Molecular Characterisation and Phylogeny of Tula Virus in Kazakhstan
Abstract
:1. Introduction
2. Materials and Methods
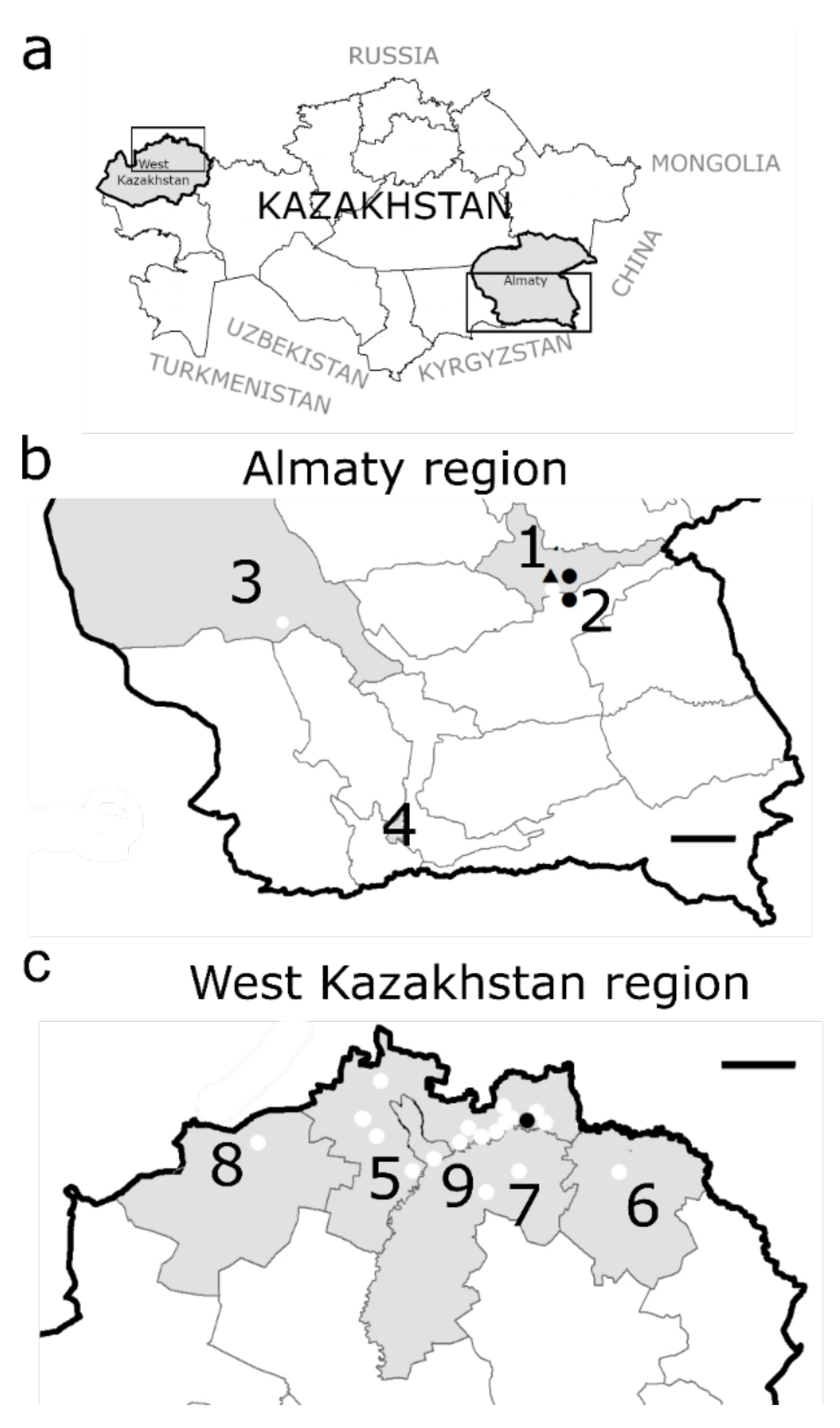
2.1. Study Setting and Rodent Sampling
2.2. RNA Extraction, PCR Amplification and Sequencing
2.3. Phylogenetic Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vaheri, A.; Henttonen, H.; Voutilainen, L.; Mustonen, J.; Sironen, T.; Vapalahti, O. Hantavirus infections in Europe and their impact on public health. Rev. Med. Virol. 2013, 23, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Laenen, L.; Vergote, V.; Calisher, C.H.; Klempa, B.; Klingström, J.; Kuhn, J.H.; Maes, P. Hantaviridae: Current Classification and Future Perspectives. Viruses 2019, 11, 788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Essbauer, S.; Krautkrämer, E. Hantaviruses—Infections, Epidemiology and Hosts. In Zoonooses: Infections Affecting Men and Animals—A Focus on Public Health Aspects; Springer: Berlin/Heidelberg, Germany, 2015; pp. 749–783. [Google Scholar]
- Schlegel, M.; Jacob, J.; Krüger, D.H.; Rang, A.; Ulrich, R.G. Hantavirus Emergence in Rodents, Insectivores and Bats. In The Role of Animals in Emerging Viral Diseases; Academic Press: Cambridge, MA, USA, 2014; pp. 235–292. [Google Scholar] [CrossRef]
- Avšič-Županc, T.; Saksida, A.; Korva, M. Hantavirus infections. Clin. Microbiol. Infect. 2019, 21S, e6–e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaheri, A.; Henttonen, H.; Mustonen, J. Hantavirus Research in Finland: Highlights and Perspectives. Viruses 2021, 13, 1452. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Numan, A.; Sharma, D.; Shukla, R.; Alexander, A.; Jain, G.K.; Ahmad, F.J.; Kesharwani, P. Epidemiology, virology and clinical aspects of hantavirus infections: An overview. Int. J. Environ. Health Res. 2021, 22, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Romero, M.G.; Anjum, F. Hemorrhagic Fever Renal Syndrome. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Ermonval, M.; Baychelier, F.; Tordo, N. What Do We Know about How Hantaviruses Interact with Their Different Hosts? Viruses 2016, 8, 223. [Google Scholar] [CrossRef] [Green Version]
- Klempa, B.; Meisel, H.; Räth, S.; Bartel, J.; Ulrich, R.; Krüger, D.H. Occurrence of Renal and Pulmonary Syndrome in a Region of Northeast Germany Where Tula Hantavirus Circulates. J. Clin. Microbiol. 2003, 41, 4894–4897. [Google Scholar] [CrossRef] [Green Version]
- Mertens, M.; Hofmann, J.; Petraityte-Burneikiene, R.; Ziller, M.; Sasnauskas, K.; Friedrich, R.; Niederstrasser, O.; Krüger, D.H.; Groschup, M.H.; Petri, E.; et al. Seroprevalence study in forestry workers of a non-endemic region in eastern Germany reveals infections by Tula and Dobrava-Belgrade hantaviruses. Med. Microbiol. Immunol. 2011, 200, 263–268. [Google Scholar] [CrossRef]
- Zelená, H.; Mrázek, J.; Kuhn, T. Tula Hantavirus Infection in Immunocompromised Host, Czech Republic. Emerg. Infect. Dis. 2013, 19, 1873–1876. [Google Scholar] [CrossRef]
- Hofmann, J.; Kramer, S.; Herrlinger, K.R.; Jeske, K.; Kuhns, M.; Weiss, S.; Ulrich, R.G.; Krüger, D.H. Tula Virus as Causative Agent of Hantavirus Disease in Immunocompetent Person, Germany. Emerg. Infect. Dis. 2021, 27, 1234–1237. [Google Scholar] [CrossRef]
- Akram, S.M.; Mangat, R.; Huang, B. Hantavirus Cardiopulmonary Syndrome. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Burek, K.A.; Rossi, C.A.; Leduc, J.W.; Yuill, T.M. Serologic and Virologic Evidence of a Prospect Hill-like Hantavirus in Wisconsin and Minnesota. Am. J. Trop. Med. Hyg. 1994, 51, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Peintner, L.; Wagner, E.; Shin, A.; Tukhanova, N.; Turebekov, N.; Abdiyeva, K.; Spaiser, O.; Serebrennikova, Y.; Tintrup, E.; Dmitrovskiy, A.; et al. Eight Years of Collaboration on Biosafety and Biosecurity Issues Between Kazakhstan and Germany as Part of the German Biosecurity Programme and the G7 Global Partnership Against the Spread of Weapons and Materials of Mass Destruction. Front. Public Health 2021, 9, 1102. [Google Scholar] [CrossRef] [PubMed]
- Aikimbayev, M. Atlas of Bacterial and Virus Zoonotic Infections Distribution in Kazakhstan; Kazakh Scientific Centre for Quarantine and Zoonotic Diseases under CSSES of the MPH of the Republic of Kazakhstan: Almaty, Kazakhstan, 2010. [Google Scholar]
- Abdiyeva, K.; Turebekov, N.; Dmitrovsky, A.; Tukhanova, N.; Shin, A.; Yeraliyeva, L.; Heinrich, N.; Hoelscher, M.; Yegemberdiyeva, R.; Shapiyeva, Z.; et al. Seroepidemiological and molecular investigations of infections with Crimean–Congo haemorrhagic fever virus in Kazakhstan. Int. J. Infect. Dis. 2019, 78, 121–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bezverhnii, A. Comprehensive Epidemiological Investigation of Zoonotic Infections in the Dzungarian Alatau. Ph.D. Thesis, Kazakhstan National University, Almaty, Kazakhstan, 1995. [Google Scholar]
- Sutyagin, V. Hemorrhagic fever with renal syndrome in Dzungarian Alatau. Quar. Zoonotic Infect. Kaz. 2010, 1, 120–121. [Google Scholar]
- Sutyagin, V.; Belyaev, A.; Kim, I.; Berdibekov, A. Distribution of hemorrhagic fever with renal syndrome in Almaty region Dzungarian Alatau. Environ. Public Health 2017, 3, 56–58. [Google Scholar]
- Plyusnina, A.; Laakkonen, J.; Niemimaa, J.; Henttonen, H.; Plyusnin, A. New Genetic Lineage of Tula Hantavirus in Microtus arvalis obscurus in Eastern Kazakhstan. Open Virol. J. 2008, 2, 32–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tukhanova, N.; Shin, A.; Abdiyeva, K.; Turebekov, N.; Yeraliyeva, L.; Yegemberdiyeva, R.; Shapiyeva, Z.; Froeschl, G.; Hoelscher, M.; Wagner, E.; et al. Serological investigation of orthohantaviruses in patients with fever of unknown origin in Kazakhstan. Zoonoses Public Health 2020, 67, 271–279. [Google Scholar] [CrossRef]
- Grazhdanov, A.; Zakharov, A.; Biryukov, A. First cases of Hemorrhagic fever with renal syndrome in Kazakhstan. J. Quar. Zoonotic Infect. Kaz. 2001, 3, 94–98. [Google Scholar]
- Bidashko, F.; Grazhdanov, A.; Rakhmankulov, R. Some aspects of the epizootology of the Ural-Ilek foci of Hemorrhagic fever with renal syndrome. J. Quar. Zoonotic Infect. Kaz. 2004, 1, 96–104. [Google Scholar]
- Grazhdanov, A.K.; Ayazbaev, T.Z.; Toporkov, A.V.; Bidashko, F.G.; Zakharov, A.V.; Belonozhkina, L.; Pak, M.V.; Andryushchenko, A.V. Concerning the Allocation of Emerging Natural Foci of the Currently Important Infectious Diseases in the West of Kazakhstan. Probl. Part. Danger. Infect. 2014, 3, 20–24. [Google Scholar] [CrossRef] [Green Version]
- Gromov, M.; Erbajeva, M. The Mammals of the Russia and Adjacent Territories (Lagomorphs and Rodents); Russian Academy of Sciences Zoological Institute: Saint Petersburg, Russia, 1995. [Google Scholar]
- Scharninghausen, J.J.; Meyer, H.; Pfeffer, M.; Davis, D.S.; Honeycutt, R.L. Genetic Evidence of Dobrava Virus in Apodemus agrarius in Hungary. Emerg. Infect. Dis. 1999, 5, 468–470. [Google Scholar] [CrossRef] [PubMed]
- Essbauer, S.; Schmidt, J.; Conraths, F.J.; Friedrich, R.; Koch, J.; Hautmann, W.; Pfeffer, M.; Wölfel, R.; Finke, J.; Dobler, G.; et al. A new Puumala hantavirus subtype in rodents associated with an outbreak of Nephropathia epidemica in South-East Germany in 2004. Epidemiol. Infect. 2006, 134, 1333–1344. [Google Scholar] [CrossRef] [PubMed]
- Mossbrugger, I.; Felder, E.; Gramsamer, B.; Wölfel, R. EvaGreen based real-time RT-PCR assay for broad-range detection of hantaviruses in the field. J. Clin. Virol. 2013, 58, 334–335. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, M.; Ali, H.S.; Stieger, N.; Groschup, M.H.; Wolf, R.; Ulrich, R.G. Molecular identification of small mammal species using novel cytochrome B gene-derived degenerated primers. Biochem. Genet. 2012, 50, 440–447. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Schmidt, S.; Saxenhofer, M.; Drewes, S.; Schlegel, M.; Wanka, K.M.; Frank, R.; Klimpel, S.; Von Blanckenhagen, F.; Maaz, D.; Herden, C.; et al. High genetic structuring of Tula hantavirus. Arch. Virol. 2016, 161, 1135–1149. [Google Scholar] [CrossRef]
- Maas, M.; de Vries, A.; van Roon, A.; Takumi, K.; van der Giessen, J.; Rockx, B. High Prevalence of Tula Hantavirus in Common Voles in The Netherlands. Vector Borne Zoonotic Dis. 2017, 17, 200–205. [Google Scholar] [CrossRef]
- Scharninghausen, J.J.; Pfeffer, M.; Meyer, H.; Davis, D.S.; Honeycutt, R.L.; Faulde, M. Genetic Evidence for Tula Virus in Microtus arvalis and Microtus agrestis Populations in Croatia. Vector-Borne Zoonotic Dis. 2002, 2, 19–27. [Google Scholar] [CrossRef]
- Bekmukhambetov, S.K. Experience of diagnosis and treatment of epidemic hemorrhagic fever in Kazakhstan. J. Med. 2012, 4, 58–66. (In Russian) [Google Scholar]
- Zakharov, A.V.; Grazhdanov, A.K.; Zakharov, V.M.; Nazhimova, G.S. Clinical manifestations of acute renal failure in hemorrhagic fever with renal syndrome. J. Quar. Zoonotic Infect. Kaz. 2010, 1–2, 18–22. (In Russian) [Google Scholar]
- Kariwa, H.; Tkachenko, E.A.; Morozov, V.G.; Seto, T.; Tanikawa, Y.; Kolominov, S.I.; Belov, S.N.; Nakamura, I.; Hashimoto, N.; Balakiev, A.E.; et al. Epidemiological Study of Hantavirus Infection in the Samara Region of European Russia. J. Vet. Med. Sci. 2009, 71, 1569–1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabenau, H.F.; Kessler, H.H.; Kortenbusch, M.; Steinhorst, A.; Raggam, R.B.; Berger, A. Verification and validation of diagnostic laboratory tests in clinical virology. J. Clin. Virol. 2007, 40, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-W.; Gligic, A.; Yanagihara, R. Identification of Tula hantavirus in Pitymys subterraneus captured in the Cacak region of Serbia-Yugoslavia. Int. J. Infect. Dis. 2002, 6, 31–36. [Google Scholar] [CrossRef] [Green Version]
- Schmidt-Chanasit, J.; Essbauer, S.; Petraityte, R.; Yoshimatsu, K.; Tackmann, K.; Conraths, F.J.; Sasnauskas, K.; Arikawa, J.; Thomas, A.; Pfeffer, M.; et al. Extensive Host Sharing of Central European Tula Virus. J. Virol. 2010, 84, 459–474. [Google Scholar] [CrossRef] [Green Version]
- Saxenhofer, M.; Labutin, A.; White, T.A.; Heckel, G. Host genetic factors associated with the range limit of a European hantavirus. Mol. Ecol. 2021, 31, 252–265. [Google Scholar] [CrossRef]
- Saxenhofer, M.; Schmidt, S.; Ulrich, R.G.; Heckel, G. Secondary contact between diverged host lineages entails ecological speciation in a European hantavirus. PLoS Biol. 2019, 10, e3000142. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, S.; Reil, D.; Jeske, K.; Drewes, S. Spatial and Temporal Dynamics and Molecular Evolution of Tula orthohantavirus in German Vole Populations. Viruses 2021, 13, 1132. [Google Scholar] [CrossRef]
- Saxenhofer, M.; de Melo, W.V.; Ulrich, R.G.; Heckel, G. Revised time scales of RNA virus evolution based on spatial information. Proc. R. Soc. B 2017, 284, 20170857. [Google Scholar] [CrossRef] [Green Version]
- Hiltbrunner, M.; Heckel, G. Assessing Genome-Wide Diversity in European Hantaviruses through Sequence Capture from Natural Host Samples. Viruses 2020, 12, 749. [Google Scholar] [CrossRef]
- Chen, J.-T.; Qin, J.; Li, K.; Xu, Q.-Y.; Wang, X.-P.; Plyusnin, A.; Hou, W.; Zhang, Y.-Z. Identification and characterization of a novel subtype of Tula virus in Microtus arvalis obscurus voles sampled from Xinjiang, China. Infect. Genet. Evol. 2019, 75, 104012. [Google Scholar] [CrossRef]
- Polat, C.; Ergünay, K.; Irmak, S.; Erdin, M.; Brinkmann, A.; Çetintaş, O.; Çoğal, M.; Sözen, M.; Matur, F.; Nitsche, A.; et al. A novel genetic lineage of Tula orthohantavirus in Altai voles (Microtus obscurus) from Turkey. Infect. Genet. Evol. 2019, 67, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Schultze, D.; Lundkvist, A.; Blauenstein, U.; Heyman, P. Tula virus infection associated with fever and exanthema after a wild rodent bite. Eur. J. Clin. Microbiol. Infect. Dis. 2002, 21, 304–306. [Google Scholar] [CrossRef] [PubMed]


| Small Mammal Species | West Kazakhstan (19 Trapping Sites) | Almaty Region (4 Trapping Sites) | Almaty City (7 Trapping Sites) |
|---|---|---|---|
| Microtus arvalis (Common vole) | 13 | 72 | 1 |
| Myodes glareolus (Bank vole) | 12 | 0 | 0 |
| Microtus kirgisorum (Tien Shan vole) | 0 | 0 | 49 |
| Apodemus uralensis (Ural or Pygmy field mouse) | 128 | 84 | 47 |
| Mus musculus (House mouse) | 62 | 27 | 39 |
| Rattus norvegicus (Brown rat) | 0 | 0 | 39 |
| Meriones meridianus (Midday jird) | 0 | 2 | 0 |
| Dryomys nitedula (Forest dormouse) | 2 | 13 | 0 |
| Sorex araneus (Common shrew) | 1 | 0 | 0 |
| Sorex minutus (Eurasian pygmy shrew) | 0 | 1 | 1 |
| Crocidura suaveolens (Lesser white-toothed shrew) | 0 | 0 | 28 |
| Total | 218 | 199 | 204 |
| Small Mammal Species | Total Collected | Sex Ratio Male/Female | Number of Positive Samples (Male/Female) | Percentage of Positive Samples [%] |
|---|---|---|---|---|
| Microtus arvalis | 86 | 40/46 | 13 (8/5) | 15.1 |
| Dryomys nitedula | 15 | 7/8 | 2 (1/1) | 13.3 |
| Myodes glareolus | 12 | 11/1 | 0 | 0 |
| Microtus kirgisorum | 49 | 26/23 | 0 | 0 |
| Apodemus uralensis | 259 | 163/96 | 0 | 0 |
| Mus musculus | 128 | 83/45 | 0 | 0 |
| Rattus norvegicus | 39 | 16/23 | 0 | 0 |
| Meriones meridianus | 2 | 2/0 | 0 | 0 |
| Sorex araneus | 1 | 0/1 | 0 | 0 |
| Sorex minutus | 2 | 1/1 | 0 | 0 |
| Crocidura suaveolens | 28 | 15/13 | 0 | 0 |
| Total | 621 | 364/257 | 15 (9/6) | 2.4 |
| S Segment Cluster | South-East Kazakhstan | China (Xinjiang)/ Russia (Siberia) | Russia (Tula and Crimea) | West Kazakhstan | Russia (Samara) |
|---|---|---|---|---|---|
| South-East Kazakhstan | 94.3–100 | 78.9–99.4 | 78.9–99.4 | 78.9–99.4 | 75.8–99.1 |
| China (Xinjiang)/ Russia (Siberia) | 84.5–87.5 | 81.6–98.5 | 82.1–87.5 | 79.9–88.9 | |
| Russia (Tula and Crimea) | 87.5–98.5 | 84.5–98.5 | 85.6–97.9 | ||
| West Kazakhstan | 100 | 93.4 | |||
| Russia (Samara) | 100 |
| L Segment Cluster | Turkey and China | South-East Kazakhstan | West Kazakhstan | Central Europe |
|---|---|---|---|---|
| Turkey and China | 85.9–100 | 80–99.3 | 81.6–85.9 | 78.3–97.2 |
| South-East Kazakhstan | 89.3–100 | 80.6–99.3 | 76.9–88.3 | |
| West Kazakhstan | 100 | 79.4–97.2 | ||
| Central Europe | 87–97.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tukhanova, N.; Shin, A.; Turebekov, N.; Nurmakhanov, T.; Abdiyeva, K.; Shevtsov, A.; Yerubaev, T.; Tokmurziyeva, G.; Berdibekov, A.; Sutyagin, V.; et al. Molecular Characterisation and Phylogeny of Tula Virus in Kazakhstan. Viruses 2022, 14, 1258. https://doi.org/10.3390/v14061258
Tukhanova N, Shin A, Turebekov N, Nurmakhanov T, Abdiyeva K, Shevtsov A, Yerubaev T, Tokmurziyeva G, Berdibekov A, Sutyagin V, et al. Molecular Characterisation and Phylogeny of Tula Virus in Kazakhstan. Viruses. 2022; 14(6):1258. https://doi.org/10.3390/v14061258
Chicago/Turabian StyleTukhanova, Nur, Anna Shin, Nurkeldi Turebekov, Talgat Nurmakhanov, Karlygash Abdiyeva, Alexandr Shevtsov, Toktasyn Yerubaev, Gulnara Tokmurziyeva, Almas Berdibekov, Vitaliy Sutyagin, and et al. 2022. "Molecular Characterisation and Phylogeny of Tula Virus in Kazakhstan" Viruses 14, no. 6: 1258. https://doi.org/10.3390/v14061258






