Conformational Changes in Ff Phage Protein gVp upon Complexation with Its Viral Single-Stranded DNA Revealed Using Magic-Angle Spinning Solid-State NMR
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Gel Electrophoresis Binding Assay
2.3. NMR Experiments
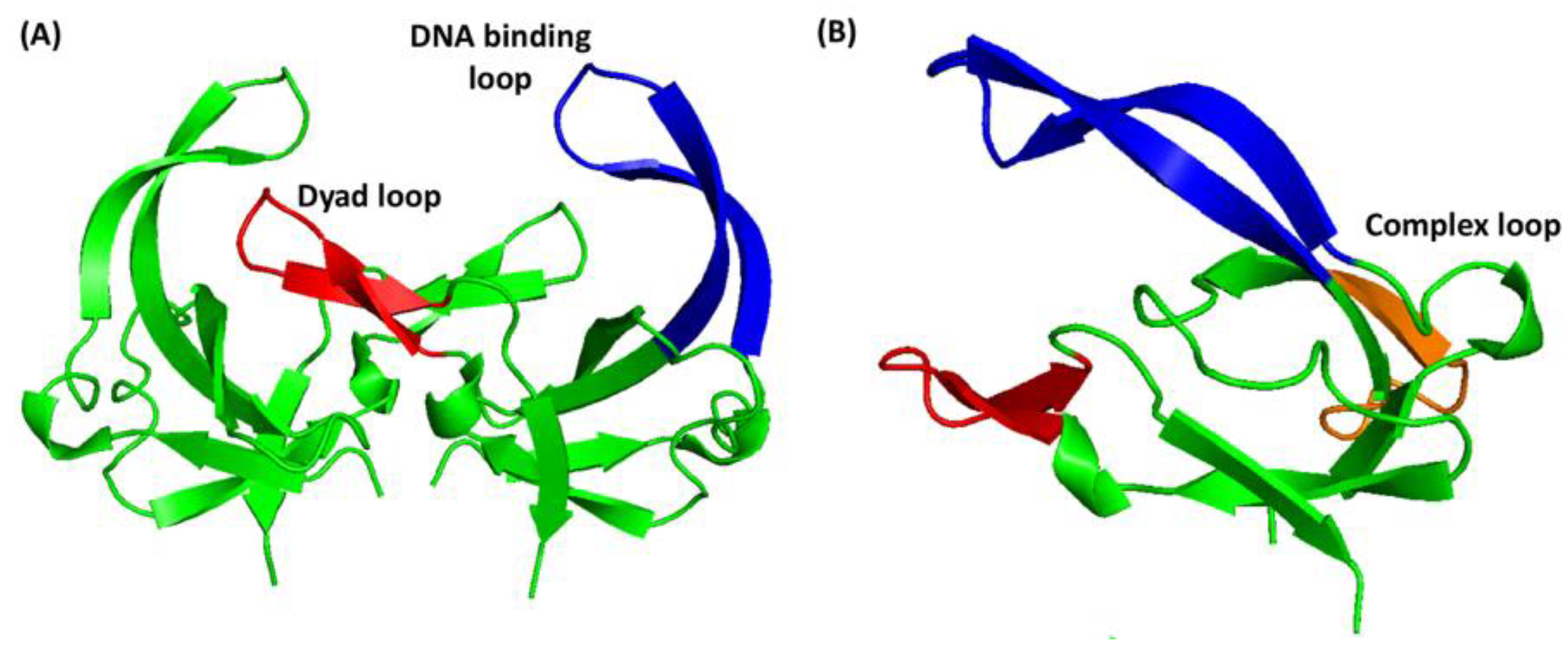
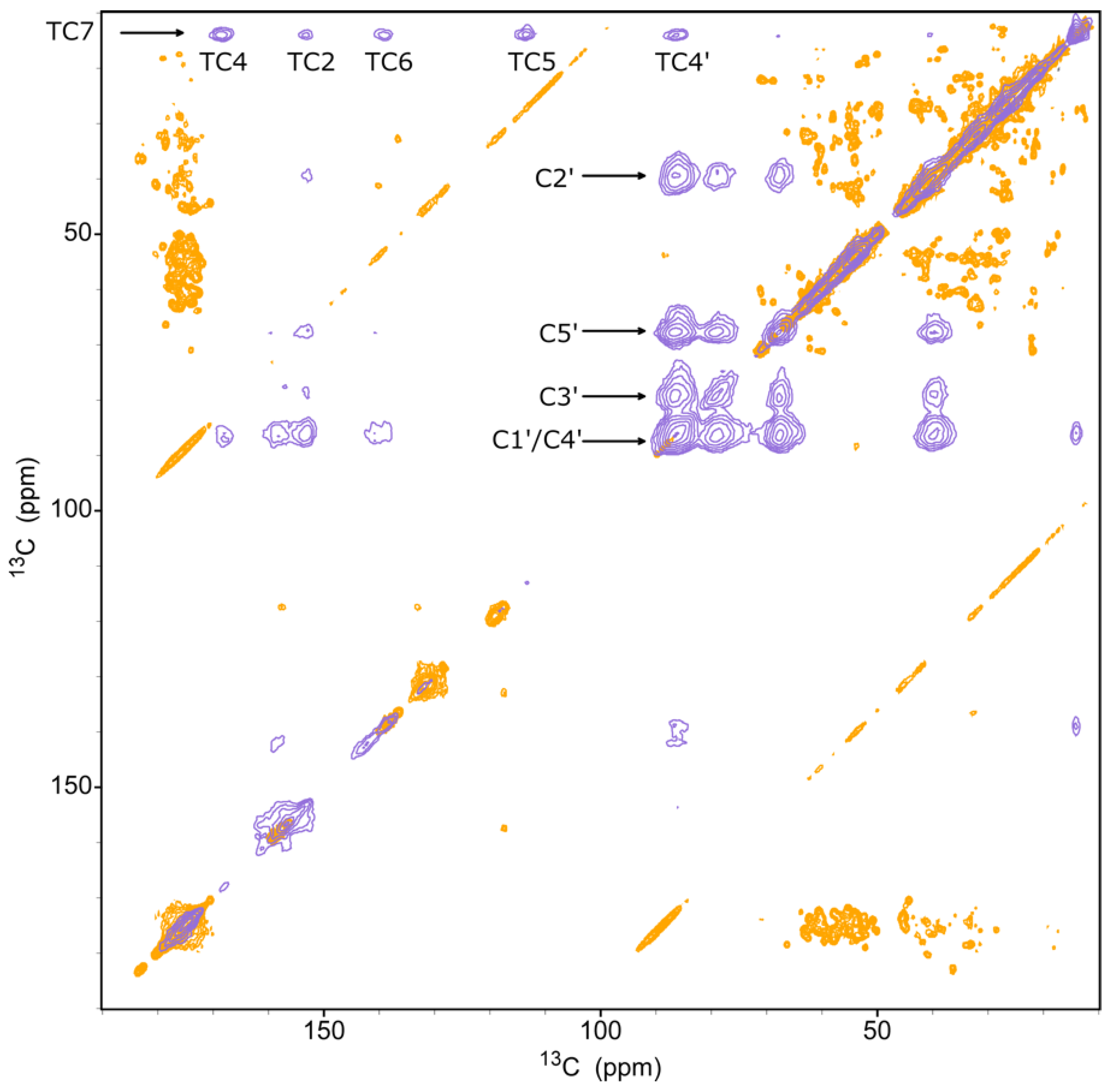
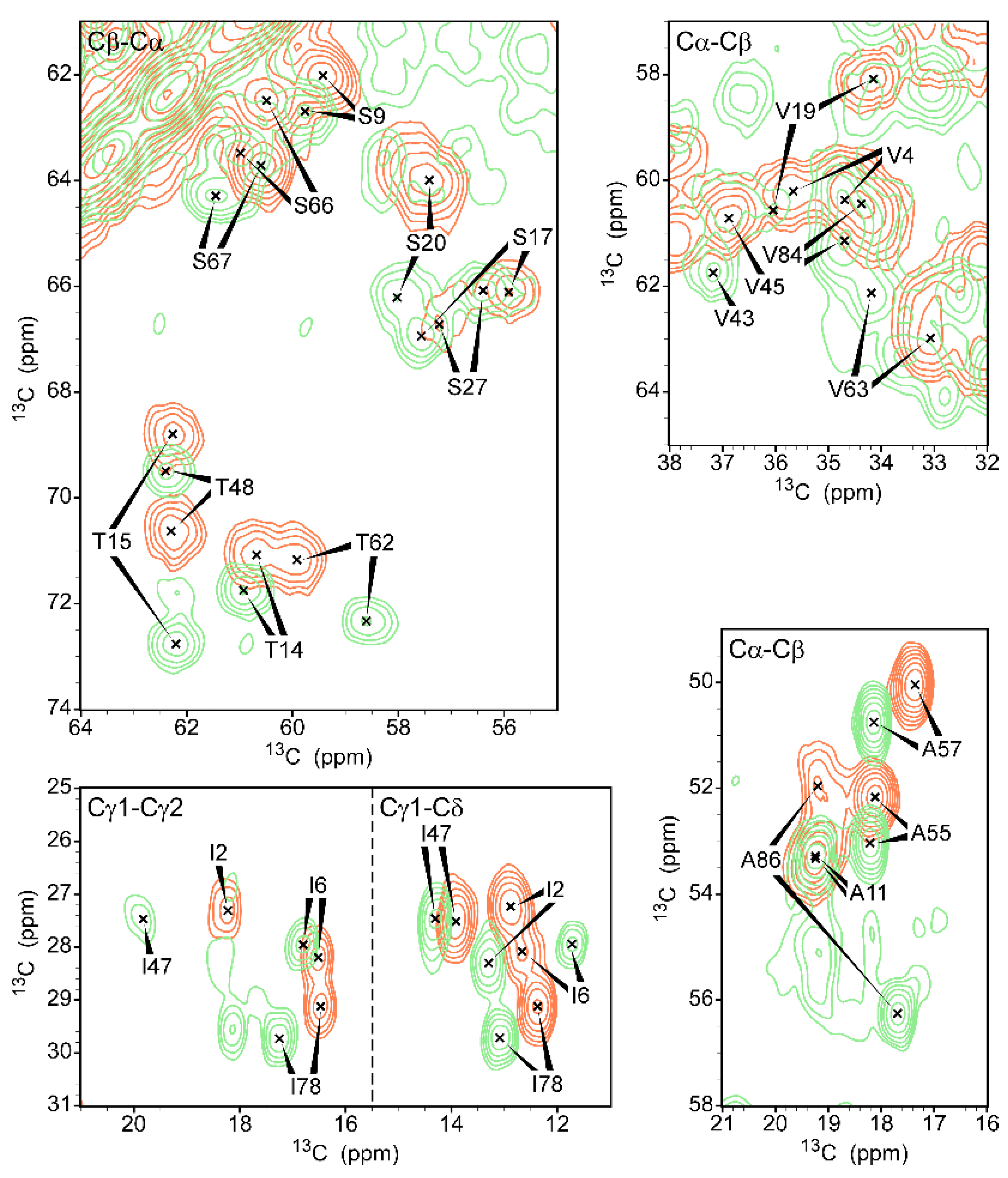
3. Results and Discussion
3.1. Complex Formation between gVp and fd Phage ssDNA
3.2. Chemical Shift Assignment of gVp
3.3. Comparison with Free gVp
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Young, R.; Gill, J.J. Phage Therapy Redux—What Is to Be Done? Science 2015, 350, 1163–1164. [Google Scholar] [CrossRef] [PubMed]
- Endersen, L.; O’Mahony, J.; Hill, C.; Ross, R.P.; McAuliffe, O.; Coffey, A. Phage Therapy in the Food Industry. Annu. Rev. Food Sci. Technol. 2014, 5, 327–349. [Google Scholar] [CrossRef] [PubMed]
- Buttimer, C.; McAuliffe, O.; Ross, R.P.; Hill, C.; O’Mahony, J.; Coffey, A. Bacteriophages and Bacterial Plant Diseases. Front. Microbiol. 2017, 8, 34. [Google Scholar] [CrossRef] [PubMed]
- Doss, J.; Culbertson, K.; Hahn, D.; Camacho, J.; Barekzi, N. A Review of Phage Therapy against Bacterial Pathogens of Aquatic and Terrestrial Organisms. Viruses 2017, 9, 50. [Google Scholar] [CrossRef] [PubMed]
- Marvin, D.A.; Welsh, L.C.; Symmons, M.F.; Scott, W.R.P.; Straus, S.K. Molecular Structure of Fd (F1, M13) Filamentous Bacteriophage Refined with Respect to X-ray Fibre Diffraction and Solid-State NMR Data Supports Specific Models of Phage Assembly at the Bacterial Membrane. J. Mol. Biol. 2006, 355, 294–309. [Google Scholar] [CrossRef] [PubMed]
- Morag, O.; Sgourakis, N.G.; Baker, D.; Goldbourt, A. The NMR-Rosetta Capsid Model of M13 Bacteriophage Reveals a Quadrupled Hydrophobic Packing Epitope. Proc. Natl. Acad. Sci. USA 2015, 112, 971–976. [Google Scholar] [CrossRef]
- Marvin, D. Filamentous Phage Structure, Infection and Assembly. Curr. Opin. Struct. Biol. 1998, 8, 150–158. [Google Scholar] [CrossRef]
- Costa, T.R.D.; Ilangovan, A.; Ukleja, M.; Redzej, A.; Santini, J.M.; Smith, T.K.; Egelman, E.H.; Waksman, G. Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex. Cell 2016, 166, 1436–1444.e10. [Google Scholar] [CrossRef]
- Bulsink, H.; Harmsen, B.J.M.; Hilbers, C.W. DNA-Binding Properties of Gene-5 Protein Encoded by Bacteriophage M 13. 1. The Kinetics of the Dissociation of Gene-5-Protein Polynucleotide Complexes upon Addition of Salt. Eur. J. Biochem. 1988, 176, 589–596. [Google Scholar] [CrossRef]
- Pratt, D.; Erdahl, W.S. Genetic Control of Bacteriophage M13 DNA Synthesis. J. Mol. Biol. 1968, 37, 181–200. [Google Scholar] [CrossRef]
- Alberts, B.; Frey, L.; Delius, H. Isolation and Characterization of Gene 5 Protein of Filamentous Bacterial Viruses. J. Mol. Biol. 1972, 68, 139–152. [Google Scholar] [CrossRef]
- Stassen, A.P.M.; Folmer, R.H.A.; Hilbers, C.W.; Konings, R.N.H. Single-Stranded DNA Binding Protein Encoded by the Filamentous Bacteriophage M13: Structural and Functional Characteristics. Mol. Biol. Rep. 1995, 20, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Cavalieri, S.J.; Neet, K.E.; Goldthwait, D.A. Gene 5 Protein of Bacteriophage Fd: A Dimer Which Interacts Co-Operatively with DNA. J. Mol. Biol. 1976, 102, 697–711. [Google Scholar] [CrossRef]
- Skinner, M.M.; Zhang, H.; Leschnitzer, D.H.; Guan, Y.; Bellamy, H.; Sweet, R.M.; Gray, C.W.; Konings, R.N.; Wang, A.H.; Terwilliger, T.C. Structure of the Gene V Protein of Bacteriophage F1 Determined by Multiwavelength X-ray Diffraction on the Selenomethionyl Protein. Proc. Natl. Acad. Sci. USA 1994, 91, 2071–2075. [Google Scholar] [CrossRef]
- Folkers, P.J.M.; Nilges, M.; Folmer, R.H.A.; Konings, R.N.H.; Hilbers, C.W. The Solution Structure of the Tyr41→His Mutant of the Single-Stranded DNA Binding Protein Encoded by Gene V of the Filamentous Bacteriophage M13. J. Mol. Biol. 1994, 236, 229–246. [Google Scholar] [CrossRef]
- Prompers, J.J.; Folmer, R.H.A.; Nilges, M.; Folkers, P.J.M.; Konings, R.N.H.; Hilbers, C.W. Refined Solution Structure of the Tyr41His Mutant of the M13 Gene V Protein. A Comparison with the Crystal Structure. Eur. J. Biochem. 1995, 232, 506–514. [Google Scholar] [CrossRef]
- Konings, R.N.H.; Folmer, R.H.A.; Folkers, P.J.M.; Nilges, M.; Hilbers, C.W. Three-Dimensional Structure of the Single-Stranded DNA-Binding Protein Encoded by Gene V of the Filamentous Bacteriophage M13 and a Model of Its Complex with Single-Stranded DNA. FEMS Microbiol. Rev. 1995, 17, 57–72. [Google Scholar] [CrossRef]
- Mou, T.-C.; Shen, M.C.; Terwilliger, T.C.; Gray, D.M. Binding and Reversible Denaturation of Double-Stranded DNA by Ff Gene 5 Protein. Biopolymers 2003, 70, 637–648. [Google Scholar] [CrossRef]
- Wen, J.-D.; Gray, D.M. Ff Gene 5 Single-Stranded DNA-Binding Protein Assembles on Nucleotides Constrained by a DNA Hairpin. Biochemistry 2004, 43, 2622–2634. [Google Scholar] [CrossRef]
- Oliver, A.W.; Bogdarina, I.; Schroeder, E.; Taylor, I.A.; Kneale, G.G. Preferential Binding of Fd Gene 5 Protein to Tetraplex Nucleic Acid Structures. J. Mol. Biol. 2000, 301, 575–584. [Google Scholar] [CrossRef]
- Terwilliger, T.C. Gene V Protein Dimerization and Cooperativity of Binding to Poly(DA). Biochemistry 1996, 35, 16652–16664. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-K.; Maye, M.M.; Zhang, Y.-B.; Gang, O.; van der Lelie, D. Controllable G5p-Protein-Directed Aggregation of SsDNA−Gold Nanoparticles. Langmuir 2009, 25, 657–660. [Google Scholar] [CrossRef] [PubMed]
- Solomun, T.; Sturm, H.; Wellhausen, R.; Seitz, H. Interaction of a Single-Stranded DNA-Binding Protein G5p with DNA Oligonucleotides Immobilised on a Gold Surface. Chem. Phys. Lett. 2012, 533, 92–94. [Google Scholar] [CrossRef]
- PyMOL. Available online: https://pymol.org/2/ (accessed on 1 June 2022).
- Torbet, J.; Gray, D.M.; Gray, C.W.; Marvin, D.A.; Siegrist, H. Structure of the Fd DNA-Gene 5 Protein Complex in Solution. J. Mol. Biol. 1981, 146, 305–320. [Google Scholar] [CrossRef]
- Gray, C.W. Three-Dimensional Structure of Complexes of Single-Stranded DNA-Binding Proteins with DNA. J. Mol. Biol. 1989, 208, 57–64. [Google Scholar] [CrossRef]
- Olah, G.A.; Gray, D.M.; Gray, C.W.; Kergil, D.L.; Sosnick, T.R.; Mark, B.L.; Vaughan, M.R.; Trewhella, J. Structures of Fd Gene 5 Proteinnucleic Acid Complexes: A Combined Solution Scattering and Electron Microscopy Study. J. Mol. Biol. 1995, 249, 576–594. [Google Scholar] [CrossRef]
- Anderson, R.A.; Nakashima, Y.; Coleman, J.E. Chemical Modifications of Functional Residues of Fd Gene 5 DNA-Binding Protein. Biochemistry 1975, 14, 907–917. [Google Scholar] [CrossRef]
- Kansy, J.W.; Clack, B.A.; Gray, D.M. The Binding of Fd Gene 5 Protein to Polydeoxynucleotides: Evidence from CD Measurements for Two Binding Modes. J. Biomol. Struct. Dyn. 1986, 3, 1079–1110. [Google Scholar] [CrossRef]
- Alma, N.C.M.; Harmsen, B.J.M.; de Jong, E.A.M.; Ven, J.V.D.; Hilbers, C.W. Fluorescence Studies of the Complex Formation between the Gene 5 Protein of Bacteriophage M13 and Polynucleotides. J. Mol. Biol. 1983, 163, 47–62. [Google Scholar] [CrossRef]
- Brayer, G.D.; McPherson, A. A Model for Intracellular Complexation between Gene-5 Protein and Bacteriophage Fd DNA. Eur. J. Biochem. 1985, 150, 287–296. [Google Scholar] [CrossRef]
- Gray, D.M.; Gray, C.W.; Carlson, R.D. Neutron Scattering Data on Reconstituted Complexes of Fd DNA and Gene 5 Protein Show That the DNA Is near the Center. Biochemistry 1982, 21, 2702–2713. [Google Scholar] [CrossRef] [PubMed]
- Dick, L.R.; Geraldes, C.F.G.C.; Sherry, D.A.; Gray, C.W.; Gray, D.M. 13C NMR of Methylated Lysines of Fd Gene 5 Protein: Evidence for a Conformational Change Involving Lysine 24 upon Binding of a Negatively Charged Lanthanide Chelate. Biochemistry 1989, 28, 7896–7904. [Google Scholar] [CrossRef] [PubMed]
- King, G.C.; Coleman, J.E. Ff Gene 5 Protein-d(PA)40-60 Complex: Proton NMR Supports a Localized Base-Binding Model. Biochemistry 1988, 27, 6947–6953. [Google Scholar] [CrossRef] [PubMed]
- King, G.C.; Coleman, J.E. Two-Dimensional Proton NMR of Gene 5 Protein Indicates That Only Two Aromatic Rings Interact Significantly with Oligodeoxynucleotide Bases. Biochemistry 1987, 26, 2929–2937. [Google Scholar] [CrossRef]
- Folkers, P.J.M.; van Duynhoven, J.P.M.; van Lieshout, H.T.M.; Harmsen, B.J.M.; van Boom, J.H.; Tesser, G.I.; Konings, R.N.H.; Hilbers, C.W. Exploring the DNA Binding Domain of Gene V Protein Encoded by Bacteriophage M13 with the Aid of Spin-Labeled Oligonucleotides in Combination with Proton NMR. Biochemistry 1993, 32, 9407–9416. [Google Scholar] [CrossRef]
- Hahn, M.B.; Solomun, T.; Wellhausen, R.; Hermann, S.; Seitz, H.; Meyer, S.; Kunte, H.-J.; Zeman, J.; Uhlig, F.; Smiatek, J.; et al. Influence of the Compatible Solute Ectoine on the Local Water Structure: Implications for the Binding of the Protein G5P to DNA. J. Phys. Chem. B 2015, 119, 15212–15220. [Google Scholar] [CrossRef]
- van der Wel, P.C.A. New Applications of Solid-State NMR in Structural Biology. Emerg. Top. Life Sci. 2018, 2, 57–67. [Google Scholar] [CrossRef]
- Demers, J.-P.; Fricke, P.; Shi, C.; Chevelkov, V.; Lange, A. Structure Determination of Supra-Molecular Assemblies by Solid-State NMR: Practical Considerations. Prog. Nucl. Magn. Reson. Spectrosc. 2018, 109, 51–78. [Google Scholar] [CrossRef]
- Quinn, C.M.; Polenova, T. Structural Biology of Supramolecular Assemblies by Magic-Angle Spinning NMR Spectroscopy. Q. Rev. Biophys. 2017, 50, e1. [Google Scholar] [CrossRef]
- Goldbourt, A. Structural Characterization of Bacteriophage Viruses by NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2019, 114–115, 192–210. [Google Scholar] [CrossRef]
- Lecoq, L.; Fogeron, M.-L.; Meier, B.H.; Nassal, M.; Böckmann, A. Solid-State NMR for Studying the Structure and Dynamics of Viral Assemblies. Viruses 2020, 12, 1069. [Google Scholar] [CrossRef] [PubMed]
- Porat-Dahlerbruch, G.; Goldbourt, A.; Polenova, T. Virus Structures and Dynamics by Magic-Angle Spinning NMR. Annu. Rev. Virol. 2021, 8, 219–237. [Google Scholar] [CrossRef] [PubMed]
- Quinn, C.M.; Lu, M.; Suiter, C.L.; Hou, G.; Zhang, H.; Polenova, T. Magic Angle Spinning NMR of Viruses. Prog. Nucl. Magn. Reson. Spectrosc. 2015, 86–87, 21–40. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hassid, R.R.; Kedem, S.; Bachar-Beck, M.; Shamir, Y.; Goldbourt, A. Solid State NMR Chemical Shift Assignment of the Non-Structural Single-Stranded DNA Binding Protein GVp from Fd Bacteriophage. Biomol. NMR Assign. 2022. [Google Scholar] [CrossRef] [PubMed]
- Marley, J.; Lu, M.; Bracken, C. A Method for Efficient Isotopic Labeling of Recombinant Proteins. J. Biomol. NMR 2001, 20, 71–75. [Google Scholar] [CrossRef]
- Day, L.A. Circular Dichroism and Ultraviolet Absorption of a Deoxyribonucleic Acid Binding Protein of Filamentous Bacteriophage. Biochemistry 1973, 12, 5329–5339. [Google Scholar] [CrossRef]
- Enshell-Seijffers, D.; Smelyanski, L.; Gershoni, J.M. The Rational Design of a “type 88” Genetically Stable Peptide Display Vector in the Filamentous Bacteriophage Fd. Nucleic Acids Res. 2001, 29, e50. [Google Scholar] [CrossRef]
- Abramov, G.; Morag, O.; Goldbourt, A. Magic-Angle Spinning NMR of a Class I Filamentous Bacteriophage Virus. J. Phys. Chem. B 2011, 115, 9671–9680. [Google Scholar] [CrossRef]
- Gullion, T.; Schaefer, J. Rotational-Echo Double-Resonance NMR. J. Magn. Reson. 1989, 81, 196–200. [Google Scholar] [CrossRef]
- Bak, M.; Rasmussen, J.T.; Nielsen, N.C. SIMPSON: A General Simulation Program for Solid-State NMR Spectroscopy. J. Magn. Reson. 2000, 147, 296–330. [Google Scholar] [CrossRef]
- Bennett, A.E.; Griffin, R.G.; Ok, J.H.; Vega, S. Chemical Shift Correlation Spectroscopy in Rotating Solids: Radio Frequency-Driven Dipolar Recoupling and Longitudinal Exchange. J. Chem. Phys. 1992, 96, 8624. [Google Scholar] [CrossRef]
- Hou, G.; Yan, S.; Trébosc, J.; Amoureux, J.-P.; Polenova, T. Broadband Homonuclear Correlation Spectroscopy Driven by Combined R2(n)(v) Sequences under Fast Magic Angle Spinning for NMR Structural Analysis of Organic and Biological Solids. J. Magn. Reson. 2013, 232, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Takegoshi, K.; Nakamura, S.; Terao, T. 13C-1H Dipolar-Assisted Rotational Resonance in Magic-Angle Spinning NMR. Chem. Phys. Lett. 2001, 344, 631–637. [Google Scholar] [CrossRef]
- Schaefer, J.; McKay, R.A.; Stejskal, E.O. Double-Cross-Polarization NMR of Solids. J. Magn. Reson. 1979, 34, 443–447. [Google Scholar] [CrossRef]
- Baldus, M.; Petkova, A.T.; Herzfeld, J.; Griffin, R.G. Cross Polarization in the Tilted Frame: Assignment and Spectral Simplification in Heteronuclear Spin Systems. Mol. Phys. 1998, 95, 1197–1207. [Google Scholar] [CrossRef]
- Igumenova, T.I.; Wand, A.J.; McDermott, A.E. Assignment of the Backbone Resonances for Microcrystalline Ubiquitin. J. Am. Chem. Soc. 2004, 126, 5323–5331. [Google Scholar] [CrossRef]
- Cadars, S.; Sein, J.; Duma, L.; Lesage, A.; Pham, T.N.; Baltisberger, J.H.; Brown, S.P.; Emsley, L. The Refocused INADEQUATE MAS NMR Experiment in Multiple Spin-Systems: Interpreting Observed Correlation Peaks and Optimising Lineshapes. J. Magn. Reson. 2007, 188, 24–34. [Google Scholar] [CrossRef]
- Shen, Y.; Bax, A. Protein Backbone and Sidechain Torsion Angles Predicted from NMR Chemical Shifts Using Artificial Neural Networks. J. Biomol. NMR 2013, 56, 227–241. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kedem, S.; Hassid, R.R.; Shamir, Y.; Goldbourt, A. Conformational Changes in Ff Phage Protein gVp upon Complexation with Its Viral Single-Stranded DNA Revealed Using Magic-Angle Spinning Solid-State NMR. Viruses 2022, 14, 1264. https://doi.org/10.3390/v14061264
Kedem S, Hassid RR, Shamir Y, Goldbourt A. Conformational Changes in Ff Phage Protein gVp upon Complexation with Its Viral Single-Stranded DNA Revealed Using Magic-Angle Spinning Solid-State NMR. Viruses. 2022; 14(6):1264. https://doi.org/10.3390/v14061264
Chicago/Turabian StyleKedem, Smadar, Roni Rene Hassid, Yoav Shamir, and Amir Goldbourt. 2022. "Conformational Changes in Ff Phage Protein gVp upon Complexation with Its Viral Single-Stranded DNA Revealed Using Magic-Angle Spinning Solid-State NMR" Viruses 14, no. 6: 1264. https://doi.org/10.3390/v14061264
APA StyleKedem, S., Hassid, R. R., Shamir, Y., & Goldbourt, A. (2022). Conformational Changes in Ff Phage Protein gVp upon Complexation with Its Viral Single-Stranded DNA Revealed Using Magic-Angle Spinning Solid-State NMR. Viruses, 14(6), 1264. https://doi.org/10.3390/v14061264





