Paraoxonase-1 Facilitates PRRSV Replication by Interacting with Viral Nonstructural Protein-9 and Inhibiting Type I Interferon Pathway
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Animal Infection Experiments
2.3. RNA Interference Assay
2.4. Construction of Plasmids and Transfection of Cells
2.5. PON1 Overexpression by Lentiviral Expression System and Activator
2.6. Virus Incubation
2.7. RNA Extraction and Quantitative PCR
2.8. Virus Titration
2.9. Coimmunoprecipitation (Co-IP) and Western Blotting
2.10. Confocal Microscopy and Immunofluorescence Analysis (IFA)
2.11. RNA Immunoprecipitation (RIP)
2.12. Statistical Analysis
3. Results
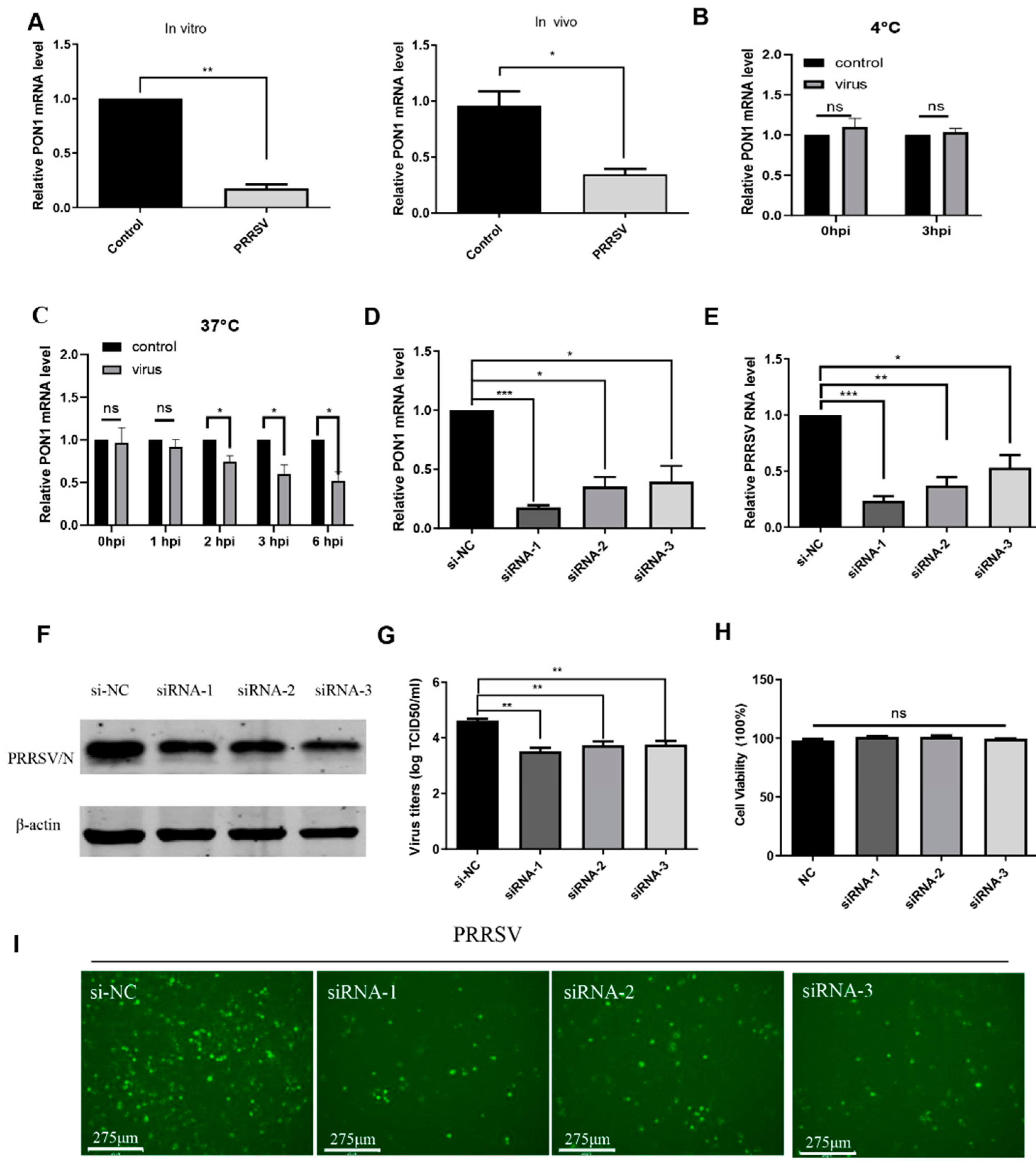
3.1. PRRSV Infection Induces PON1 Downregulation
3.2. PON1 Positively Regulates PRRSV Infection
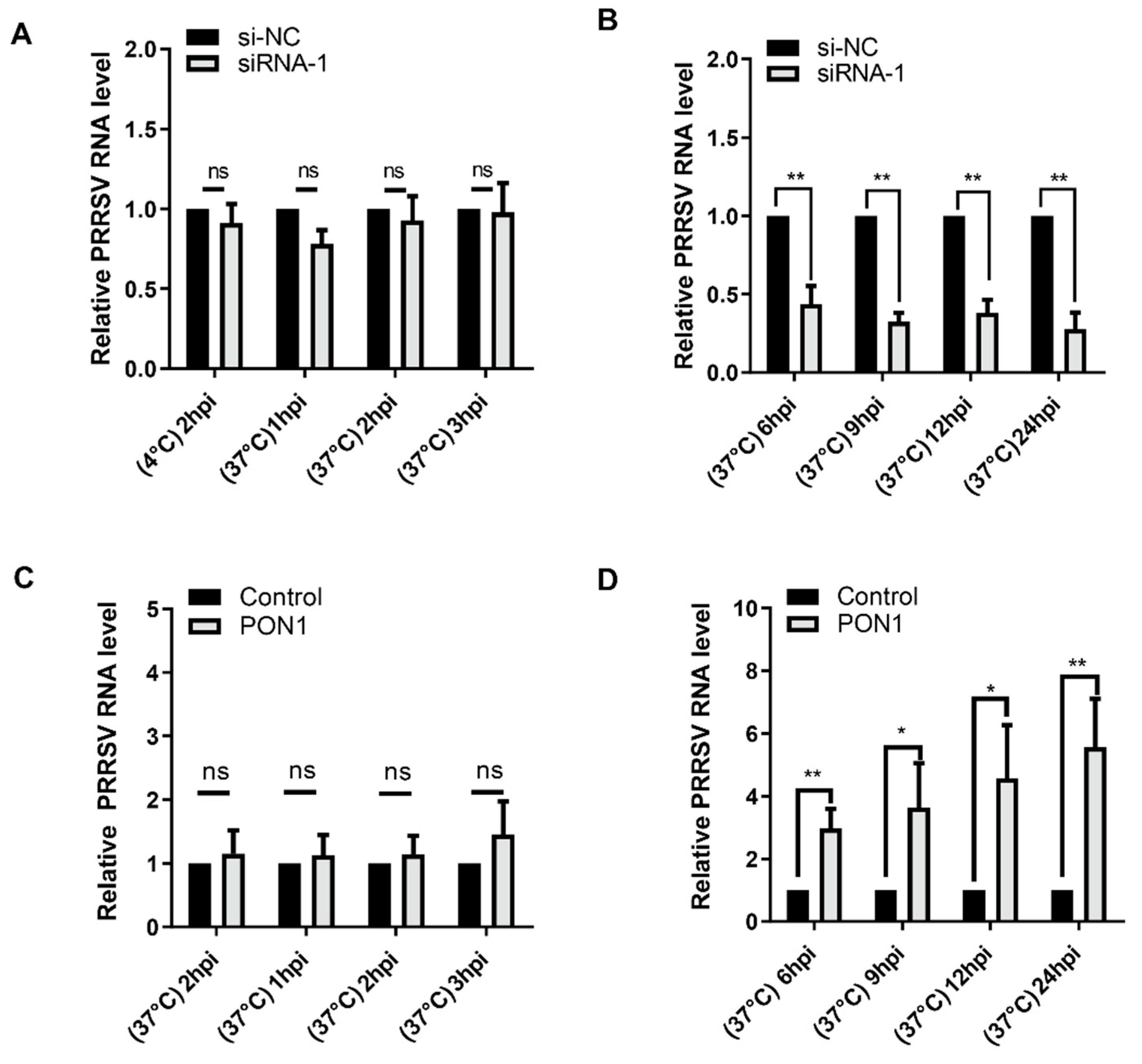
3.3. PON1 Regulates PRRSV Infection at the Stage of Replication
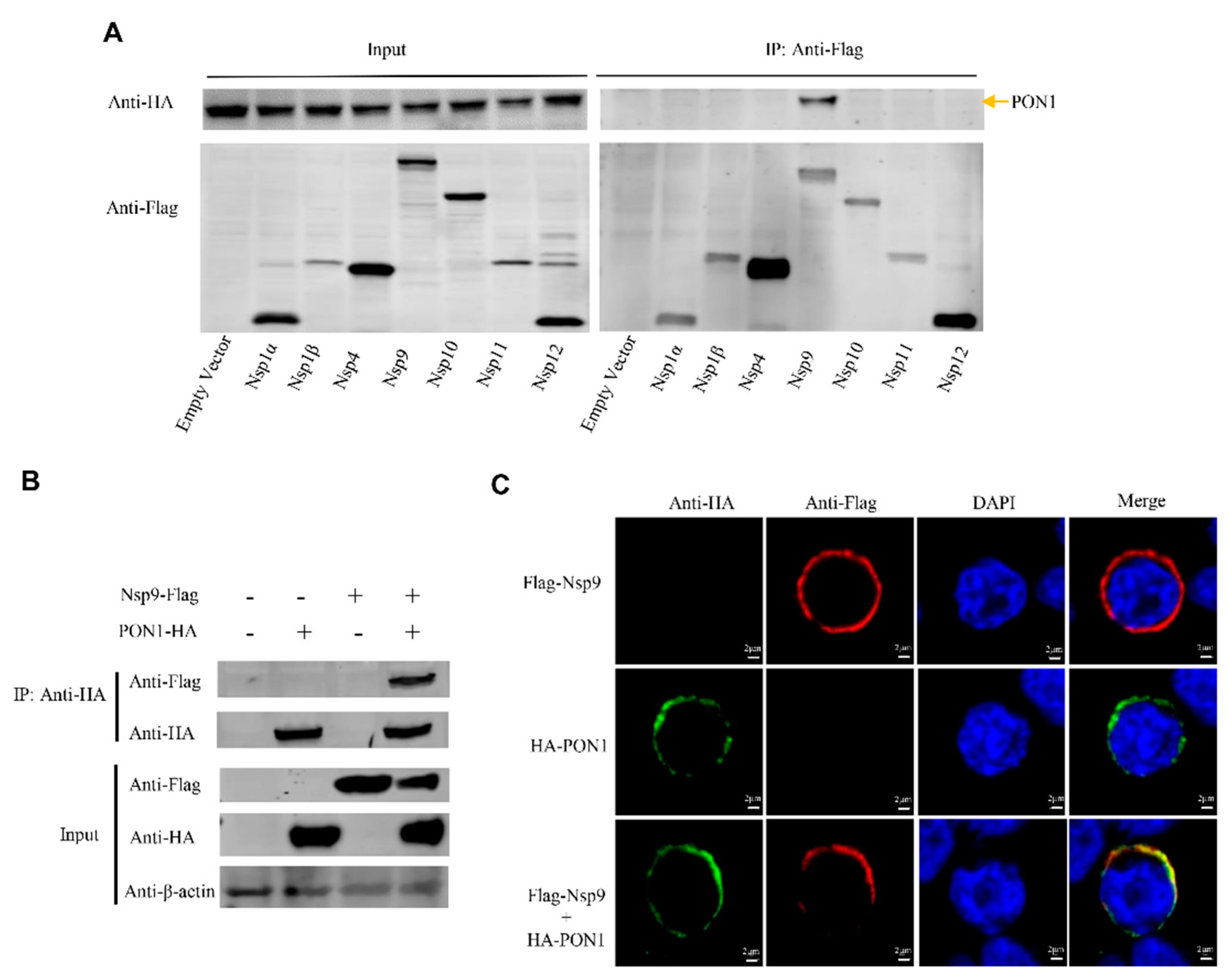
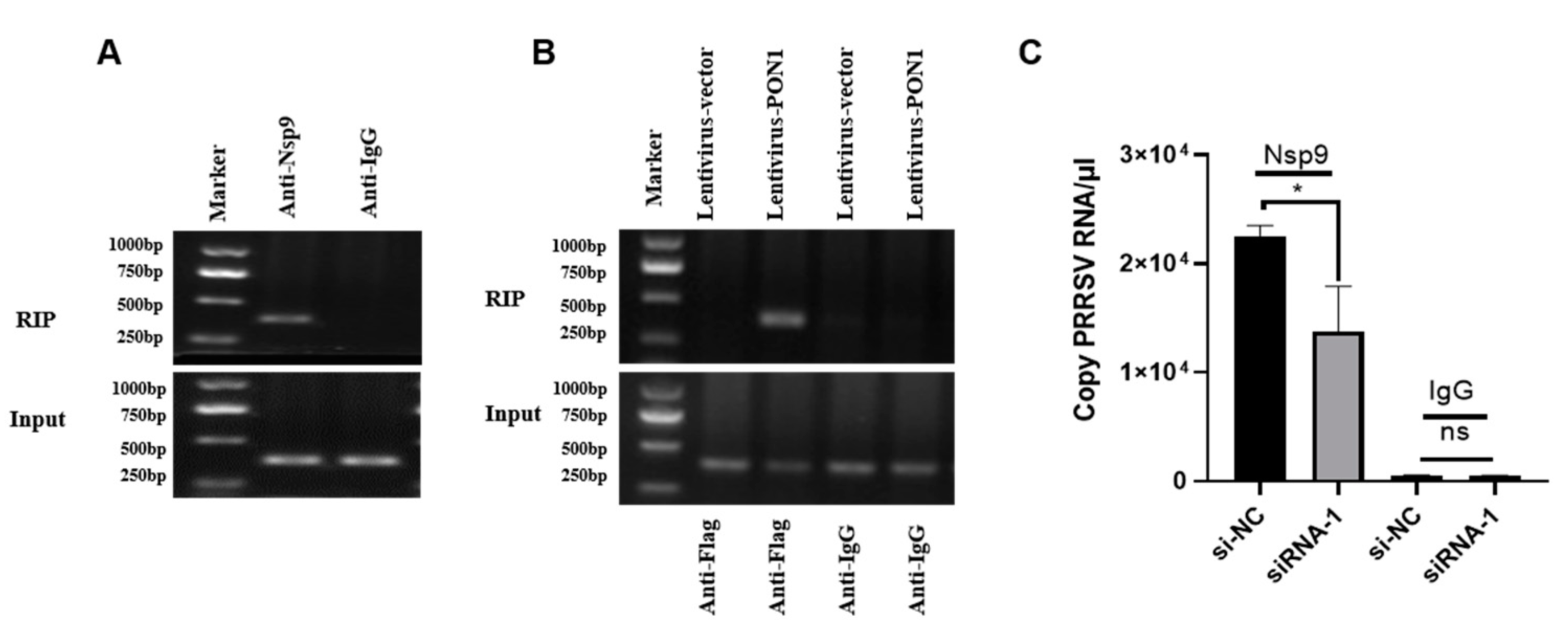
3.4. PON1 Interacts with PRRSV Nsp9
3.5. The Interaction of PON1 with Nsp9 Assists PRRSV Replication
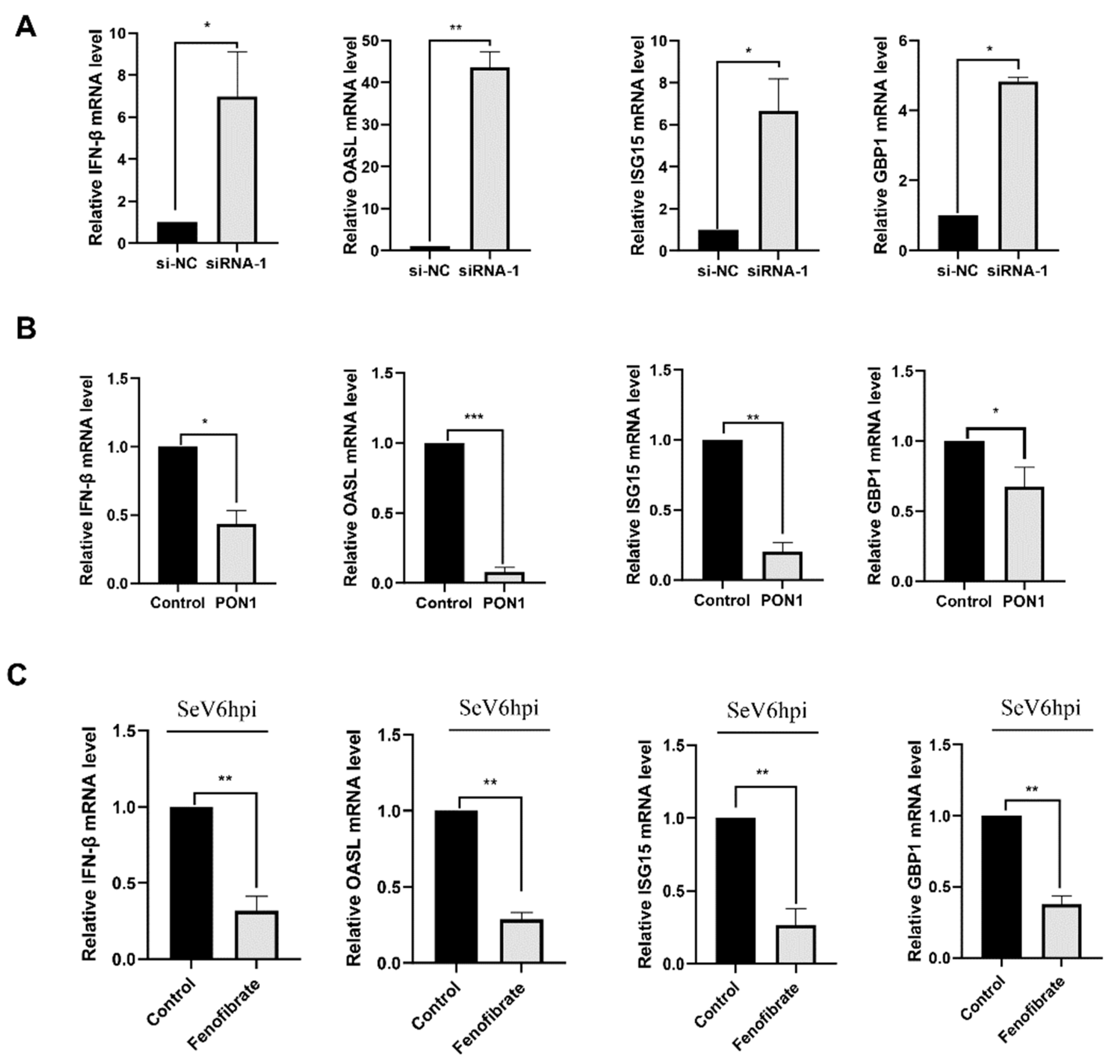
3.6. PON1 Negatively Regulates the Type I IFN Signaling Pathway
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mahrooz, A.; Mackness, M.; Bagheri, A.; Ghaffari-Cherati, M.; Masoumi, P. The epigenetic regulation of paraoxonase 1 (PON1) as an important enzyme in HDL function: The missing link between environmental and genetic regulation. Clin. Biochem. 2019, 73, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Grdic Rajkovic, M.; Rumora, L.; Barisic, K. The paraoxonase 1, 2 and 3 in humans. Biochem. Med. 2011, 21, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Mahrooz, A. Pharmacological interactions of paraoxonase 1 (PON1): A HDL-bound antiatherogenic enzyme. Curr. Clin. Pharmacol. 2016, 11, 259–264. [Google Scholar] [CrossRef] [PubMed]
- Seres, I.; Paragh, G.; Deschene, E.; Fulop Jr, T.; Khalil, A. Study of factors influencing the decreased HDL associated PON1 activity with aging. Exp. Gerontol. 2004, 39, 59–66. [Google Scholar] [CrossRef]
- Murillo-González, F.; Ponce-Ruiz, N.; Rojas-García, A.; Rothenberg, S.; Bernal-Hernández, Y.; Cerda-Flores, R.; Mackness, M.; Barrón-Vivanco, B.; González-Arias, C.; Ponce-Gallegos, J. PON1 lactonase activity and its association with cardiovascular disease. Clin. Chim. Acta 2020, 500, 47–53. [Google Scholar] [CrossRef]
- Zargari, M.; Sharafeddin, F.; Mahrooz, A.; Alizadeh, A.; Masoumi, P. The common variant Q192R at the paraoxonase 1 (PON1) gene and its activity are responsible for a portion of the altered antioxidant status in type 2 diabetes. Exp. Biol. Med. 2016, 241, 1489–1496. [Google Scholar] [CrossRef]
- Schulpis, K.H.; Barzeliotou, A.; Papadakis, M.; Rodolakis, A.; Antsaklis, A.; Papassotiriou, I.; Vlachos, G.D. Maternal chronic hepatitis B virus is implicated with low neonatal paraoxonase/arylesterase activities. Clin. Biochem. 2008, 41, 282–287. [Google Scholar] [CrossRef]
- Ali, E.M.; Shehata, H.H.; Ali-Labib, R.; Zahra, L.M.E. Oxidant and antioxidant of arylesterase and paraoxonase as biomarkers in patients with hepatitis C virus. Clin. Biochem. 2009, 42, 1394–1400. [Google Scholar] [CrossRef]
- Rodríguez-Tomàs, E.; Iftimie, S.; Castañé, H.; Baiges-Gaya, G.; Hernández-Aguilera, A.; González-Viñas, M.; Castro, A.; Camps, J.; Joven, J. Clinical performance of paraoxonase-1-related variables and novel markers of inflammation in coronavirus disease-19. A machine learning approach. Antioxidants 2021, 10, 991. [Google Scholar] [CrossRef]
- Dias, C.; Marinho, A.; Morello, J.; Almeida, G.; Caixas, U.; Soto, K.; Monteiro, E.; Pereira, S. Monitoring of the lactonase activity of paraoxonase-1 enzyme in HIV-1-infection. J. Int. AIDS Soc. 2014, 17, 19682. [Google Scholar] [CrossRef] [PubMed]
- Van Lenten, B.J.; Wagner, A.C.; Nayak, D.P.; Hama, S.; Navab, M.; Fogelman, A.M. High-density lipoprotein loses its anti-inflammatory properties during acute influenza a infection. Circulation 2001, 103, 2283–2288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- CAVANAGE, D. Nidovirales: A new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 1997, 142, 629–633. [Google Scholar]
- Kappes, M.A.; Faaberg, K.S. PRRSV structure, replication and recombination: Origin of phenotype and genotype diversity. Virology 2015, 479, 475–486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Music, N.; Gagnon, C.A. The role of porcine reproductive and respiratory syndrome (PRRS) virus structural and non-structural proteins in virus pathogenesis. Anim. Health Res. Rev. 2010, 11, 135–163. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Snijder, E.J. The PRRSV replicase: Exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 2010, 154, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Snijder, E.J.; Wassenaar, A.; Spaan, W. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J. Virol. 1994, 68, 5755–5764. [Google Scholar] [CrossRef] [Green Version]
- Wassenaar, A.; Spaan, W.; Gorbalenya, A.E.; Snijder, E.J. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: Evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J. Virol. 1997, 71, 9313–9322. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Treffers, E.E.; Li, Y.; Tas, A.; Sun, Z.; Van Der Meer, Y.; De Ru, A.H.; Van Veelen, P.A.; Atkins, J.F.; Snijder, E.J. Efficient− 2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc. Natl. Acad. Sci. USA 2012, 109, E2920–E2928. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, K.C.; Gulyaeva, A.; Zevenhoven-Dobbe, J.C.; Janssen, G.M.; Ruben, M.; Overkleeft, H.S.; Van Veelen, P.A.; Samborskiy, D.V.; Kravchenko, A.A.; Leontovich, A.M. Discovery of an essential nucleotidylating activity associated with a newly delineated conserved domain in the RNA polymerase-containing protein of all nidoviruses. Nucleic Acids Res. 2015, 43, 8416–8434. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, K.C.; Gorbalenya, A.E.; Snijder, E.J.; Posthuma, C.C. Arterivirus RNA-dependent RNA polymerase: Vital enzymatic activity remains elusive. Virology 2016, 487, 68–74. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.-Y.; Fang, Q.-Q.; Cong, F.; Liu, Y.-G.; Wang, H.-M.; Zhang, H.-L.; Tian, Z.-J.; Tang, Y.-D.; Cai, X.-H. The Nsp12-coding region of type 2 PRRSV is required for viral subgenomic mRNA synthesis. Emerg. Microbes Infect. 2019, 8, 1501–1510. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.-Y.; Sun, M.-X.; Zhang, H.-L.; Wang, G.; Zhan, G.; Tian, Z.-J.; Cai, X.-H.; Su, C.; Tang, Y.-D. Evasion of Antiviral Innate Immunity by Porcine Reproductive and Respiratory Syndrome Virus. Front. Microbiol. 2021, 12, 693799. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Nauwynck, H.; Pensaert, M. Effects of origin and state of differentiation and activation of monocytes/macrophages on their susceptibility to porcine reproductive and respiratory syndrome virus (PRRSV). Arch. Virol. 1997, 142, 2483–2497. [Google Scholar] [CrossRef] [PubMed]
- Van Breedam, W.; Delputte, P.L.; Van Gorp, H.; Misinzo, G.; Vanderheijden, N.; Duan, X.; Nauwynck, H.J. Porcine reproductive and respiratory syndrome virus entry into the porcine macrophage. J. Gen. Virol. 2010, 91, 1659–1667. [Google Scholar] [CrossRef]
- Goyal, S.M. Porcine reproductive and respiratory syndrome. J. Vet. Diagn. Invest. 1993, 5, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Zhou, L.; Ge, X.; Guo, X.; Yang, H. Pathogenesis and control of the Chinese highly pathogenic porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 2017, 209, 30–47. [Google Scholar] [CrossRef]
- Deakin, S.P.; Bioletto, S.; Bochaton-Piallat, M.-L.; James, R.W. HDL-associated paraoxonase-1 can redistribute to cell membranes and influence sensitivity to oxidative stress. Free Radic. Biol. Med. 2011, 50, 102–109. [Google Scholar] [CrossRef] [Green Version]
- Aharoni, S.; Aviram, M.; Fuhrman, B. Paraoxonase 1 (PON1) reduces macrophage inflammatory responses. Atherosclerosis 2013, 228, 353–361. [Google Scholar] [CrossRef]
- García-Heredia, A.; Marsillach, J.; Rull, A.; Triguero, I.; Fort, I.; Mackness, B.; Mackness, M.; Shih, D.M.; Joven, J.; Camps, J. Paraoxonase-1 inhibits oxidized low-density lipoprotein-induced metabolic alterations and apoptosis in endothelial cells: A nondirected metabolomic study. Mediators Inflamm. 2013, 2013, 156053. [Google Scholar] [CrossRef] [Green Version]
- Mackness, M.; Mackness, B. Targeting paraoxonase-1 in atherosclerosis. Expert Opin. Ther. Targets. 2013, 17, 829–837. [Google Scholar] [CrossRef]
- Xie, S.; Li, J.; Chen, Y.; Wang, C.; Zhang, H.; Mo, D. Sequence identification, chromosomal mapping and tissue specific expression of the porcine paraoxonase 1 (PON1) gene. Mol. Biol. Rep. 2010, 37, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Pan, Y.; Xu, Y.; Zhang, W.; Zhang, L.; Li, X.; Tian, Z.; Chen, H.; Wang, Y. Unveiling the long non-coding RNA profile of porcine reproductive and respiratory syndrome virus-infected porcine alveolar macrophages. BMC Genom. 2021, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Pan, Y.; Gao, J.; Xu, Y.; Li, X.; Tian, Z.; Chen, H.; Wang, Y. Downregulation of miR-218 by porcine reproductive and respiratory syndrome virus facilitates viral replication via inhibition of type I interferon responses. J. Biol. Chem. 2021, 296, 100683. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Guo, L.; Yang, L.; Xu, J.; Zhang, L.; Feng, L.; Chen, H.; Wang, Y. Metalloprotease ADAM17 regulates porcine epidemic diarrhea virus infection by modifying aminopeptidase N. Virology 2018, 517, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, L.; Xu, Y.; Zhang, H.; Gao, J.; Wang, Q.; Tian, Z.; Xuan, L.; Chen, H.; Wang, Y. PP2A facilitates porcine reproductive and respiratory syndrome virus replication by Deactivating irf3 and limiting type I interferon production. Viruses 2019, 11, 948. [Google Scholar] [CrossRef] [Green Version]
- Chua, B.H.; Phuektes, P.; Sanders, S.A.; Nicholls, P.K.; McMinn, P.C. The molecular basis of mouse adaptation by human enterovirus 71. J. Gen. Virol. 2008, 89, 1622–1632. [Google Scholar] [CrossRef]
- Ramírez, M.; Bauermann, F.V.; Navarro, D.; Rojas, M.; Manchego, A.; Nelson, E.A.; Diel, D.G.; Rivera, H. Detection of porcine reproductive and respiratory syndrome virus (PRRSV) 1-7-4-type strains in Peru. Transbound. Emerg. Dis. 2019, 66, 1107–1113. [Google Scholar] [CrossRef]
- Papatsiros, V.; Stylianaki, I.; Papakonstantinou, G.; Tsekouras, N.; Bitchava, D.; Christodoulopoulos, G.; Papaioannou, N. Histopathological lesions accompanied with first-time isolation of a PRRSV-2 strain in Greece. Viral Immunol. 2020, 33, 565–570. [Google Scholar] [CrossRef]
- Wang, H.M.; Liu, Y.G.; Tang, Y.D.; Liu, T.X.; Zheng, L.L.; Wang, T.Y.; Liu, S.G.; Wang, G.; Cai, X.H. A natural recombinant PRRSV between HP-PRRSV JXA 1-like and NADC 30-like strains. Transbound. Emerg. Dis. 2018, 65, 1078–1086. [Google Scholar] [CrossRef]
- Montaner-Tarbes, S.; Del Portillo, H.; Montoya, M.; Fraile, L. Key gaps in the knowledge of the porcine respiratory reproductive syndrome virus (PRRSV). Front. Vet. Sci. 2019, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotur-Stevuljevic, J.; Spasic, S.; Jelic-Ivanovic, Z.; Spasojevic-Kalimanovska, V.; Stefanovic, A.; Vujovic, A.; Memon, L.; Kalimanovska-Ostric, D. PON1 status is influenced by oxidative stress and inflammation in coronary heart disease patients. Clin. Biochem. 2008, 41, 1067–1073. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Meng, J.; Li, H.; Wei, H.; Bi, F.; Liu, S.; Tang, K.; Guo, H.; Liu, W. Resveratrol exhibits an effect on attenuating retina inflammatory condition and damage of diabetic retinopathy via PON1. Exp. Eye Res. 2019, 181, 356–366. [Google Scholar] [CrossRef]
- Bhattacharyya, T.; Nicholls, S.J.; Topol, E.J.; Zhang, R.; Yang, X.; Schmitt, D.; Fu, X.; Shao, M.; Brennan, D.M.; Ellis, S.G. Relationship of paraoxonase 1 (PON1) gene polymorphisms and functional activity with systemic oxidative stress and cardiovascular risk. Jama 2008, 299, 1265–1276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, D.; Song, C.; Sun, Y.; Du, Y.; Kim, O.; Liu, H.-C. Modulation of host cell responses and evasion strategies for porcine reproductive and respiratory syndrome virus. Virus Res. 2010, 154, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Zhang, Q.; Feng, W.-h. Regulation and evasion of antiviral immune responses by porcine reproductive and respiratory syndrome virus. Virus Res. 2015, 202, 101–111. [Google Scholar] [CrossRef]
- Zhao, Y.; Song, Z.; Bai, J.; Liu, X.; Nauwynck, H.; Jiang, P. ZAP, a CCCH-type zinc finger protein, inhibits porcine reproductive and respiratory syndrome virus replication and interacts with viral Nsp9. J. Virol. 2019, 93, e00001–e00019. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.; Gao, J.-C.; Xiong, J.-Y.; Guo, J.-C.; Yang, Y.-B.; Jiang, C.-G.; Tang, Y.-D.; Tian, Z.-J.; Cai, X.-H.; Tong, G.-Z. Two residues in NSP9 contribute to the enhanced replication and pathogenicity of highly pathogenic porcine reproductive and respiratory syndrome virus. J. Virol. 2018, 92, e02209–e02217. [Google Scholar] [CrossRef] [Green Version]
- Knoops, K.; Bárcena, M.; Limpens, R.W.; Koster, A.J.; Mommaas, A.M.; Snijder, E.J. Ultrastructural characterization of arterivirus replication structures: Reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J. Virol. 2012, 86, 2474–2487. [Google Scholar] [CrossRef] [Green Version]
- Dong, J.; Zhang, N.; Ge, X.; Zhou, L.; Guo, X.; Yang, H. The interaction of nonstructural protein 9 with retinoblastoma protein benefits the replication of genotype 2 porcine reproductive and respiratory syndrome virus in vitro. Virology 2014, 464, 432–440. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Guo, D.; Huang, L.; Yin, M.; Liu, Q.; Wang, Y.; Yang, C.; Liu, Y.; Zhang, L.; Tian, Z. The interaction between host Annexin A2 and viral Nsp9 is beneficial for replication of porcine reproductive and respiratory syndrome virus. Virus Res. 2014, 189, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Ge, X.; Wang, X.; Liu, A.; Guo, X.; Zhou, L.; Yu, K.; Yang, H. The DEAD-box RNA helicase 5 positively regulates the replication of porcine reproductive and respiratory syndrome virus by interacting with viral Nsp9 in vitro. Virus Res. 2015, 195, 217–224. [Google Scholar] [CrossRef] [PubMed]
- González-Navajas, J.M.; Lee, J.; David, M.; Raz, E. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 2012, 12, 125–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, X.-J.; Liu, L.-C.; Guo, C.; Shen, W.-W.; Cao, J.; Du, F.; Wu, D.-F.; Yu, H. Hepatic paraoxonase 1 ameliorates dysfunctional high-density lipoprotein and atherosclerosis in scavenger receptor class B type I deficient mice. Ann. Transl. Med. 2021, 9, 1063. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Fang, L.; Jin, H.; Jiang, Y.; Wang, D.; Chen, H.; Xiao, S. Antiviral activity of type I and type III interferons against porcine reproductive and respiratory syndrome virus (PRRSV). Antivira Res. 2011, 91, 99–101. [Google Scholar] [CrossRef]

| RNA Oligo Name | Sequence (Positive Strand) (5′-3′) |
|---|---|
| Negative Control | ACGUGACACGUUCGGAGAATT |
| siRNA-1 | AUUAUCUUCAUCUGUGAAG |
| siRNA-2 | UAGUAAACAGCAUAUGACC |
| siRNA-3 | AAUCUAGAGACUUCAAUGG |
| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| pCAGGS-PON1-CDS-F-KpnI | CGGGGTACCATGGCGAAGCTGATGGTGCT |
| pCAGGS-PON1-CDS-R-XhoI | CCGCTCGAGGAGCTCACAGTAAAGAGCTC |
| pLVX-PON1-CDS-F-XhoI | CCGCTCGAGATGGCGAAGCTGATGGTGCT |
| pLVX-PON1-CDS -R- BamHI | CGCGGATCCGAGCTCACAGTAAAGAGCTC |
| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| PRRSV-ORF7-F | AGATCATCGCCCAACAAAAC |
| PRRSV-ORF7-R | GACACAATTGCCGCTCACTA |
| Porcine-β-actin-F | CTTCCTGGGCATGGAGTCC |
| Porcine-β-actin-R | GGCGCGATGATCTTGATCTTC |
| Porcine-PON1-F | CCATCAAACACAAACTTCTGCC |
| Porcine-PON1-R | CTCCCAGAATGTCAGGTAAGTG |
| Porcine-IFN-β-F | GCTAACAAGTGCATCCTCCAAA |
| Porcine-IFN-β-R | CCAGGAGCTTCTGACATGCCA |
| Porcine-ISG15-F | GATGCTGGGAGGCAAGGA |
| Porcine-ISG15-R | CAGGATGCTCAGTGGGTCTCT |
| Porcine-OASL-F | TCCCTGGGAAGAATGTGCAG |
| Porcine-OASL-R | CCCTGGCAAGAGCATAGTGT |
| Porcine-GBP1-F | GAAGGGTGACAACCAGAACGAC |
| Porcine-GBP1-R | AGGTTCCGACTTTGCCCTGATT |
| Primer Name | Primer Sequence (5′-3′) |
|---|---|
| PRRSV-ORF7-CDS-F | ATGCCAAATAACAACGGCAAGCAG |
| PRRSV-ORF7-CDS-R | TCATGCTGAGGGTGATGCTGTGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Pan, Y.; Xu, Y.; Zhang, W.; Ma, W.; Ibrahim, Y.M.; Werid, G.M.; Zhang, H.; Xia, C.; Wei, P.; et al. Paraoxonase-1 Facilitates PRRSV Replication by Interacting with Viral Nonstructural Protein-9 and Inhibiting Type I Interferon Pathway. Viruses 2022, 14, 1203. https://doi.org/10.3390/v14061203
Zhang L, Pan Y, Xu Y, Zhang W, Ma W, Ibrahim YM, Werid GM, Zhang H, Xia C, Wei P, et al. Paraoxonase-1 Facilitates PRRSV Replication by Interacting with Viral Nonstructural Protein-9 and Inhibiting Type I Interferon Pathway. Viruses. 2022; 14(6):1203. https://doi.org/10.3390/v14061203
Chicago/Turabian StyleZhang, Lin, Yu Pan, Yunfei Xu, Wenli Zhang, Wenjie Ma, Yassein M. Ibrahim, Gebremeskel Mamu Werid, He Zhang, Changyou Xia, Ping Wei, and et al. 2022. "Paraoxonase-1 Facilitates PRRSV Replication by Interacting with Viral Nonstructural Protein-9 and Inhibiting Type I Interferon Pathway" Viruses 14, no. 6: 1203. https://doi.org/10.3390/v14061203








