Transcriptomic Analysis of MDBK Cells Infected with Cytopathic and Non-Cytopathic Strains of Bovine Viral Diarrhea Virus (BVDV)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viruses and Cells
2.2. Microarray Analysis
2.3. Statistical Analyses
2.4. Differential Expression Analysis
2.5. Quantitative Real-Time RT-PCR (RT-qPCR)
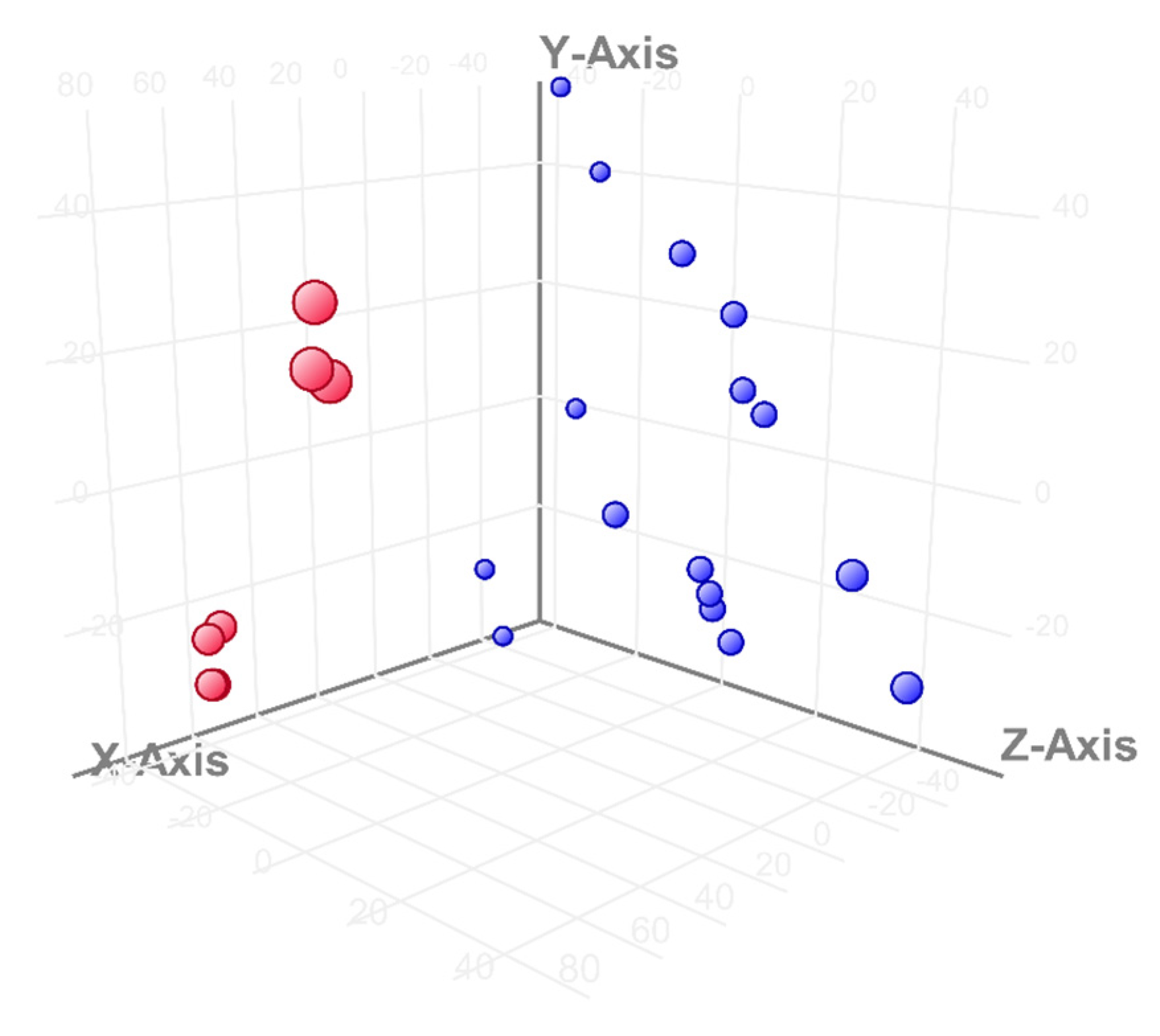
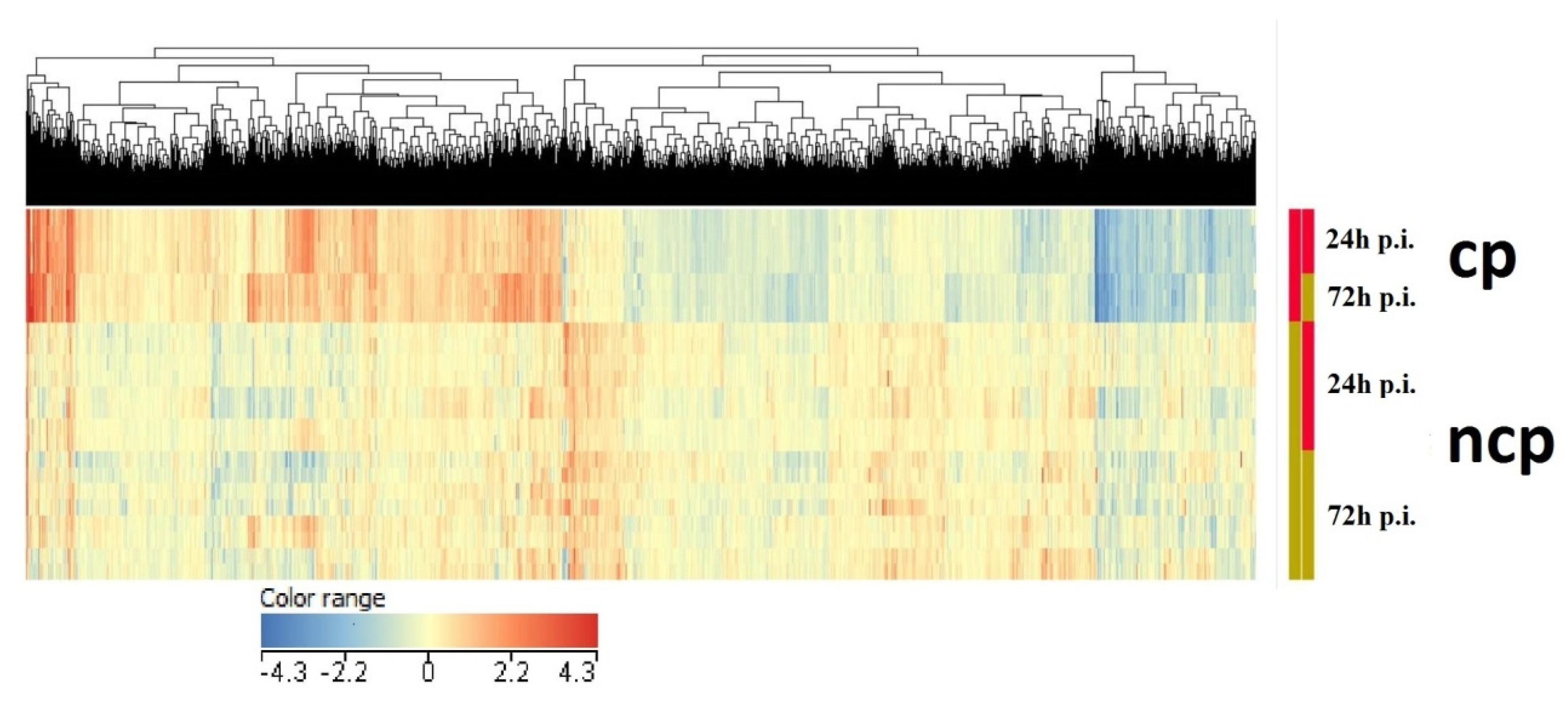
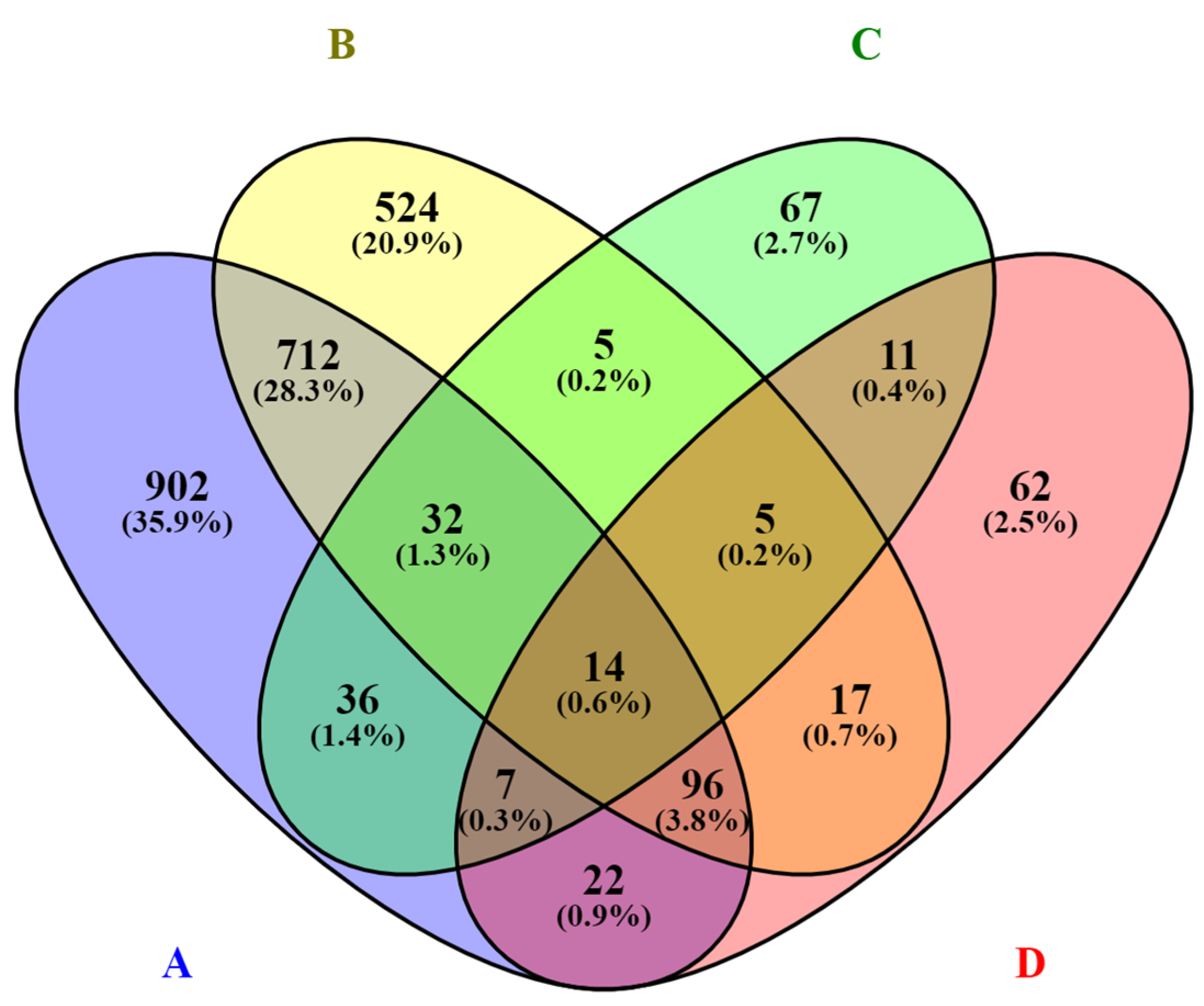
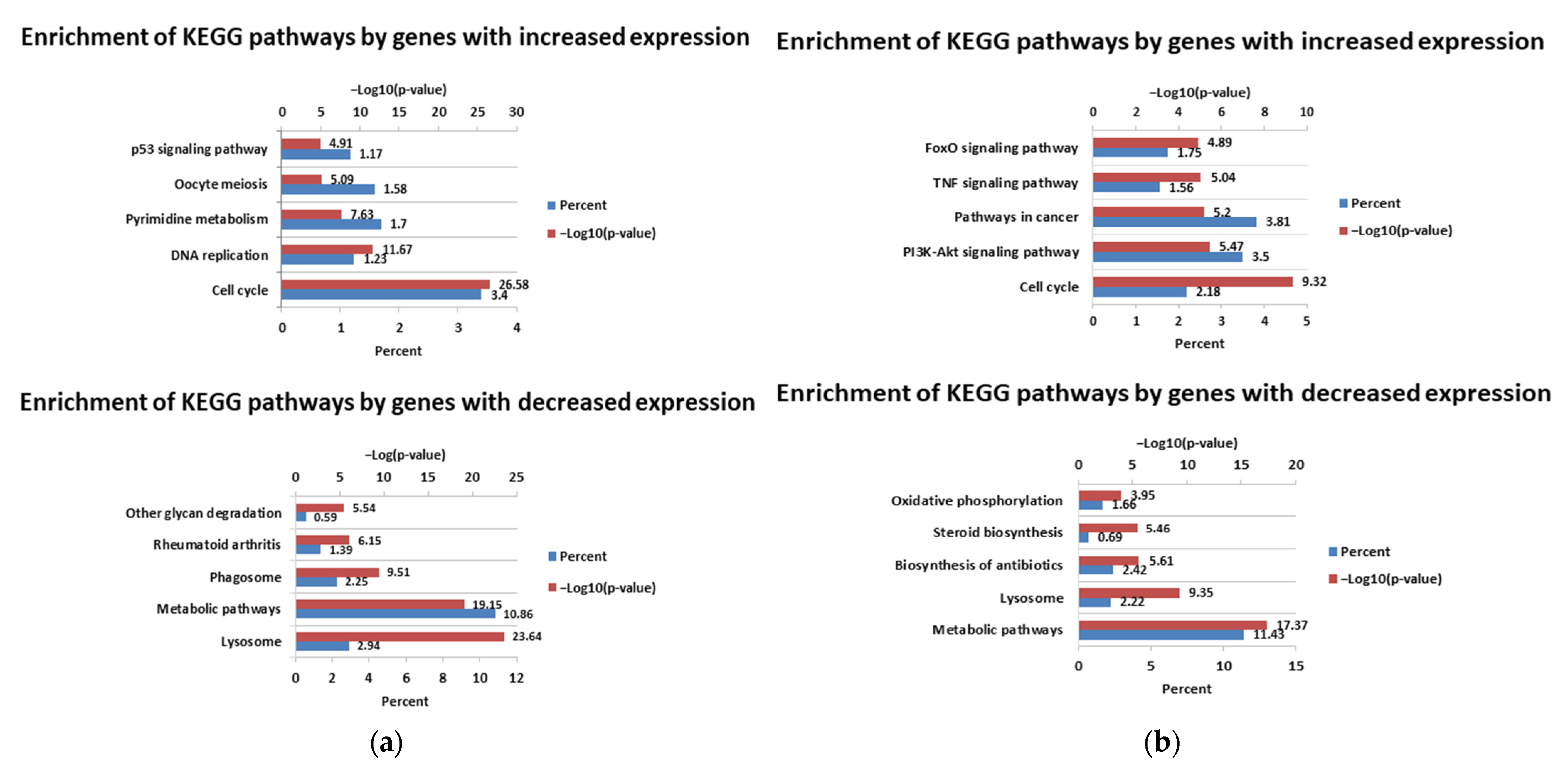
3. Results
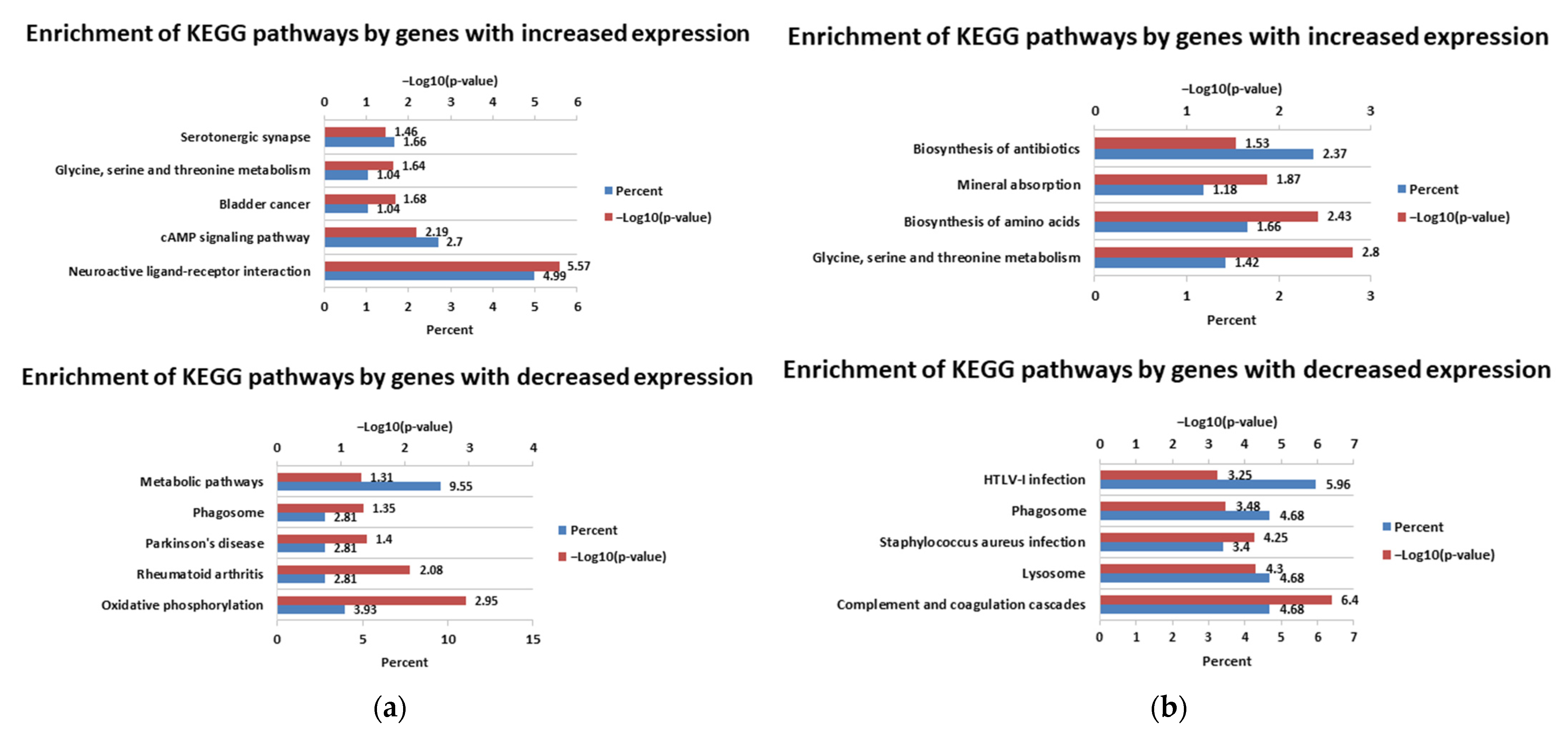
3.1. Differential Expression Analysis
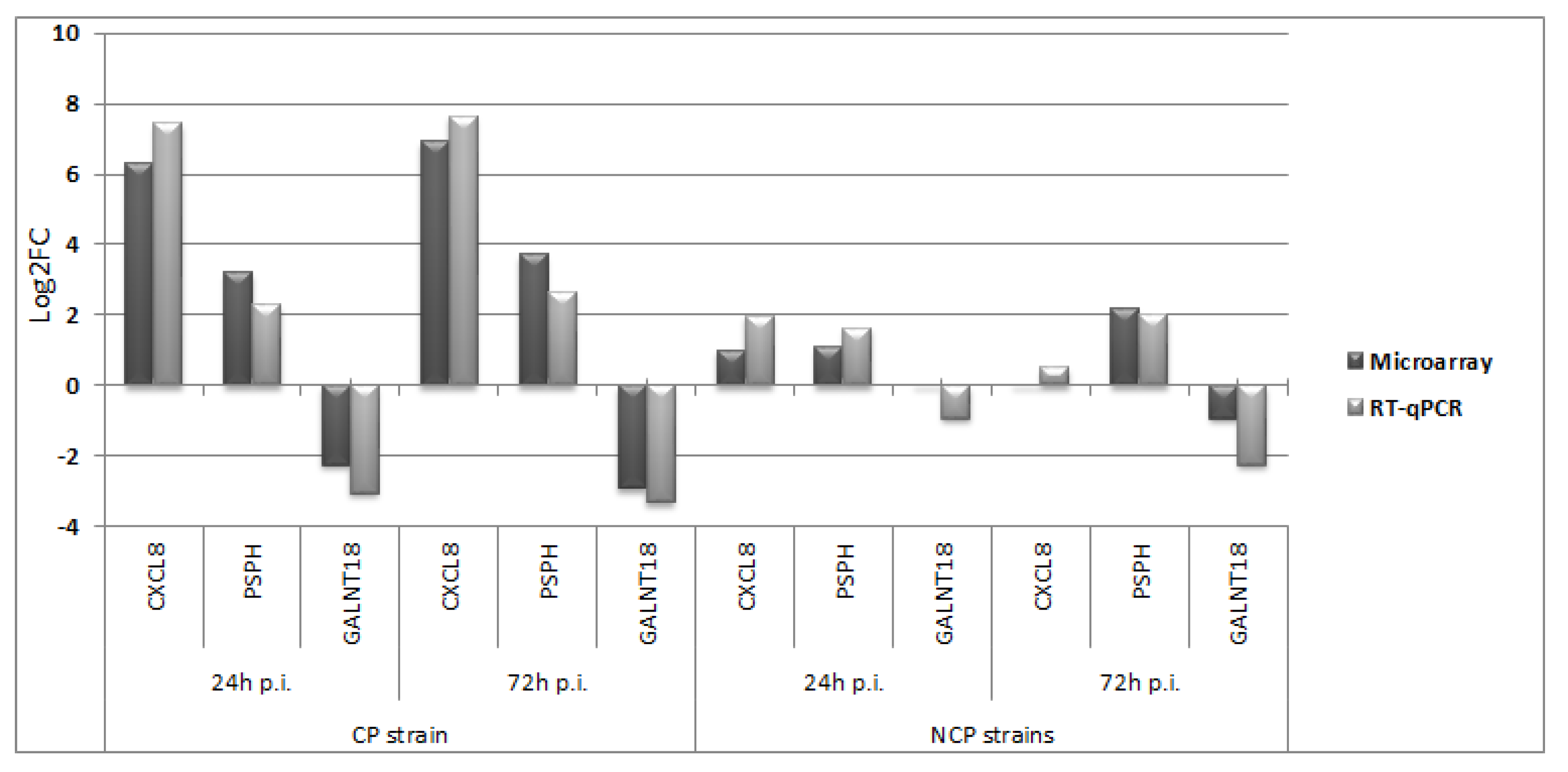
3.2. Quantitative Real-Time RT-PCR (RT-qPCR)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Houe, H. Economic impact of BVDV infection in dairies. Biologicals 2003, 31, 137–143. [Google Scholar] [CrossRef]
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Meyling, A.; Houe, H.; Jensen, A. Epidemiology of bovine virus diarrhoea virus. Rev. Sci. Technol. OIE 1990, 9, 75–93. [Google Scholar] [CrossRef] [PubMed]
- Reid, E.; Juleff, N.; Windsor, M.; Gubbins, S.; Roberts, L.; Morgan, S.; Meyers, G.; Perez-Martin, E.; Tchilian, E.; Charleston, B.; et al. Type I and III IFNs Produced by Plasmacytoid Dendritic Cells in Response to a Member of theFlaviviridaeSuppress Cellular Immune Responses. J. Immunol. 2016, 196, 4214–4226. [Google Scholar] [CrossRef]
- Ridpath, J.F. Immunology of BVDV vaccines. Biologicals 2013, 41, 14–19. [Google Scholar] [CrossRef]
- Tautz, N.; Meyers, G.; Thiel, H.-J. Pathogenesis of mucosal disease, a deadly disease of cattle caused by a pestivirus. Clin. Diagn. Virol. 1998, 10, 121–127. [Google Scholar] [CrossRef]
- Peterhans, E.; Schweizer, M. BVDV: A pestivirus inducing tolerance of the innate immune response. Biologicals 2013, 41, 39–51. [Google Scholar] [CrossRef]
- Bauhofer, O.; Summerfield, A.; Sakoda, Y.; Tratschin, J.-D.; Hofmann, M.A.; Ruggli, N. Classical Swine Fever Virus N pro Interacts with Interferon Regulatory Factor 3 and Induces Its Proteasomal Degradation. J. Virol. 2007, 81, 3087–3096. [Google Scholar] [CrossRef]
- Fiebach, A.R.; Guzylack-Piriou, L.; Python, S.; Summerfield, A.; Ruggli, N. Classical Swine Fever Virus N pro Limits Type I Interferon Induction in Plasmacytoid Dendritic Cells by Interacting with Interferon Regulatory Factor 7. J. Virol. 2011, 85, 8002–8011. [Google Scholar] [CrossRef]
- Johns, H.L.; Doceul, V.; Everett, H.; Crooke, H.; Charleston, B.; Seago, J. The classical swine fever virus N-terminal protease Npro binds to cellular HAX-1. J. Gen. Virol. 2010, 91, 2677–2686. [Google Scholar] [CrossRef]
- Luo, X.; Pan, R.; Wan, C.; Liu, X.; Wu, J.; Pan, Z. Glycosylation of classical swine fever virus Erns is essential for binding double-stranded RNA and preventing interferon-beta induction. Virus Res. 2009, 146, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Zahoor, M.A.; Yamane, D.; Mohamed, Y.M.; Suda, Y.; Kobayashi, K.; Kato, K.; Tohya, Y.; Akashi, H. Bovine viral diarrhea virus non-structural protein 5A interacts with NIK- and IKKβ-binding protein. J. Gen. Virol. 2010, 91, 1939–1948. [Google Scholar] [CrossRef] [PubMed]
- Brazma, A.; Hingamp, P.; Quackenbush, J.; Sherlock, G.; Spellman, P.; Stoeckert, C.; Aach, J.; Ansorge, W.; Ball, C.A.; Causton, H.C.; et al. Minimum information about a microarray experiment (MIAME)—Toward standards for microarray data. Nat. Genet. 2001, 29, 365–371. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Oliveros, J.C. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 6 January 2022).
- Elgendy, R.; Giantin, M.; Dacasto, M. Transcriptomic characterization of bovine primary cultured hepatocytes; a cross-comparison with a bovine liver and the Madin-Darby bovine kidney cells. Res. Vet. Sci. 2017, 113, 40–49. [Google Scholar] [CrossRef]
- Yamane, D.; Zahoor, M.A.; Mohamed, Y.M.; Azab, W.; Kato, K.; Tohya, Y.; Akashi, H. Microarray analysis reveals distinct signaling pathways transcriptionally activated by infection with bovine viral diarrhea virus in different cell types. Virus Res. 2009, 142, 188–199. [Google Scholar] [CrossRef]
- Liu, C.; Liu, Y.; Liang, L.; Cui, S.; Zhang, Y. RNA-Seq based transcriptome analysis during bovine viral diarrhoea virus (BVDV) infection. BMC Genom. 2019, 20, 1–18. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Mirosław, P.; Polak, M. Increased genetic variation of bovine viral diarrhea virus in dairy cattle in Poland. BMC Vet. Res. 2019, 15, 1–12. [Google Scholar] [CrossRef]
- Li, S.; Hu, X.; Tian, R.; Guo, Y.; Chen, J.; Li, Z.; Zhao, X.; Kuang, L.; Ran, D.; Zhao, H.; et al. RNA-Seq-based transcriptomic profiling of primary interstitial cells of Cajal in response to bovine viral diarrhea virus infection. Vet. Res. Commun. 2019, 43, 143–153. [Google Scholar] [CrossRef]
- Behura, S.K.; Tizioto, P.C.; Kim, J.; Grupioni, N.V.; Seabury, C.M.; Schnabel, R.D.; Gershwin, L.J.; Van Eenennaam, A.L.; Toaff-Rosenstein, R.; Neibergs, H.L.; et al. Tissue Tropism in Host Transcriptional Response to Members of the Bovine Respiratory Disease Complex. Sci. Rep. 2017, 7, 17938. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Mao, L.; Shu, X.; Liu, R.; Hao, F.; Li, J.; Liu, M.; Yang, L.; Zhang, W.; Sun, M.; et al. Transcriptome analysis reveals differential immune related genes expression in bovine viral diarrhea virus-2 infected goat peripheral blood mononuclear cells (PBMCs). BMC Genom. 2019, 20, 516. [Google Scholar] [CrossRef] [PubMed]
- Emmett, S.R.; Dove, B.; Mahoney, L.; Wurm, T.; Hiscox, J.A. The Cell Cycle and Virus Infection. In Cell Cycle Control; Humana Press: Totowa, NJ, USA, 2005; pp. 197–218. [Google Scholar]
- Davy, C.; Doorbar, J. G2/M cell cycle arrest in Athe life cycle of viruses. Virology 2007, 368, 219–226. [Google Scholar] [CrossRef]
- Ohtsubo, M.; Theodoras, A.M.; Schumacher, J.; Roberts, J.M.; Pagano, M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 1995, 15, 2612–2624. [Google Scholar] [CrossRef]
- Tizioto, P.C.; Kim, J.; Seabury, C.M.; Schnabel, R.D.; Gershwin, L.J.; Van Eenennaam, A.; Toaff-Rosenstein, R.; Neibergs, H.L.; Taylor, J.F. Bovine Respiratory Disease Complex Coordinated Agricultural Project Research Team Immunological Response to Single Pathogen Challenge with Agents of the Bovine Respiratory Disease Complex: An RNA-Sequence Analysis of the Bronchial Lymph Node Transcriptome. PLoS ONE 2015, 10, e0131459. [Google Scholar] [CrossRef]
- Zhu, L.; Ding, X.; Zhu, X.; Meng, S.; Wang, J.; Zhou, H.; Duan, Q.; Tao, J.; Schifferli, D.M.; Zhu, G. Biphasic activation of PI3K/Akt and MAPK/Erk1/2 signaling pathways in bovine herpesvirus type 1 infection of MDBK cells. Vet. Res. 2011, 42, 57. [Google Scholar] [CrossRef]
- Nguyen, H.; Mudryj, M.; Guadalupe, M.; Dandekar, S. Hepatitis C virus core protein expression leads to biphasic regulation of the p21 cdk inhibitor and modulation of hepatocyte cell cycle. Virology 2003, 312, 245–253. [Google Scholar] [CrossRef]
- Hilbe, M.; Girao, V.; Bachofen, C.; Schweizer, M.; Zlinszky, K.; Ehrensperger, F. Apoptosis in Bovine Viral Diarrhea Virus (BVDV)–Induced Mucosal Disease Lesions. Vet.-Pathol. 2012, 50, 46–55. [Google Scholar] [CrossRef]
- Warburg, O. The Metabolism of Carcinoma Cells. J. Cancer Res. 1925, 9, 148–163. [Google Scholar] [CrossRef]
- Ramière, C.; Rodriguez, J.; Enache, L.S.; Lotteau, V.; André, P.; Diaz, O. Activity of Hexokinase Is Increased by Its Interaction with Hepatitis C Virus Protein NS5A. J. Virol. 2014, 88, 3246–3254. [Google Scholar] [CrossRef]
- Gerresheim, G.K.; Bathke, J.; Michel, A.M.; Andreev, D.E.; Shalamova, L.A.; Rossbach, O.; Hu, P.; Glebe, D.; Fricke, M.; Marz, M.; et al. Cellular Gene Expression during Hepatitis C Virus Replication as Revealed by Ribosome Profiling. Int. J. Mol. Sci. 2019, 20, 1321. [Google Scholar] [CrossRef]
- Amelio, I.; Cutruzzolá, F.; Antonov, A.; Agostini, M.; Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci. 2014, 39, 191–198. [Google Scholar] [CrossRef]
- Jung, G.-S.; Jeon, J.-H.; Choi, Y.-K.; Jang, S.Y.; Park, S.Y.; Kim, S.-W.; Byun, J.-K.; Kim, M.-K.; Lee, S.; Shin, E.-C.; et al. Pyruvate dehydrogenase kinase regulates hepatitis C virus replication. Sci. Rep. 2016, 6, 30846. [Google Scholar] [CrossRef]
- Gaunt, E.R.; Cheung, W.; Richards, J.E.; Lever, A.; Desselberger, U. Inhibition of rotavirus replication by downregulation of fatty acid synthesis. J. Gen. Virol. 2013, 94, 1310–1317. [Google Scholar] [CrossRef]
- Heaton, N.S.; Perera, R.; Berger, K.L.; Khadka, S.; LaCount, D.J.; Kuhn, R.J.; Randall, G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 17345–17350. [Google Scholar] [CrossRef]
- York, A.G.; Williams, K.J.; Argus, J.P.; Zhou, Q.; Brar, G.; Vergnes, L.; Gray, E.E.; Zhen, A.; Wu, N.; Yamada, D.H.; et al. Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type I IFN Signaling. Cell 2015, 163, 1716–1729. [Google Scholar] [CrossRef]
- Li, Y.; Guo, T.; Wang, X.; Ni, W.; Hu, R.; Cui, Y.; Mi, T.; Hu, S. ITRAQ-based quantitative proteomics reveals the proteome profiles of MDBK cells infected with bovine viral diarrhea virus. Virol. J. 2021, 18, 1–13. [Google Scholar] [CrossRef]
- Wang, L.; Ou, J.-H.J. Regulation of Autophagy by Hepatitis C Virus for Its Replication. DNA Cell Biol. 2018, 37, 287–290. [Google Scholar] [CrossRef]
- Grummer, B.; Grotha, S.; Greiser-Wilke, I. Bovine Viral Diarrhoea Virus is Internalized by Clathrin-dependent Receptor-mediated Endocytosis. J. Vet. Med. Ser. B 2004, 51, 427–432. [Google Scholar] [CrossRef]
- Kim, Y.K.; Shin, J.-S.; Nahm, M.H. NOD-Like Receptors in Infection, Immunity, and Diseases. Yonsei Med. J. 2016, 57, 5–14. [Google Scholar] [CrossRef]
- Listen, P.; Roy, N.; Tamai, K.; Lefebvre, C.; Baird, S.; Cherton-Horvat, G.; Farahani, R.; McLean, M.; Lkeda, J.-E.; MacKenzie, A.; et al. Suppression of apoptosis in mammalian cells by NAIP and a related family of IAP genes. Nature 1996, 379, 349–353. [Google Scholar] [CrossRef]
- Prasad, A.S.; Bao, B.; Beck, F.W.; Sarkar, F.H. Zinc-suppressed inflammatory cytokines by induction of A20-mediated inhibition of nuclear factor-κB. Nutrition 2011, 27, 816–823. [Google Scholar] [CrossRef]
- Villalba, M.; Fredericksen, F.; Otth, C.; Olavarría, V. Transcriptomic analysis of responses to cytopathic bovine viral diarrhea virus-1 (BVDV-1) infection in MDBK cells. Mol. Immunol. 2016, 71, 192–202. [Google Scholar] [CrossRef]
- Lee, S.-R.; Pharr, G.; Boyd, B.; Pinchuk, L. Bovine viral diarrhea viruses modulate toll-like receptors, cytokines and co-stimulatory molecules genes expression in bovine peripheral blood monocytes. Comp. Immunol. Microbiol. Infect. Dis. 2008, 31, 403–418. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. Regulation of NF-κB by TNF family cytokines. Semin. Immunol. 2014, 26, 253–266. [Google Scholar] [CrossRef]
- Murdoch, C.; Finn, A. Chemokine receptors and their role in inflammation and infectious diseases. Blood 2000, 95, 3032–3043. [Google Scholar] [CrossRef]
- Ganheim, C.; Hulten, C.; Carlsson, U.; Kindahl, H.; Niskanen, R.; Waller, K.P. The Acute Phase Response in Calves Experimentally Infected with Bovine Viral Diarrhoea Virus and/or Mannheimia haemolytica. J. Vet. Med. Ser. B 2003, 50, 183–190. [Google Scholar] [CrossRef]
- Levy, D.E.; Marie, I.J.; Durbin, J.E. Induction and function of type I and III interferon in response to viral infection. Curr. Opin. Virol. 2011, 1, 476–486. [Google Scholar] [CrossRef]
- Ostachuk, A. Bovine viral diarrhea virus structural protein E2 as a complement regulatory protein. Arch. Virol. 2016, 161, 1769–1782. [Google Scholar] [CrossRef]
- Mawatari, S.; Uto, H.; Ido, A.; Nakashima, K.; Suzuki, T.; Kanmura, S.; Kumagai, K.; Oda, K.; Tabu, K.; Tamai, T.; et al. Hepatitis C Virus NS3/4A Protease Inhibits Complement Activation by Cleaving Complement Component 4. PLoS ONE 2013, 8, e82094. [Google Scholar] [CrossRef]
- Mazumdar, B.; Kim, H.; Meyer, K.; Bose, S.K.; Di Bisceglie, A.M.; Ray, R.B.; Ray, R. Hepatitis C Virus Proteins Inhibit C3 Complement Production. J. Virol. 2012, 86, 2221–2228. [Google Scholar] [CrossRef]
- Oldenburger, A.; Maarsingh, H.; Schmidt, M. Multiple Facets of cAMP Signalling and Physiological Impact: cAMP Compartmentalization in the Lung. Pharmaceuticals 2012, 5, 1291–1331. [Google Scholar] [CrossRef]
- Orr, C.L.; Hutcheson, D.P.; Grainger, R.B.; Cummins, J.M.; Mock, R.E. Serum copper, zinc, calcium and phosphorus concentrations of calves stressed by bovine respiratory disease and infectious bovine rhinotracheitis. J. Anim. Sci. 1990, 68, 2893–2900. [Google Scholar] [CrossRef]
- Yin, X.; Knecht, D.A.; Lynes, M.A. Metallothionein mediates leukocyte chemotaxis. BMC Immunol. 2005, 6, 21. [Google Scholar] [CrossRef]
- Ghoshal, K.; Majumder, S.; Zhu, Q.; Hunzeker, J.; Datta, J.; Shah, M.; Sheridan, J.F.; Jacob, S.T. Influenza Virus Infection Induces Metallothionein Gene Expression in the Mouse Liver and Lung by Overlapping but Distinct Molecular Mechanisms. Mol. Cell. Biol. 2001, 21, 8301–8317. [Google Scholar] [CrossRef]
- Cancellieri, M.; Bassetto, M.; Widjaja, I.; van Kuppeveld, F.; de Haan, C.A.; Brancale, A. In silico structure-based design and synthesis of novel anti-RSV compounds. Antivir. Res. 2015, 122, 46–50. [Google Scholar] [CrossRef]
- Read, S.A.; Parnell, G.; Booth, D.; Douglas, M.W.; George, J.; Ahlenstiel, G. The antiviral role of zinc and metallothioneins in hepatitis C infection. J. Viral Hepat. 2017, 25, 491–501. [Google Scholar] [CrossRef]
- Wilson, B.K.; Vazquez-Anon, M.; Step, D.L.; Moyer, K.D.; Haviland, C.L.; Maxwell, C.L.; O’Neill, C.F.; Gifford, C.A.; Krehbiel, C.R.; Richards, C.J. Effect of copper, manganese, and zinc supplementation on the performance, clinical signs, and mineral status of calves following exposure to bovine viral diarrhea virus type 1b and subsequent Mannheimia haemolytica infection1. J. Anim. Sci. 2016, 94, 1123–1140. [Google Scholar] [CrossRef]


| Gene | Primer | Sequence | Reference |
|---|---|---|---|
| B2M * | B2M-F | TCGTGGCCTTGGTCCTTCT | Elgendy et al., 2017 [16] |
| B2M-R | AATCTTTGGAGGACGCTGGAT | ||
| IL-8 | IL-8-F | TGAAGCTGCAGTTCTGTC | Yamane et al., 2009 [17] |
| IL-8-R | ATTTGGGGTGGAAAGGTG | ||
| PSPH | PSPH-F | TCAAGGCTGCCCTCACACA | Liu et al., 2019 [18] |
| PSPH-R | AGGAGCCTCTGCACCTGTTC | ||
| GALNT18 | GALNT18-F | GTGCCGCAACCTCTCGTT | |
| GALNT18-R | CACGGAGAGTGCTTCATTGACA |
| Enrichment of GO Terms by Genes with Changed Expression 24 h p.i. | Enrichment of GO Terms by Genes with Changed Expression 72 h p.i. | |||||
|---|---|---|---|---|---|---|
| Enrichment of GO terms by genes with increased expression | ||||||
| Category | Term | Count | p-value | Term | Count | p-value |
| biological process | cell division | 60 | 2.17 × 10−22 | cell division | 36 | 1.64 × 10−7 |
| mitotic nuclear division | 38 | 9.40 × 10−13 | regulation of cell proliferation | 35 | 2.58 × 10−7 | |
| cell cycle | 29 | 3.85 × 10−8 | apoptotic process | 43 | 4.11 × 10−7 | |
| cellular component | cytoplasm | 413 | 2.25 × 10−15 | cytoplasm | 369 | 1.23 × 10−9 |
| nucleus | 385 | 9.02 × 10−15 | nucleus | 332 | 2.19 × 10−7 | |
| spindle pole | 31 | 1.48 × 10−12 | spindle pole | 22 | 1.73 × 10−6 | |
| molecular function | ATP binding | 172 | 2.85 × 10−8 | microtubule binding | 20 | 9.85 × 10−6 |
| DNA binding | 101 | 1.75 × 10−7 | ATP binding | 146 | 1.86 × 10−4 | |
| histone binding | 16 | 5.94 × 10−6 | protein kinase binding | 19 | 1.97 × 10−4 | |
| Enrichment of GO terms by genes with decreased expression | ||||||
| biological process | sphingolipid biosynthetic process | 9 | 8.48 × 10−6 | metabolic process | 20 | 8.70 × 10−5 |
| antigen processing and presentation of peptide or polysaccharide antigen via MHC class II | 10 | 2.19 × 10−5 | ATP synthesis coupled electron transport | 5 | 5.12 × 10−4 | |
| protein glycosylation | 21 | 4.46 × 10−5 | response to oxidative stress | 14 | 8.06 × 10−4 | |
| cellular component | lysosomal membrane | 62 | 2.11 × 10−18 | extracellular exosome | 247 | 1.29 × 10−10 |
| extracellular exosome | 336 | 5.33 × 10−18 | lysosome | 33 | 2.92 × 10−8 | |
| lysosome | 44 | 1.47 × 10−11 | lysosomal membrane | 36 | 3.92 × 10−7 | |
| molecular function | monooxygenase activity | 12 | 1.38 × 10−4 | ion binding | 287 | 3.85 × 10−4 |
| heme binding | 26 | 2.01 × 10−4 | oxidoreductase activity, acting on NAD(P)H | 16 | 7.17 × 10−4 | |
| serine-type carboxypeptidase activity | 7 | 2.57 × 10−4 | metal ion binding | 272 | 7.26 × 10−4 | |
| Enrichment of GO Terms by Genes with Changed Expression 24 h p.i. | Enrichment of GO Terms by Genes with Changed Expression 72 h p.i. | |||||
|---|---|---|---|---|---|---|
| Enrichment of GO terms by genes with increased expression | ||||||
| Category | Term | Count | p-value | Term | Count | p-value |
| biological process | G-protein coupled receptor signaling pathway | 40 | 2.43 × 10−5 | L-serine biosynthetic process | 3 | 0.001510151 |
| sensory perception of smell | 16 | 2.13 × 10−4 | reverse cholesterol transport | 4 | 0.001657372 | |
| cell maturation | 6 | 0.001282379 | cholesterol efflux | 5 | 0.001914924 | |
| cellular component | integral component of membrane | 168 | 1.20 × 10−10 | neuron projection | 10 | 0.002921923 |
| integral component of plasma membrane | 50 | 4.05 × 10−8 | integral component of plasma membrane | 29 | 0.021268826 | |
| plasma membrane | 93 | 4.63 × 10−6 | adherens junction | 4 | 0.032851986 | |
| molecular function | G-protein coupled receptor activity | 47 | 2.61 × 10−5 | cholesterol transporter activity | 5 | 1.07 × 10−04 |
| olfactory receptor activity | 35 | 0.002688453 | hormone activity | 5 | 0.038843617 | |
| chemokine activity | 5 | 0.014352821 | sterol-transporting ATPase activity | 2 | 0.044400249 | |
| Enrichment of GO terms by genes with decreased expression | ||||||
| biological process | positive regulation of interferon-gamma production | 4 | 0.005107499 | proteolysis | 10 | 0.001012604 |
| - | 10 | 2.19 × 10−5 | immune response | 11 | 0.001159766 | |
| - | 21 | 4.46 × 10−5 | pulmonary artery morphogenesis | 3 | 0.002023761 | |
| cellular component | respiratory chain | 4 | 3.32 × 10−4 | extracellular exosome | 66 | 5.00 × 10−10 |
| extracellular exosome | 29 | 0.042136161 | extracellular space | 34 | 1.01 × 10−6 | |
| - | 44 | 1.47 × 10−11 | lysosome | 10 | 1.58 × 10−4 | |
| molecular function | NADH dehydrogenase (ubiquinone) activity | 5 | 2.09 × 10−4 | serine-type endopeptidase activity | 11 | 8.04 × 10−5 |
| GTPase activator activity | 6 | 0.024894456 | serine-type carboxypeptidase activity | 4 | 4.69 × 10−4 | |
| growth factor activity | 4 | 0.027131177 | calcium ion binding | 20 | 7.32 × 10−4 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mirosław, P.; Rola-Łuszczak, M.; Kuźmak, J.; Polak, M.P. Transcriptomic Analysis of MDBK Cells Infected with Cytopathic and Non-Cytopathic Strains of Bovine Viral Diarrhea Virus (BVDV). Viruses 2022, 14, 1276. https://doi.org/10.3390/v14061276
Mirosław P, Rola-Łuszczak M, Kuźmak J, Polak MP. Transcriptomic Analysis of MDBK Cells Infected with Cytopathic and Non-Cytopathic Strains of Bovine Viral Diarrhea Virus (BVDV). Viruses. 2022; 14(6):1276. https://doi.org/10.3390/v14061276
Chicago/Turabian StyleMirosław, Paweł, Marzena Rola-Łuszczak, Jacek Kuźmak, and Mirosław P. Polak. 2022. "Transcriptomic Analysis of MDBK Cells Infected with Cytopathic and Non-Cytopathic Strains of Bovine Viral Diarrhea Virus (BVDV)" Viruses 14, no. 6: 1276. https://doi.org/10.3390/v14061276
APA StyleMirosław, P., Rola-Łuszczak, M., Kuźmak, J., & Polak, M. P. (2022). Transcriptomic Analysis of MDBK Cells Infected with Cytopathic and Non-Cytopathic Strains of Bovine Viral Diarrhea Virus (BVDV). Viruses, 14(6), 1276. https://doi.org/10.3390/v14061276






