan Horse leukocytes for HIV prevention. AIDS 2010, 24, 163–187. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, D.; Krempels, R.; Ryfe, J.; Weitzman, K.; Stephenson, D.; Gigley, J.P. NK cells in mucosal defense against infection. Biomed. Res. Int. 2014, 2014, 413982. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.T.; Zhang, T.H.; Carmona, C.; Lee, B.; Seet, C.S.; Kostelny, M.; Shah, N.; Chen, H.; Farrell, K.; Soliman, M.S.A.; et al. Latency reversal plus natural killer cells diminish HIV reservoir in vivo. Nat. Commun. 2022, 13, 121. [Google Scholar] [CrossRef] [PubMed]
- Barker, E.; Evans, D.T. HLA-C Downmodulation by HIV-1 Vpu. Cell Host. Microbe 2016, 19, 570–571. [Google Scholar] [CrossRef] [Green Version]
- Jennes, W.; Verheyden, S.; Mertens, J.W.; Camara, M.; Seydi, M.; Dieye, T.N.; Mboup, S.; Demanet, C.; Kestens, L. Inhibitory KIR/HLA incompatibility between sexual partners confers protection against HIV-1 transmission. Blood 2013, 121, 1157–1164. [Google Scholar] [CrossRef] [Green Version]
- MacDonald, K.S.; Embree, J.; Njenga, S.; Nagelkerke, N.J.; Ngatia, I.; Mohammed, Z.; Barber, B.H.; Ndinya-Achola, J.; Bwayo, J.; Plummer, F.A. Mother-child class I HLA concordance increases perinatal human immunodeficiency virus type 1 transmission. J. Infect. Dis. 1998, 177, 551–556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackelprang, R.D.; John-Stewart, G.; Carrington, M.; Richardson, B.; Rowland-Jones, S.; Gao, X.; Mbori-Ngacha, D.; Mabuka, J.; Lohman-Payne, B.; Farquhar, C. Maternal HLA homozygosity and mother-child HLA concordance increase the risk of vertical transmission of HIV-1. J. Infect. Dis. 2008, 197, 1156–1161. [Google Scholar] [CrossRef] [Green Version]
- Lockett, S.F.; Robertson, J.R.; Brettle, R.P.; Yap, P.L.; Middleton, D.; Leigh Brown, A.J. Mismatched human leukocyte antigen alleles protect against heterosexual HIV transmission. J. Acquir. Immune Defic. Syndr. 2001, 27, 277–280. [Google Scholar] [CrossRef]
- Dorak, M.T.; Tang, J.; Penman-Aguilar, A.; Westfall, A.O.; Zulu, I.; Lobashevsky, E.S.; Kancheya, N.G.; Schaen, M.M.; Allen, S.A.; Kaslow, R.A. Transmission of HIV-1 and HLA-B allele-sharing within serodiscordant heterosexual Zambian couples. Lancet 2004, 363, 2137–2139. [Google Scholar] [CrossRef]
- Polycarpou, A.; Ntais, C.; Korber, B.T.; Elrich, H.A.; Winchester, R.; Krogstad, P.; Wolinsky, S.; Rostron, T.; Rowland-Jones, S.L.; Ammann, A.J.; et al. Association between maternal and infant class I and II HLA alleles and of their concordance with the risk of perinatal HIV type 1 transmission. AIDS Res. Hum. Retrovir. 2002, 18, 741–746. [Google Scholar] [CrossRef]
- Jennes, W.; Verheyden, S.; Demanet, C.; dje-Toure, C.A.; Vuylsteke, B.; Nkengasong, J.N.; Kestens, L. Cutting edge: Resistance to HIV-1 infection among African female sex workers is associated with inhibitory KIR in the absence of their HLA ligands. J. Immunol. 2006, 177, 6588–6592. [Google Scholar] [CrossRef] [Green Version]
- Hens, J.; Goovaerts, O.; Ceulemans, A.; Jennes, W.; Kestens, L. Impact of the Variable Killer Ig-Like Receptor-Human Leukocyte Antigen Interactions on Natural Killer Cell Cytotoxicity Toward Foreign CD4 T Cells. Front. Immunol. 2018, 9, 1588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bournazos, S.; Klein, F.; Pietzsch, J.; Seaman, M.S.; Nussenzweig, M.C.; Ravetch, J.V. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 2014, 158, 1243–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomescu, C.; Seaton, K.E.; Smith, P.; Taylor, M.; Tomaras, G.D.; Metzger, D.S.; Montaner, L.J. Innate activation of MDC and NK cells in high-risk HIV-1-exposed seronegative IV-drug users who share needles when compared with low-risk nonsharing IV-drug user controls. J. Acquir. Immune Defic. Syndr. 2015, 68, 264–273. [Google Scholar] [CrossRef] [Green Version]
- Colon, K.; Speicher, D.W.; Smith, P.; Taylor, M.; Metzger, D.S.; Montaner, L.J.; Tomescu, C. S100A14 Is Increased in Activated NK Cells and Plasma of HIV-Exposed Seronegative People Who Inject Drugs and Promotes Monocyte-NK Crosstalk. J. Acquir. Immune Defic. Syndr. 2019, 80, 234–241. [Google Scholar] [CrossRef] [PubMed]
- Card, C.M.; Ball, T.B.; Fowke, K.R. Immune quiescence: A model of protection against HIV infection. Retrovirology 2013, 10, 141. [Google Scholar] [CrossRef] [Green Version]
- Camara, M.; Dieye, T.N.; Seydi, M.; Diallo, A.A.; Fall, M.; Diaw, P.A.; Sow, P.S.; Mboup, S.; Kestens, L.; Jennes, W. Low-level CD4+ T cell activation in HIV-exposed seronegative subjects: Influence of gender and condom use. J. Infect. Dis. 2010, 201, 835–842. [Google Scholar] [CrossRef] [Green Version]
- Tran, H.K.; Chartier, L.; Troung, L.X.; Nguyen, N.N.; Fontanet, A.; Barre-Sinoussi, F.E.; Pancino, G.; Scott-Algara, D. Systemic immune activation in HIV-1-exposed uninfected Vietnamese intravascular drug users. AIDS Res. Hum. Retrovir. 2006, 22, 255–261. [Google Scholar] [CrossRef]
- Restrepo, C.; Rallon, N.I.; del Romero, J.; Rodriguez, C.; Hernando, V.; Lopez, M.; Peris, A.; Lozano, S.; Sempere-Ortells, J.M.; Soriano, V.; et al. Low-level exposure to HIV induces virus-specific T cell responses and immune activation in exposed HIV-seronegative individuals. J. Immunol. 2010, 185, 982–989. [Google Scholar] [CrossRef] [Green Version]
- Saulle, I.; Biasin, M.; Gnudi, F.; Rainone, V.; Ibba, S.V.; Lo Caputo, S.; Mazzotta, F.; Trabattoni, D.; Clerici, M. Short Communication: Immune Activation Is Present in HIV-1-Exposed Seronegative Individuals and Is Independent of Microbial Translocation. AIDS Res. Hum. Retrovir. 2016, 32, 129–133. [Google Scholar] [CrossRef] [Green Version]
- Biasin, M.; Caputo, S.L.; Speciale, L.; Colombo, F.; Racioppi, L.; Zagliani, A.; Ble, C.; Vichi, F.; Cianferoni, L.; Masci, A.M.; et al. Mucosal and systemic immune activation is present in human immunodeficiency virus-exposed seronegative women. J. Infect. Dis. 2000, 182, 1365–1374. [Google Scholar] [CrossRef] [Green Version]
- Tomescu, C.; Duh, F.M.; Lanier, M.A.; Kapalko, A.; Mounzer, K.C.; Martin, M.P.; Carrington, M.; Metzger, D.S.; Montaner, L.J. Increased plasmacytoid dendritic cell maturation and natural killer cell activation in HIV-1 exposed, uninfected intravenous drug users. AIDS 2010, 24, 2151–2160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowke, K.R.; Nagelkerke, N.J.; Kimani, J.; Simonsen, J.N.; Anzala, A.O.; Bwayo, J.J.; MacDonald, K.S.; Ngugi, E.N.; Plummer, F.A. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet 1996, 348, 1347–1351. [Google Scholar] [CrossRef]
- Guma, M.; Cabrera, C.; Erkizia, I.; Bofill, M.; Clotet, B.; Ruiz, L.; Lopez-Botet, M. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J. Infect. Dis. 2006, 194, 38–41. [Google Scholar] [CrossRef]
- Lopez-Verges, S.; Milush, J.M.; Schwartz, B.S.; Pando, M.J.; Jarjoura, J.; York, V.A.; Houchins, J.P.; Miller, S.; Kang, S.M.; Norris, P.J.; et al. Expansion of a unique CD57(+)NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 14725–14732. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.; Rodríguez, A.; Carretero, M.; López-Botet, M.; Phillips, J.H.; Lanier, L.L. Molecular characterization of human CD94: A type II membrane glycoprotein related to the C-type lectin superfamily. Eur. J. Immunol. 1995, 25, 2433–2437. [Google Scholar] [CrossRef] [PubMed]
- Llano, M.; Lee, N.; Navarro, F.; Garcia, P.; Albar, J.P.; Geraghty, D.E.; Lopez-Botet, M. HLA-E-bound peptides influence recognition by inhibitory and triggering CD94/NKG2 receptors: Preferential response to an HLA-G-derived nonamer. Eur. J. Immunol. 1998, 28, 2854–2863. [Google Scholar] [CrossRef]
- Hammer, Q.; Ruckert, T.; Borst, E.M.; Dunst, J.; Haubner, A.; Durek, P.; Heinrich, F.; Gasparoni, G.; Babic, M.; Tomic, A.; et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 2018, 19, 453–463. [Google Scholar] [CrossRef]
- Lanier, L.L.; Corliss, B.; Wu, J.; Phillips, J.H. Association of DAP12 with activating CD94/NKG2C NK cell receptors. Immunity 1998, 8, 693–701. [Google Scholar] [CrossRef] [Green Version]
- Schlums, H.; Cichocki, F.; Tesi, B.; Theorell, J.; Beziat, V.; Holmes, T.D.; Han, H.; Chiang, S.C.; Foley, B.; Mattsson, K.; et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 2015, 42, 443–456. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Zhang, T.; Hwang, I.; Kim, A.; Nitschke, L.; Kim, M.; Scott, J.M.; Kamimura, Y.; Lanier, L.L.; Kim, S. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 2015, 42, 431–442. [Google Scholar] [CrossRef] [Green Version]
- Hikami, K.; Tsuchiya, N.; Yabe, T.; Tokunaga, K. Variations of human killer cell lectin-like receptors: Common occurrence of NKG2-C deletion in the general population. Genes Immun. 2003, 4, 160–167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyashita, R.; Tsuchiya, N.; Hikami, K.; Kuroki, K.; Fukazawa, T.; Bijl, M.; Kallenberg, C.G.; Hashimoto, H.; Yabe, T.; Tokunaga, K. Molecular genetic analyses of human NKG2C (KLRC2) gene deletion. Int. Immunol. 2004, 16, 163–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.L.; Landskron, J.; Ask, E.H.; Enqvist, M.; Sohlberg, E.; Traherne, J.A.; Hammer, Q.; Goodridge, J.P.; Larsson, S.; Jayaraman, J.; et al. Critical Role of CD2 Co-stimulation in Adaptive Natural Killer Cell Responses Revealed in NKG2C-Deficient Humans. Cell Rep. 2016, 15, 1088–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moraru, M.; Canizares, M.; Muntasell, A.; de Pablo, R.; Lopez-Botet, M.; Vilches, C. Assessment of copy-number variation in the NKG2C receptor gene in a single-tube and characterization of a reference cell panel, using standard polymerase chain reaction. Tissue Antigens 2012, 80, 184–187. [Google Scholar] [CrossRef]
- Goncalves, A.; Makalo, P.; Joof, H.; Burr, S.; Ramadhani, A.; Massae, P.; Malisa, A.; Mtuy, T.; Derrick, T.; Last, A.R.; et al. Differential frequency of NKG2C/KLRC2 deletion in distinct African populations and susceptibility to Trachoma: A new method for imputation of KLRC2 genotypes from SNP genotyping data. Hum. Genet. 2016, 135, 939–951. [Google Scholar] [CrossRef] [Green Version]
- Thomas, R.; Low, H.Z.; Kniesch, K.; Jacobs, R.; Schmidt, R.E.; Witte, T. NKG2C deletion is a risk factor of HIV infection. AIDS Res. Hum. Retrovir. 2012, 28, 844–851. [Google Scholar] [CrossRef] [Green Version]
- Alsulami, K.; Bolastig, N.; Dupuy, F.P.; Mabanga, T.; Gilbert, L.; Kiani, Z.; Routy, J.P.; Bruneau, J.; Thomas, R.; Tremblay, C.; et al. Influence of NKG2C Genotypes on HIV Susceptibility and Viral Load Set Point. J. Virol. 2021, 95, e0041721. [Google Scholar] [CrossRef]
- Rerks-Ngarm, S.; Pitisuttithum, P.; Nitayaphan, S.; Kaewkungwal, J.; Chiu, J.; Paris, R.; Premsri, N.; Namwat, C.; de, S.M.; Adams, E.; et al. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 2009, 361, 2209–2220. [Google Scholar] [CrossRef]
- Haynes, B.F.; Gilbert, P.B.; McElrath, M.J.; Zolla-Pazner, S.; Tomaras, G.D.; Alam, S.M.; Evans, D.T.; Montefiori, D.C.; Karnasuta, C.; Sutthent, R.; et al. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 2012, 366, 1275–1286. [Google Scholar] [CrossRef] [Green Version]
- Gray, G.E.; Bekker, L.G.; Laher, F.; Malahleha, M.; Allen, M.; Moodie, Z.; Grunenberg, N.; Huang, Y.; Grove, D.; Prigmore, B.; et al. Vaccine Efficacy of ALVAC-HIV and Bivalent Subtype C gp120-MF59 in Adults. N. Engl. J. Med. 2021, 384, 1089–1100. [Google Scholar] [CrossRef]
- Goodier, M.R.; Riley, E.M. Regulation of the human NK cell compartment by pathogens and vaccines. Clin. Trans. Immunol. 2021, 10, e1244. [Google Scholar] [CrossRef]
- Rydyznski, C.E.; Waggoner, S.N. Boosting vaccine efficacy the natural (killer) way. Trends Immunol. 2015, 36, 536–546. [Google Scholar] [CrossRef] [Green Version]
- Martin-Fontecha, A.; Thomsen, L.L.; Brett, S.; Gerard, C.; Lipp, M.; Lanzavecchia, A.; Sallusto, F. Induced recruitment of NK cells to lymph nodes provides IFN-gamma for T(H)1 priming. Nat. Immunol. 2004, 5, 1260–1265. [Google Scholar] [CrossRef] [PubMed]
- Farsakoglu, Y.; Palomino-Segura, M.; Latino, I.; Zanaga, S.; Chatziandreou, N.; Pizzagalli, D.U.; Rinaldi, A.; Bolis, M.; Sallusto, F.; Stein, J.V.; et al. Influenza Vaccination Induces NK-Cell-Mediated Type-II IFN Response that Regulates Humoral Immunity in an IL-6-Dependent Manner. Cell Rep. 2019, 26, 2307–2315.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochoa, M.C.; Minute, L.; Rodriguez, I.; Garasa, S.; Perez-Ruiz, E.; Inoges, S.; Melero, I.; Berraondo, P. Antibody-dependent cell cytotoxicity: Immunotherapy strategies enhancing effector NK cells. Immunol. Cell Biol. 2017, 95, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Li, A.P.Y.; Cohen, C.A.; Leung, N.H.L.; Fang, V.J.; Gangappa, S.; Sambhara, S.; Levine, M.Z.; Iuliano, A.D.; Perera, R.; Ip, D.K.M.; et al. Immunogenicity of standard, high-dose, MF59-adjuvanted, and recombinant-HA seasonal influenza vaccination in older adults. NPJ Vaccines 2021, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Coccia, M.; Collignon, C.; Herve, C.; Chalon, A.; Welsby, I.; Detienne, S.; van Helden, M.J.; Dutta, S.; Genito, C.J.; Waters, N.C.; et al. Cellular and molecular synergy in AS01-adjuvanted vaccines results in an early IFNgamma response promoting vaccine immunogenicity. NPJ Vaccines 2017, 2, 25. [Google Scholar] [CrossRef] [PubMed]
- Howard, L.M.; Hoek, K.L.; Goll, J.B.; Samir, P.; Galassie, A.; Allos, T.M.; Niu, X.; Gordy, L.E.; Creech, C.B.; Prasad, N.; et al. Cell-Based Systems Biology Analysis of Human AS03-Adjuvanted H5N1 Avian Influenza Vaccine Responses: A Phase I Randomized Controlled Trial. PLoS ONE 2017, 12, e0167488. [Google Scholar] [CrossRef] [PubMed]
- Cox, A.; Cevik, H.; Feldman, H.A.; Canaday, L.M.; Lakes, N.; Waggoner, S.N. Targeting natural killer cells to enhance vaccine responses. Trends Pharmacol. Sci. 2021, 42, 789–801. [Google Scholar] [CrossRef]
- Luetke-Eversloh, M.; Hammer, Q.; Durek, P.; Nordstrom, K.; Gasparoni, G.; Pink, M.; Hamann, A.; Walter, J.; Chang, H.D.; Dong, J.; et al. Human cytomegalovirus drives epigenetic imprinting of the IFNG locus in NKG2Chi natural killer cells. PLoS Pathog 2014, 10, e1004441. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Scott, J.M.; Hwang, I.; Kim, S. Cutting edge: Antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. J. Immunol. 2013, 190, 1402–1406. [Google Scholar] [CrossRef] [Green Version]
- Gyurova, I.E.; Ali, A.; Waggoner, S.N. Natural Killer Cell Regulation of B Cell Responses in the Context of Viral Infection. Viral Immunol 2020, 33, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Rydyznski, C.; Daniels, K.A.; Karmele, E.P.; Brooks, T.R.; Mahl, S.E.; Moran, M.T.; Li, C.; Sutiwisesak, R.; Welsh, R.M.; Waggoner, S.N. Generation of cellular immune memory and B-cell immunity is impaired by natural killer cells. Nat. Commun. 2015, 6, 6375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rydyznski, C.E.; Cranert, S.A.; Zhou, J.Q.; Xu, H.; Kleinstein, S.H.; Singh, H.; Waggoner, S.N. Affinity Maturation Is Impaired by Natural Killer Cell Suppression of Germinal Centers. Cell Rep. 2018, 24, 3367–3373.e4. [Google Scholar] [CrossRef] [Green Version]
- Dupuy, F.P.; Kant, S.; Barbe, A.; Routy, J.P.; Bruneau, J.; Lebouche, B.; Tremblay, C.; Pazgier, M.; Finzi, A.; Bernard, N.F. Antibody-Dependent Cellular Cytotoxicity-Competent Antibodies against HIV-1-Infected Cells in Plasma from HIV-Infected Subjects. MBio 2019, 10, e02690-19. [Google Scholar] [CrossRef] [Green Version]
- Richard, J.; Prevost, J.; Alsahafi, N.; Ding, S.; Finzi, A. Impact of HIV-1 Envelope Conformation on ADCC Responses. Trends Microbiol. 2018, 26, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Richard, J.; Prevost, J.; Baxter, A.E.; von Bredow, B.; Ding, S.; Medjahed, H.; Delgado, G.G.; Brassard, N.; Sturzel, C.M.; Kirchhoff, F.; et al. Uninfected Bystander Cells Impact the Measurement of HIV-Specific Antibody-Dependent Cellular Cytotoxicity Responses. MBio 2018, 9, e00358-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| HLA | Receptor |
|---|---|
| HLA-C1 (Asn 77) | KIR2DL2 (major), KIR2DL3 |
| HLA-C2 (Lys 80) | KIR2DL1, KIR2DL2 (minor), KIR2DS1 |
| HLA-Bw4 | KIR3DL1 |
| HLA-F (open conformation) | KIR3DS1 |
| HLA-A*03, HLA-A*11 | KIR3DL2 |
| HLA-E presenting epitopes | NKG2A |
| from the HLA leader sequences |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bernard, N.F.; Alsulami, K.; Pavey, E.; Dupuy, F.P. NK Cells in Protection from HIV Infection. Viruses 2022, 14, 1143. https://doi.org/10.3390/v14061143
Bernard NF, Alsulami K, Pavey E, Dupuy FP. NK Cells in Protection from HIV Infection. Viruses. 2022; 14(6):1143. https://doi.org/10.3390/v14061143
Chicago/Turabian StyleBernard, Nicole F., Khlood Alsulami, Erik Pavey, and Franck P. Dupuy. 2022. "NK Cells in Protection from HIV Infection" Viruses 14, no. 6: 1143. https://doi.org/10.3390/v14061143
APA StyleBernard, N. F., Alsulami, K., Pavey, E., & Dupuy, F. P. (2022). NK Cells in Protection from HIV Infection. Viruses, 14(6), 1143. https://doi.org/10.3390/v14061143






