Research Progress on Emerging Viral Pathogens of Small Ruminants in China during the Last Decade
Abstract
:1. Introduction
2. Peste des Petits Ruminants Virus (PPRV)
2.1. Etiology and Epidemiology
2.2. Virus–Host Interaction
2.2.1. Interaction of PPRV and IFN-Related Pathways
2.2.2. PPRV Infection and Autophagy
2.2.3. Additional Virus–Host Interactions
2.3. Diagnosis
2.3.1. Etiological Detection Methods
2.3.2. Serological Detection Methods
2.4. PPRV Vaccines
2.4.1. Live Attenuated Vaccines
2.4.2. Vector Vaccines
2.4.3. Virus-like Particle Vaccines
2.5. Prevention and Control of PPR
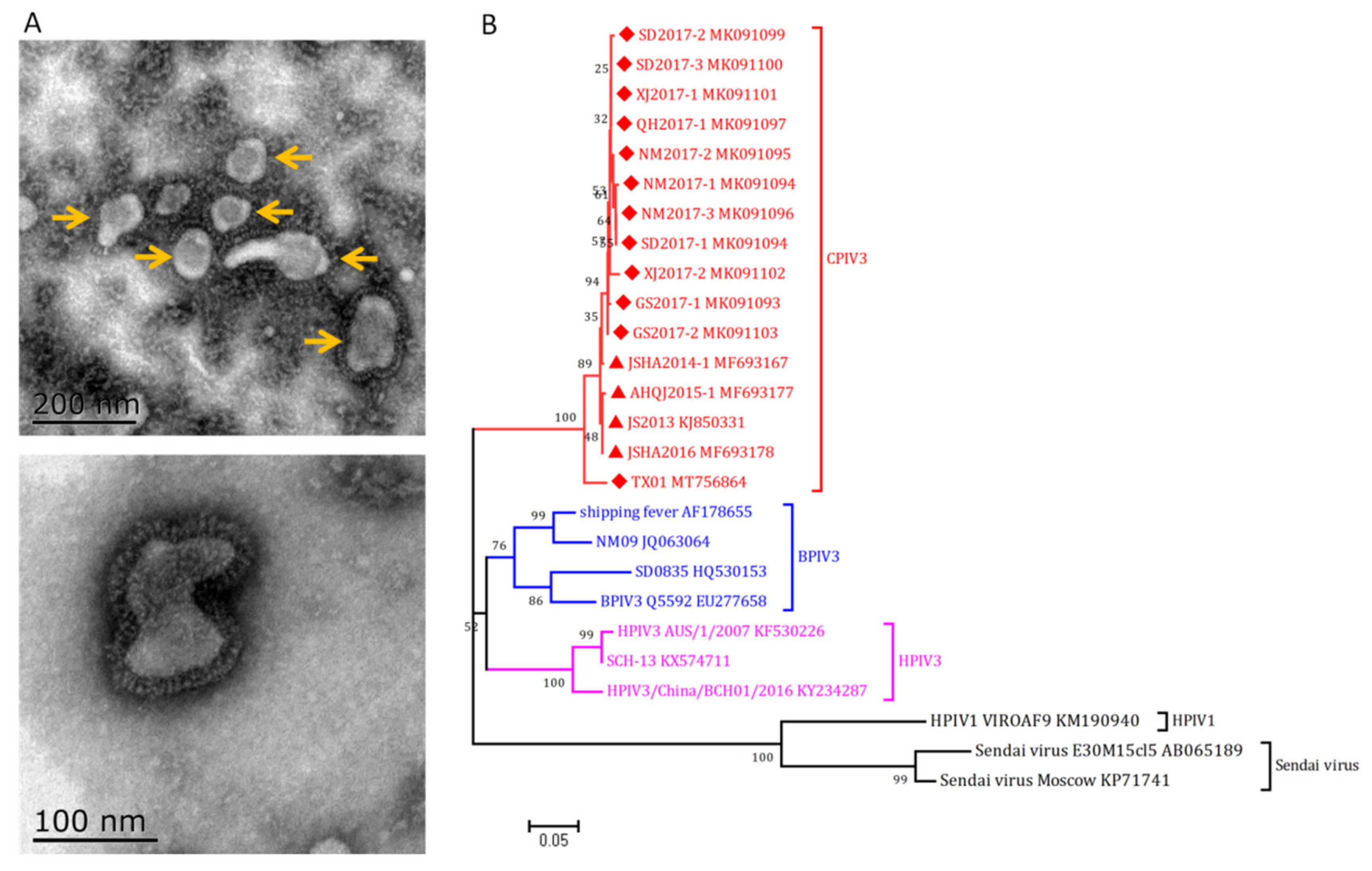
3. Caprine Parainfluenza Virus Type 3 (CPIV3)
3.1. Etiology and Pathogenesis
3.2. Epidemiology
3.3. Virus–Host Interaction
3.4. Diagnosis
3.4.1. Virus Isolation and Identification
3.4.2. Molecular Tests
3.4.3. Serological Tests
3.5. Prevention and Control
4. Border Disease Virus (BDV)
4.1. Etiology and Genotyping
4.2. Epidemiology and Cross-Species Transmission
4.3. Diagnosis
4.4. Prevention and Control
5. Other Emerging Viral Pathogens
5.1. Enzootic asal Tumor Virus (ENTV)
5.1.1. Etiology
5.1.2. Epidemiology
5.1.3. Diagnosis
5.2. Caprine Herpesvirus 1 (CpHV-1)
5.3. Enterovirus
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, F.; Li, J.; Li, L.; Liu, Y.; Wu, X.; Wang, Z. Peste des petits ruminants in China since its first outbreak in 2007: A 10-year review. Transbound. Emerg. Dis. 2018, 65, 638–648. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Mao, L.; Cheng, S.; Wang, Q.; Huang, J.; Deng, J.; Wang, Z.; Zhang, W.; Yang, L.; Hao, F.; et al. A novel parainfluenza virus type 3 (PIV3) identified from goat herds with respiratory diseases in eastern China. Vet. Microbiol. 2014, 174, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Liu, X.; Li, W.; Yang, L.; Zhang, W.; Jiang, J. Characterization of one sheep border disease virus in China. Virol. J. 2015, 12, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Mao, L.; Zhao, Y.; Sun, Y.; He, K.; Jiang, J. Detection of border disease virus (BDV) in goat herds suffering diarrhea in eastern China. Virol. J. 2013, 10, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, F.; Mao, L.; Li, W.; Li, J.; Yang, L.; Zhang, W.; Jiang, J.; Sun, M.; Xie, X.; Liu, M. Epidemiological investigation and genomic characterization of Caprine herpesvirus 1 from goats in China. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2020, 79, 104168. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Xing, Z.; Gai, X.; Li, S.; San, Z.; Wang, X. Identification of a novel enterovirus E isolates HY12 from cattle with severe respiratory and enteric diseases. PLoS ONE 2014, 9, e97730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, G.; Kaiyu, W.; Qigui, Y.; Zhongqiong, Y.; Yingdong, Y.; Defang, C.; Jinlu, H. Descriptive study of enzootic nasal adenocarcinoma in goats in southwestern China. Transbound. Emerg. Dis. 2010, 57, 197–200. [Google Scholar] [CrossRef]
- Adams, M.J.; Lefkowitz, E.J.; King, A.M.Q.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch. Virol. 2017, 162, 2505–2538. [Google Scholar] [CrossRef]
- FAO; OIE. Global Control and Eradication of Peste Des Petits Ruminants; FAO: Rome, Italy, 2015. [Google Scholar]
- Wang, Z.L.; Bao, J.Y.; Wu, X.D.; Liu, Y.T.; Li, L.; Liu, P.L.; Zhao, Y.G.; Liu, C.J.; Xiao, X. Diagnosis of the first outbreak of peste des petits ruminants in Tibet. China Anim. Health Insp. 2007, 24, 24–26. (In Chinese) [Google Scholar]
- Wang, Z.; Bao, J.; Wu, X.; Liu, Y.; Li, L.; Liu, C.; Suo, L.; Xie, Z.; Zhao, W.; Zhang, W.; et al. Peste des petits ruminants virus in Tibet, China. Emerg. Infect. Dis. 2009, 15, 299–301. [Google Scholar] [CrossRef]
- Xia, J.; Zheng, X.G.; Adili, G.Z.; Wei, Y.R.; Ma, W.G.; Xue, X.M.; Mi, X.Y.; Yi, Z.; Chen, S.J.; Du, W.; et al. Sequence analysis of peste des petits ruminants virus from ibexes in Xinjiang, China. Genet. Mol. Res. GMR 2016, 15, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Wang, Q.; Li, L.; Liu, C.; Zhang, Z.; Li, J.; Wang, S.; Wu, X.; Wang, Z. Evolutionary dynamics of recent peste des petits ruminants virus epidemic in China during 2013–2014. Virology 2017, 510, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Bao, J.; Wang, Q.; Zhang, Y.; Liu, C.; Li, L.; Wang, Z. Complete Genome Sequence of a Novel Variant Strain of Peste des Petits Ruminants Virus, China/XJYL/2013. Genome Announc. 2014, 2, e00762-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Li, L.; Wu, X.; Liu, F.; Zou, Y.; Wang, Q.; Liu, C.; Bao, J.; Wang, W.; Ma, W.; et al. Diagnosis of Peste des Petits Ruminants in Wild and Domestic Animals in Xinjiang, China, 2013-2016. Transbound. Emerg. Dis. 2017, 64, e43–e47. [Google Scholar] [CrossRef]
- Zhu, Z.; Zhang, X.; Adili, G.; Huang, J.; Du, X.; Zhang, X.; Li, P.; Zheng, X.; Liu, X.; Zheng, H.; et al. Genetic Characterization of a Novel Mutant of Peste Des Petits Ruminants Virus Isolated from Capra ibex in China during 2015. BioMed Res. Int. 2016, 2016, 7632769. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Cao, X.; Wu, J.; Dou, Y.; Meng, X.; Liu, D.; Liu, Y.; Shang, Y.; Liu, X. Epidemic and evolutionary characteristics of peste des petits ruminants virus infecting Procapra przewalskii in Western China. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019, 75, 104004. [Google Scholar] [CrossRef]
- Ying, L.; Yang, Y.Q.; Li, L.; Zhao, Q.B.; Wang, X.Z.; La, H.; Liu, F.X.; Tan, S.K.; Song, L.Z.; Wu, X.D.; et al. Diagnosis and control of the first peste des petits ruminants infecting wild Pseudois nayaur in the Qinghai plateau area. Anim. Husb. Vet. Med. 2019, 51, 112–117. (In Chinese) [Google Scholar]
- Ma, J.; Gao, X.; Liu, B.; Chen, H.; Xiao, J.; Wang, H. Peste des petits ruminants in China: Spatial risk analysis. Transbound. Emerg. Dis. 2019, 66, 1784–1788. [Google Scholar] [CrossRef]
- Miao, Q.; Qi, R.; Meng, C.; Zhu, J.; Tang, A.; Dong, D.; Guo, H.; van Oers, M.M.; Pijlman, G.P.; Liu, G. Caprine MAVS Is a RIG-I Interacting Type I Interferon Inducer Downregulated by Peste des Petits Ruminants Virus Infection. Viruses 2021, 13, 409. [Google Scholar] [CrossRef]
- Ma, P.; Li, L.; Jin, L.; Zhang, D.; Cao, X.; Guo, F.; Zhao, Y.; Bai, J.; Ma, Z.; Shang, Y.; et al. Antiviral responses of ATG13 to the infection of peste des petits ruminants virus through activation of interferon response. Gene 2020, 754, 144858. [Google Scholar] [CrossRef]
- Chen, S.; Yang, F.; Cao, W.; Liu, H.; Wen, B.; Sun, Y.; Zheng, H.; Wang, J.; Zhu, Z. Quantitative Proteomics Reveals a Novel Role of the E3 Ubiquitin-Protein Ligase FANCL in the Activation of the Innate Immune Response through Regulation of TBK1 Phosphorylation during Peste des Petits Ruminants Virus Infection. J. Proteome Res. 2021, 20, 4113–4130. [Google Scholar] [CrossRef]
- Li, P.; Zhu, Z.; Zhang, X.; Dang, W.; Li, L.; Du, X.; Zhang, M.; Wu, C.; Xue, Q.; Liu, X.; et al. The Nucleoprotein and Phosphoprotein of Peste des Petits Ruminants Virus Inhibit Interferons Signaling by Blocking the JAK-STAT Pathway. Viruses 2019, 11, 629. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Z.; Li, P.; Yang, F.; Cao, W.; Zhang, X.; Dang, W.; Ma, X.; Tian, H.; Zhang, K.; Zhang, M.; et al. Peste des Petits Ruminants Virus Nucleocapsid Protein Inhibits Beta Interferon Production by Interacting with IRF3 To Block Its Activation. J. Virol. 2019, 93, e00362-19. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Zhu, Z.; Cao, W.; Yang, F.; Ma, X.; Tian, H.; Zhang, K.; Liu, X.; Zheng, H. Dysregulation of the RIG-I-like Receptor Pathway Signaling by Peste des Petits Ruminants Virus Phosphoprotein. J. Immunol. 2021, 206, 566–579. [Google Scholar] [CrossRef]
- Li, L.; Shi, X.; Ma, X.; Cao, X.; Ali, A.; Bai, J. Peste des petits ruminants virus non-structural C protein inhibits the induction of interferon-beta by potentially interacting with MAVS and RIG-I. Virus Genes 2021, 57, 60–71. [Google Scholar] [CrossRef]
- Li, H.; Xue, Q.; Wan, Y.; Chen, Y.; Zeng, W.; Wei, S.; Zhang, Y.; Wang, J.; Qi, X. PPRV-induced novel miR-3 contributes to inhibit type I IFN production by targeting IRAK1. J. Virol. 2021, 95, e02045-20. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, S.; Lv, J.; Feng, C.; Deng, J.; Wang, C.; Yuan, X.; Zhang, T.; Lin, X. Peste des petits ruminants virus exploits cellular autophagy machinery for replication. Virology 2013, 437, 28–38. [Google Scholar] [CrossRef]
- Yang, B.; Xue, Q.; Guo, J.; Wang, X.; Zhang, Y.; Guo, K.; Li, W.; Chen, S.; Xue, T.; Qi, X.; et al. Autophagy induction by the pathogen receptor NECTIN4 and sustained autophagy contribute to peste des petits ruminants virus infectivity. Autophagy 2020, 16, 842–861. [Google Scholar] [CrossRef]
- Yang, B.; Xue, Q.; Qi, X.; Wang, X.; Jia, P.; Chen, S.; Wang, T.; Xue, T.; Wang, J. Autophagy enhances the replication of Peste des petits ruminants virus and inhibits caspase-dependent apoptosis in vitro. Virulence 2018, 9, 1176–1194. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Chen, Y.; Wang, T.; Zhao, B.; Zeng, W.; Zhang, L.; Zhang, Y.; Cao, S.; Wang, J.; Xue, Q.; et al. PPRV-Induced Autophagy Facilitates Infectious Virus Transmission by the Exosomal Pathway. J. Virol. 2022, 96, e0024422. [Google Scholar] [CrossRef]
- Xue, Q.; Liu, H.; Sun, M.; Zhao, W.; Chen, Y.; Chen, J.; Wei, C.; Cai, X.; Xue, Q. Peste des petits ruminants virus hemagglutinin (H) induces lysosomal degradation of host cyclophilin A to facilitate viral replication. Virus Res. 2020, 277, 197844. [Google Scholar] [CrossRef]
- Li, L.; Yang, W.; Ma, X.; Wu, J.; Qin, X.; Cao, X.; Zhou, J.; Jin, L.; He, J.; Zheng, H.; et al. Peste Des Petits Ruminants Virus N Protein Is a Critical Proinflammation Factor That Promotes MyD88 and NLRP3 Complex Assembly. J. Virol. 2022, 96, e00309-22. [Google Scholar] [CrossRef]
- Qi, X.; Li, Z.; Li, H.; Wang, T.; Zhang, Y.; Wang, J. MicroRNA-1 Negatively Regulates Peripheral NK Cell Function via Tumor Necrosis Factor-Like Weak Inducer of Apoptosis (TWEAK) Signaling Pathways During PPRV Infection. Front. Immunol. 2019, 10, 3066. [Google Scholar] [CrossRef]
- Kinimi, E.; Odongo, S.; Muyldermans, S.; Kock, R.; Misinzo, G. Paradigm shift in the diagnosis of peste des petits ruminants: Scoping review. Acta Vet. Scand. 2020, 62, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wu, X.; Liu, F.; Wang, Z.; Liu, C.; Wang, Q.; Bao, J. Rapid detection of lineage IV peste des petits ruminants virus by real-time RT-PCR. J. Virol. Methods 2016, 235, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Bao, J.; Wu, X.; Wang, Z.; Wang, J.; Gong, M.; Liu, C.; Li, J. Rapid detection of peste des petits ruminants virus by a reverse transcription loop-mediated isothermal amplification assay. J. Virol. Methods 2010, 170, 37–41. [Google Scholar] [CrossRef]
- Yang, Y.; Qin, X.; Song, Y.; Zhang, W.; Hu, G.; Dou, Y.; Li, Y.; Zhang, Z. Development of real-time and lateral flow strip reverse transcription recombinase polymerase Amplification assays for rapid detection of peste des petits ruminants virus. Virol. J. 2017, 14, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Li, L.; Fan, X.; Zou, Y.; Zhang, Y.; Wang, Q.; Sun, C.; Pan, S.; Wu, X.; Wang, Z. Development of real-time reverse transcription recombinase polymerase amplification (RPA) for rapid detection of peste des petits ruminants virus in clinical samples and its comparison with real-time PCR test. Sci. Rep. 2018, 8, 17760. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, J.; Zhang, Z.; Mei, L.; Wang, J.; Wu, S.; Lin, X. Development of recombinase polymerase amplification assays for the rapid detection of peste des petits ruminants virus. J. Virol. Methods 2018, 254, 35–39. [Google Scholar] [CrossRef]
- Yang, M.F.; Ma, Y.Y.; Yu, Z.Y.; Liu, J.S.; Kang, H.T.; Jiang, Q.; Qu, L.D. Development and application of a chip digital PCR assay for PPRV. China Anim. Health Insp. 2021, 38, 91–98. (In Chinese) [Google Scholar]
- Prajapati, M.; Dou, Y.; Zhu, X.; Zhao, S.; Alfred, N.; Li, Y.; Zhang, Z. Development of an Enzyme-Linked Immunosorbent Assay Based on CD150/SLAM for the Detection of Peste des Petits Ruminant Virus. Front. Vet. Sci. 2020, 7, 196. [Google Scholar] [CrossRef]
- Ruan, Z.Y.; Hu, B.; Song, Y.H.; Wei, H.J.; Fan, Z.Y.; Wu, X.D.; Wang, F. Establishment and preliminary application of a simplex suspension array for detection of the peste des petits ruminants virus. Anim. Husb. Vet. Med. 2018, 50, 69–73. (In Chinese) [Google Scholar]
- Sun, Y.; Song, X.; Xiao, Y.; Li, X.; Lv, Y.; Wang, R.; Jiang, F.; Sun, H.; Yang, L.; Wang, C. A comparative study on double-antigen S-ELISA for the detections of antibodies based on different recombinant N protein of peste des petits ruminants virus. Chin. Vet. Sci. 2019, 49, 1484–1491. (In Chinese) [Google Scholar]
- Qian, B.; Li, Y.M.; Zhu, X.L.; Y, Z.X.; Niyokwishimira, A.; Dou, Y.X.; Zhang, Z.D. Establishment of an iELISA method for detection of antibody against Peste des petits ruminants virus (PPRV) based on H protein epitope peptide. Acta Vet. Et Zootech. Sin. 2021, 52, 144–153. (In Chinese) [Google Scholar]
- Zhang, G.R.; Yu, R.S.; Zeng, J.Y.; Zhu, Y.M.; Dong, S.J.; Dunzhu, L.; Zhu, S.; Duoji, C.; Lei, Z.H.; Li, Z. Development of an epitope-based competitive ELISA for the detection of antibodies against Tibetan peste des petits ruminants virus. Intervirology 2013, 56, 55–59. [Google Scholar] [CrossRef]
- Cheng, S.; Sun, J.; Yang, J.; Lv, J.; Wu, F.; Lin, Y.; Liao, L.; Ye, Y.; Cao, C.; Fang, L.; et al. A new immunoassay of serum antibodies against Peste des petits ruminants virus using quantum dots and a lateral-flow test strip. Anal. Bioanal. Chem. 2017, 409, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Song, X.H.; Xiao, Y.; Wang, R.N.; Li, X.M.; Ren, X.J.; Li, X.G.; Wei, W.; Yang, L.; Wang, C.B. Establishment of chemiluminescent immunoassay for detection of antibody against peste des petits ruminant virus. Anim. Husb. Vet. Med. 2020, 52, 109–114. (In Chinese) [Google Scholar]
- Hodgson, S.; Moffat, K.; Hill, H.; Flannery, J.T.; Graham, S.P.; Baron, M.D.; Darpel, K.E. Comparison of the Immunogenicities and Cross-Lineage Efficacies of Live Attenuated Peste des Petits Ruminants Virus Vaccines PPRV/Nigeria/75/1 and PPRV/Sungri/96. J. Virol. 2018, 92, e01471-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, F.; Li, W.L.; Mao, L.; Li, J.Z.; Yang, L.L.; Zhang, W.W.; Sun, M.; Liu, M.J.; Jiang, J.Y. Influence of PPR maternal antibody and vaccination of goat pox vaccine and FMD vaccine on efficiency of PPR vaccine. Chin. J. Vet. Sci. 2018, 38, 2306–2310. (In Chinese) [Google Scholar]
- Chen, W.; Hu, S.; Qu, L.; Hu, Q.; Zhang, Q.; Zhi, H.; Huang, K.; Bu, Z. A goat poxvirus-vectored peste-des-petits-ruminants vaccine induces long-lasting neutralization antibody to high levels in goats and sheep. Vaccine 2010, 28, 4742–4750. [Google Scholar] [CrossRef]
- Hu, Q.; Chen, W.; Huang, K.; Baron, M.D.; Bu, Z. Rescue of recombinant peste des petits ruminants virus: Creation of a GFP-expressing virus and application in rapid virus neutralization test. Vet. Res. 2012, 43, 48. [Google Scholar] [CrossRef] [Green Version]
- Yin, C.; Chen, W.; Hu, Q.; Wen, Z.; Wang, X.; Ge, J.; Yin, Q.; Zhi, H.; Xia, C.; Bu, Z. Induction of protective immune response against both PPRV and FMDV by a novel recombinant PPRV expressing FMDV VP1. Vet. Res. 2014, 45, 62. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Liu, G.; Shi, L.; Li, W.; Li, C.; Chen, Z.; Jin, H.; Xu, B.; Li, G. Immune responses in mice vaccinated with a suicidal DNA vaccine expressing the hemagglutinin glycoprotein from the peste des petits ruminants virus. J. Virol. Methods 2013, 193, 525–530. [Google Scholar] [CrossRef]
- Li, W.; Jin, H.; Sui, X.; Zhao, Z.; Yang, C.; Wang, W.; Li, J.; Li, G. Self-assembly and release of peste des petits ruminants virus-like particles in an insect cell-baculovirus system and their immunogenicity in mice and goats. PLoS ONE 2014, 9, e104791. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.; Banadyga, L.; Zhao, Y.; Zhao, Z.; Schiffman, Z.; Huang, P.; Li, E.; Wang, C.; Gao, Y.; Feng, N.; et al. Peste des Petits Ruminants Virus-Like Particles Induce a Potent Humoral and Cellular Immune Response in Goats. Viruses 2019, 11, 918. [Google Scholar] [CrossRef] [Green Version]
- Yan, F.; Li, E.; Li, L.; Schiffman, Z.; Huang, P.; Zhang, S.; Li, G.; Jin, H.; Wang, H.; Zhang, X.; et al. Virus-Like Particles Derived From a Virulent Strain of Pest des Petits Ruminants Virus Elicit a More Vigorous Immune Response in Mice and Small Ruminants Than Those From a Vaccine Strain. Front. Microbiol. 2020, 11, 609. [Google Scholar] [CrossRef]
- Liu, F.; Wu, X.; Zou, Y.; Li, L.; Wang, Z. Peste des petits ruminants virus-like particles induce both complete virus-specific antibodies and virus neutralizing antibodies in mice. J. Virol. Methods 2015, 213, 45–49. [Google Scholar] [CrossRef]
- Njeumi, F.; Bailey, D.; Soula, J.J.; Diop, B.; Tekola, B.G. Eradicating the Scourge of Peste Des Petits Ruminants from the World. Viruses 2020, 12, 313. [Google Scholar] [CrossRef] [Green Version]
- Wu, H.; ManChu, R.; Wang, F.; Li, C.; Jin, N.; Chen, Q. Epidimiological situation of research progress on molecular detection of peste des petits ruminants virus. Prog. Vet. Med. 2022, 43, 122–126. (In Chinese) [Google Scholar]
- Yang, L.; Li, W.; Mao, L.; Hao, F.; Wang, Z.; Zhang, W.; Deng, J.; Jiang, J. Analysis on the complete genome of a novel caprine parainfluenza virus 3. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2016, 38, 29–34. [Google Scholar] [CrossRef]
- Makoschey, B.; Berge, A.C. Review on bovine respiratory syncytial virus and bovine parainfluenza-usual suspects in bovine respiratory disease-a narrative review. BMC Vet. Res. 2021, 17, 261. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, Y.; Zan, X.; Wu, Y.; Wang, J.; Li, G.; Chai, C.; Fu, C.; Wang, S.; Yin, H.; et al. Phylogenetic and pathogenicity analysis of a novel lineage of caprine parainfluenza virus type 3. Microb. Pathog. 2021, 154, 104854. [Google Scholar] [CrossRef]
- Zhu, Y.M.; Chen, R.H.; Lin, J.; Yang, M.J.; Xue, F. Isolation and identification of ovine parainfluenza virus type 3 strain PE2019. Chin. J. Prev. Vet. Med. 2020, 42, 434–438. (In Chinese) [Google Scholar]
- Mao, L.; Yang, L.; Li, W.; Liang, P.; Zhang, S.; Li, J.; Sun, M.; Zhang, W.; Wang, L.; Zhong, C.; et al. Epidemiological investigation and phylogenetic analysis of caprine parainfluenza virus type 3 in sheep of China. Transbound. Emerg. Dis. 2019, 66, 1411–1416. [Google Scholar] [CrossRef]
- Mao, L.; Li, W.; Zhou, T.; Yang, L.; Hao, F.; Li, J.; Zhang, W.; Luo, X.; Jiang, J. Development of a blocking ELISA for Caprine parainfluenza virus type 3. J. Virol. Methods 2017, 250, 59–65. [Google Scholar] [CrossRef]
- Li, J.; Li, W.; Mao, L.; Hao, F.; Yang, L.; Zhang, W.; Jiang, J. Rapid detection of novel caprine parainfluenza virus type 3 (CPIV3) using a TaqMan-based RT-qPCR. J. Virol. Methods 2016, 236, 126–131. [Google Scholar] [CrossRef]
- Li, W.; Hao, F.; Mao, L.; Wang, Z.; Zhou, T.; Deng, J.; Li, J.; Zhang, W.; Yang, L.; Lv, Y.; et al. Pathogenicity and horizontal transmission studies of caprine parainfluenza virus type 3 JS2013 strain in goats. Virus Res. 2016, 223, 80–87. [Google Scholar] [CrossRef]
- Bi, J.S.; Wang, W.X.; Wei, X.K.; Su, J.X.; Zheng, M. Serological investigation of antibody against Caprine Parainfluenza virus type 3 in Guangxi. Heilongjiang Anim. Sci. Vet. Med. 2018, 6, 104–105. (In Chinese) [Google Scholar]
- Chen, J.L.; Huang, X.J.; Hao, S.N.; Zhang, D.P.; Wang, J.L.; Shen, Z.Q. Truncated expression of Caprine Parainfluenza virus type 3 NP protein and establishment of indirect ELISA. Chin. J. Vet. Sci. 2020, 40, 1449–1453. (In Chinese) [Google Scholar]
- Li, W.; Yang, L.; Mao, L.; Liu, M.; Li, J.; Zhang, W.; Sun, M. Cholesterol-rich lipid rafts both in cellular and viral membrane are critical for caprine parainfluenza virus type3 entry and infection in host cells. Vet. Microbiol. 2020, 248, 108794. [Google Scholar] [CrossRef]
- Li, J.; Mao, L.; Xiao, F.; Liao, Z.; Yin, J.; Li, W.; Sun, M.; Liu, M.; Ji, X.; Liu, C.; et al. Interferon-stimulated genes inhibit caprine parainfluenza virus type 3 replication in Madin-Darby bovine kidney cells. Vet. Microbiol. 2020, 241, 108573. [Google Scholar] [CrossRef]
- Mao, L.; Liang, P.; Li, W.; Zhang, S.; Liu, M.; Yang, L.; Li, J.; Li, H.; Hao, F.; Sun, M.; et al. Exosomes promote caprine parainfluenza virus type 3 infection by inhibiting autophagy. J. Gen. Virol. 2020, 101, 717–734. [Google Scholar] [CrossRef]
- Li, J.; Yang, L.; Mao, L.; Li, W.; Sun, M.; Liu, C.; Xue, T.; Zhang, W.; Liu, M.; Li, B. Caprine parainfluenza virus type 3 N protein promotes viral replication via inducing apoptosis. Vet. Microbiol. 2021, 259, 109129. [Google Scholar] [CrossRef]
- Li, J.; Zhong, C.; Liao, Z.; Mao, L.; Li, W.; Sun, M.; Liu, M.; Ji, X.; Liu, C.; Xue, T.; et al. Bta-miR-98 Suppresses Replication of Caprine Parainfluenza Virus Type 3 Through Inhibiting Apoptosis by Targeting Caspase-3. Front. Immunol. 2020, 11, 1575. [Google Scholar] [CrossRef]
- Sun, M.; Li, W.; Zhang, W.; Yang, L.; Hao, F.; Li, J.; Mao, L.; Jiang, J.; Liu, M. Screening interferon antagonists from accessory proteins encoded by P gene for immune escape of Caprine parainfluenza virus 3. Vet. Microbiol. 2021, 254, 108980. [Google Scholar] [CrossRef]
- Li, J.; Mao, L.; Zhong, C.; Li, W.; Hao, F.; Sun, M.; Zhu, X.; Ji, X.; Xiao, F.; Yang, L.; et al. Cellular microRNA bta-miR-222 suppresses caprine parainfluenza virus type 3 replication via downregulation of interferon regulatory factor 2. Vet. Microbiol. 2018, 224, 58–65. [Google Scholar] [CrossRef]
- Ellis, J.A. Bovine parainfluenza-3 virus. Vet. Clin. N. Am. Food Anim. Pract. 2010, 26, 575–593. [Google Scholar] [CrossRef]
- Maidana, S.S.; Lomonaco, P.M.; Combessies, G.; Craig, M.I.; Diodati, J.; Rodriguez, D.; Parreno, V.; Zabal, O.; Konrad, J.L.; Crudelli, G.; et al. Isolation and characterization of bovine parainfluenza virus type 3 from water buffaloes (Bubalus bubalis) in Argentina. BMC Vet. Res. 2012, 8, 83. [Google Scholar] [CrossRef] [Green Version]
- Yun, J.L.; He, M.F.; Wang, S.H.; Mao, L.; Yang, L.L.; Zhang, W.W.; Sun, M.; Liu, M.J.; Li, W.L. Diagnosis and pathogenic identification of the respiratory disease in a fattening goat farm. Anim. Husb. Vet. Med. 2022, 54, 94–99. (In Chinese) [Google Scholar]
- Wang, M.; Li, W.L.; Hao, F.; Mao, L.; Yang, L.L.; Zhang, W.W.; Jiang, J.Y. Prokaryotic expression of N protein of Caprine Parainfluenza virus type 3 and establishment of indirect ELISA antibody detection method. Acta Vet. Et Zootech. Sin. 2017, 48, 150–156. (In Chinese) [Google Scholar]
- Nettleton, P.F.; Gilray, J.A.; Russo, P.; Dlissi, E. Border disease of sheep and goats. Vet. Res. 1998, 29, 327–340. [Google Scholar] [PubMed]
- Nettleton, P.F.; Entrican, G. Ruminant pestiviruses. Br. Vet. J. 1995, 151, 615–642. [Google Scholar] [CrossRef]
- Oguzoglu, T.C.; Tan, M.T.; Toplu, N.; Demir, A.B.; Bilge-Dagalp, S.; Karaoglu, T.; Ozkul, A.; Alkan, F.; Burgu, I.; Haas, L.; et al. Border disease virus (BDV) infections of small ruminants in Turkey: A new BDV subgroup? Vet. Microbiol. 2009, 135, 374–379. [Google Scholar] [CrossRef] [PubMed]
- King, A.M.Q.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; Knowles, N.J.; et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Arch. Virol. 2018, 163, 2601–2631. [Google Scholar] [CrossRef] [Green Version]
- Righi, C.; Petrini, S.; Pierini, I.; Giammarioli, M.; De Mia, G.M. Global Distribution and Genetic Heterogeneity of Border Disease Virus. Viruses 2021, 13, 950. [Google Scholar] [CrossRef]
- Peletto, S.; Caruso, C.; Cerutti, F.; Modesto, P.; Zoppi, S.; Dondo, A.; Acutis, P.L.; Masoero, L. A new genotype of border disease virus with implications for molecular diagnostics. Arch. Virol. 2016, 161, 471–477. [Google Scholar] [CrossRef]
- Giammarioli, M.; La Rocca, S.A.; Steinbach, F.; Casciari, C.; De Mia, G.M. Genetic and antigenic typing of border disease virus (BDV) isolates from Italy reveals the existence of a novel BDV group. Vet. Microbiol. 2011, 147, 231–236. [Google Scholar] [CrossRef]
- Sozzi, E.; Lavazza, A.; Gaffuri, A.; Bencetti, F.C.; Prosperi, A.; Lelli, D.; Chiapponi, C.; Moreno, A. Isolation and Full-Length Sequence Analysis of a Pestivirus from Aborted Lamb Fetuses in Italy. Viruses 2019, 11, 744. [Google Scholar] [CrossRef] [Green Version]
- Casciari, C.; Sozzi, E.; Bazzucchi, M.; Moreno Martin, A.M.; Gaffuri, A.; Giammarioli, M.; Lavazza, A.; De Mia, G.M. Serological relationship between a novel ovine pestivirus and classical swine fever virus. Transbound. Emerg. Dis. 2020, 67, 1406–1410. [Google Scholar] [CrossRef]
- Li, W.L.; Mao, L.; Zhang, W.W.; Zhang, Z.B.; Zhao, Y.Q.; He, K.W.; Jiang, J.Y. Isolation and identification of border disease virus (BDV) from a goat herd suffering unremitting diarrhea. Jiangsu J. Agric. Sci. 2013, 29, 222–224. (In Chinese) [Google Scholar]
- Chen, Y.Z.; Bao, G.C.; Zhang, S.X.; Han, M. Epidemiological investigation of bovine viral diarrhea virus and sheep boundary virus in Tibetan sheep in Haibei area, Qinghai province. Chin. J. Vet. Drug 2017, 51, 7–11. (In Chinese) [Google Scholar]
- Wang, Y.L.; Xu, S.R.; Zhang, X.Z.; Xu, C.F. Etiological investigation and analysis of bovine viral diarrhea virus, border disease virus and enterovirus in Tibetan sheep in Haidong City, Qinghai Province. Anim. Husb. Vet. Med. 2018, 50, 87–90. (In Chinese) [Google Scholar]
- Peng, C.; Wang, S.C.; Zhuang, Q.Y.; Hou, G.Y.; Huang, J.; Shan, H.; Chen, J.M. Detection of border disease virus (BDV) in apparently healthy sheep in Shandong. China Anim. Health Insp. 2015, 32, 1–3. (In Chinese) [Google Scholar]
- Vilcek, S.; Belak, S. Genetic identification of pestivirus strain Frijters as a border disease virus from pigs. J. Virol. Methods 1996, 60, 103–108. [Google Scholar] [CrossRef]
- Nagai, M.; Aoki, H.; Sakoda, Y.; Kozasa, T.; Tominaga-Teshima, K.; Mine, J.; Abe, Y.; Tamura, T.; Kobayashi, T.; Nishine, K.; et al. Molecular, biological, and antigenic characterization of a Border disease virus isolated from a pig during classical swine fever surveillance in Japan. J. Vet. Diagn. Investig. Off. Publ. Am. Assoc. Vet. Lab. Diagn. Inc. 2014, 26, 547–552. [Google Scholar] [CrossRef] [Green Version]
- Kawanishi, N.; Tsuduku, S.; Shimizu, H.; Ohtani, Y.; Kameyama, K.; Yamakawa, M.; Tsutsui, T.; Matsuura, K.; Ohashi, S.; Isobe, T.; et al. First isolation of border disease virus in Japan is from a pig farm with no ruminants. Vet. Microbiol. 2014, 171, 210–214. [Google Scholar] [CrossRef]
- Rosell, R.; Cabezon, O.; Pujols, J.; Domingo, M.; Munoz, I.; Nunez, J.I.; Ganges, L. Identification of a porcine pestivirus as a border disease virus from naturally infected pigs in Spain. Vet. Rec. 2014, 174, 18. [Google Scholar] [CrossRef]
- Hornberg, A.; Fernandez, S.R.; Vogl, C.; Vilcek, S.; Matt, M.; Fink, M.; Kofer, J.; Schopf, K. Genetic diversity of pestivirus isolates in cattle from Western Austria. Vet. Microbiol. 2009, 135, 205–213. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Romero, N.; Basurto-Alcantara, F.J.; Verdugo-Rodriguez, A.; Lagunes-Quintanilla, R.; Bauermann, F.V.; Ridpath, J.F. Detection of border disease virus in Mexican cattle. Transbound. Emerg. Dis. 2018, 65, 267–271. [Google Scholar] [CrossRef] [Green Version]
- Braun, U.; Hilbe, M.; Peterhans, E.; Schweizer, M. Border disease in cattle. Vet. J. 2019, 246, 12–20. [Google Scholar] [CrossRef]
- Becher, P.; Orlich, M.; Kosmidou, A.; Konig, M.; Baroth, M.; Thiel, H.J. Genetic diversity of pestiviruses: Identification of novel groups and implications for classification. Virology 1999, 262, 64–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, C.; Duquesne, V.; Adam, G.; Belleau, E.; Gauthier, D.; Champion, J.L.; Saegerman, C.; Thiery, R.; Dubois, E. Pestiviruses infections at the wild and domestic ruminants interface in the French Southern Alps. Vet. Microbiol. 2015, 175, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Li, W.; Liu, X.; Hao, F.; Yang, L.; Deng, J.; Zhang, W.; Wei, J.; Jiang, J. Chinese border disease virus strain JSLS12-01 infects piglets and down-regulates the antibody responses of classical swine fever virus C strain vaccination. Vaccine 2015, 33, 3918–3922. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.H.; He, B.; Li, K.R.; Li, F.; Zhang, L.Y.; Li, X.Q.; Zhao, L. First report of border disease virus in Melophagus ovinus (sheep ked) collected in Xinjiang, China. PLoS ONE 2019, 14, e0221435. [Google Scholar] [CrossRef] [PubMed]
- Marco, I.; Rosell, R.; Cabezon, O.; Beneria, M.; Mentaberre, G.; Casas, E.; Hurtado, A.; Lopez-Olvera, J.R.; Lavin, S. Serologic and virologic investigations into pestivirus infection in wild and domestic ruminants in the Pyrenees (NE Spain). Res. Vet. Sci. 2009, 87, 149–153. [Google Scholar] [CrossRef]
- Thabti, F.; Letellier, C.; Hammami, S.; Pepin, M.; Ribiere, M.; Mesplede, A.; Kerkhofs, P.; Russo, P. Detection of a novel border disease virus subgroup in Tunisian sheep. Arch. Virol. 2005, 150, 215–229. [Google Scholar] [CrossRef]
- Newcomer, B.W.; Givens, M.D. Approved and experimental countermeasures against pestiviral diseases: Bovine viral diarrhea, classical swine fever and border disease. Antivir. Res. 2013, 100, 133–150. [Google Scholar] [CrossRef]
- Vilcek, S.; Herring, A.J.; Herring, J.A.; Nettleton, P.F.; Lowings, J.P.; Paton, D.J. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch. Virol. 1994, 136, 309–323. [Google Scholar] [CrossRef]
- Vilbek, S.; Paton, D.J. A RT-PCR assay for the rapid recognition of border disease virus. Vet. Res. 2000, 31, 437–445. [Google Scholar] [CrossRef] [Green Version]
- Strong, R.; La Rocca, S.A.; Ibata, G.; Sandvik, T. Antigenic and genetic characterisation of border disease viruses isolated from UK cattle. Vet. Microbiol. 2010, 141, 208–215. [Google Scholar] [CrossRef] [Green Version]
- Vilcek, S.; Nettleton, P.F.; Paton, D.J.; Belak, S. Molecular characterization of ovine pestiviruses. J. Gen. Virol. 1997, 78 Pt 4, 725–735. [Google Scholar] [CrossRef] [Green Version]
- Willoughby, K.; Valdazo-Gonzalez, B.; Maley, M.; Gilray, J.; Nettleton, P.F. Development of a real time RT-PCR to detect and type ovine pestiviruses. J. Virol. Methods 2006, 132, 187–194. [Google Scholar] [CrossRef]
- De las Heras, M.; Ortin, A.; Cousens, C.; Minguijon, E.; Sharp, J.M. Enzootic nasal adenocarcinoma of sheep and goats. Curr. Top. Microbiol. Immunol. 2003, 275, 201–223. [Google Scholar] [CrossRef]
- Apostolidi, E.D.; Psalla, D.; Chassalevris, T.; Chaintoutis, S.C.; Giadinis, N.D.; Psychas, V.; Dovas, C.I. Development of real-time PCR-based methods for the detection of enzootic nasal tumor virus 2 in goats. Arch. Virol. 2019, 164, 707–716. [Google Scholar] [CrossRef]
- Cousens, C.; Minguijon, E.; Dalziel, R.G.; Ortin, A.; Garcia, M.; Park, J.; Gonzalez, L.; Sharp, J.M.; de las Heras, M. Complete sequence of enzootic nasal tumor virus, a retrovirus associated with transmissible intranasal tumors of sheep. J. Virol. 1999, 73, 3986–3993. [Google Scholar] [CrossRef] [Green Version]
- Ortin, A.; Cousens, C.; Minguijon, E.; Pascual, Z.; Villarreal, M.P.; Sharp, J.M.; Heras, M.L. Characterization of enzootic nasal tumour virus of goats: Complete sequence and tissue distribution. J. Gen. Virol. 2003, 84, 2245–2252. [Google Scholar] [CrossRef]
- De Las Heras, M.; Borobia, M.; Ortin, A. Neoplasia-Associated Wasting Diseases with Economic Relevance in the Sheep Industry. Anim. Open Access J. 2021, 11, 381. [Google Scholar] [CrossRef]
- Walsh, S.R.; Linnerth-Petrik, N.M.; Laporte, A.N.; Menzies, P.I.; Foster, R.A.; Wootton, S.K. Full-length genome sequence analysis of enzootic nasal tumor virus reveals an unusually high degree of genetic stability. Virus Res. 2010, 151, 74–87. [Google Scholar] [CrossRef]
- Walsh, S.R.; Stinson, K.J.; Menzies, P.I.; Wootton, S.K. Development of an ante-mortem diagnostic test for enzootic nasal tumor virus and detection of neutralizing antibodies in host serum. J. Gen. Virol. 2014, 95, 1843–1854. [Google Scholar] [CrossRef]
- Sid, N.; Belalmi, N.E.H.; Benhamza, L.; Ouhida, S.; Zebiri, M.E.; Aydogan, A.; Leroux, C. First case report of enzootic nasal adenocarcinoma in "Ouled Djellal" ewe in Algeria. Open Vet. J. 2018, 8, 9–12. [Google Scholar] [CrossRef] [Green Version]
- Lin, X.; Hao, X.; Zhao, Z.; Yu, W.; Gu, Y.; Bao, G. Pathological study of goat nasal adenoma and adenocarcinoma. Acta Vcterinaria Et Zootechhnica Sin. 1995, 5, 456–461. (In Chinese) [Google Scholar]
- He, Y.; Zhang, Q.; Wang, J.; Zhou, M.; Fu, M.; Xu, X. Full-length genome sequence analysis of enzootic nasal tumor virus isolated from goats in China. Virol. J. 2017, 14, 141. [Google Scholar] [CrossRef] [Green Version]
- Ye, C.; Huang, Q.; Chen, T.; Jiang, J.; Hou, F.; Xu, D.; Peng, Y.; Fang, R.; Chen, J. First detection and genotypic analysis of goat enzootic nasal tumor virus 2 in Chongqing, China. Arch. Virol. 2019, 164, 1647–1650. [Google Scholar] [CrossRef]
- Zhai, S.L.; Lv, D.H.; Xu, Z.H.; Yu, J.S.; Wen, X.H.; Zhang, H.; Chen, Q.L.; Jia, C.L.; Zhou, X.R.; Zhai, Q.; et al. A Novel Enzootic Nasal Tumor Virus Circulating in Goats from Southern China. Viruses 2019, 11, 956. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Feng, Y.C.; Yan, Q.G.; Han, G.Q. Diagnosis of an enzootic nasal tumor of goat in Sichuan. Anim. Husb. Vet. Med. 2011, 43, 83–86. (In Chinese) [Google Scholar]
- Jiang, J.X.; Lin, Y.S.; Jiang, B.; Mao, K.M.; You, W.; Zhang, J.P.; Hu, Q.L. Molecular epidemiology of enzootic nasal tumor virus on goats in Fujian. Fujian J. Agric. Sci. 2017, 32, 837–841. (In Chinese) [Google Scholar]
- Yu, Y.D.; Wei, L.F.; Huang, X.J.; Zhang, B.; Yu, X.H.; Tang, C. Diagnosis of four cases of enzootic nasal adenocarcinoma of goats. Prog. Vet. Med. 2014, 35, 129–131. (In Chinese) [Google Scholar]
- Hou, H.Z.; Zhang, D.; Hu, X.; Zhao, R.; Dai, Y. Cloning and analysis of gag gene of enzootic nasal tumor virus in goats. China Herbiv. Sci. 2018, 38, 53–55. (In Chinese) [Google Scholar]
- Xu, G.; Xian, S.; Zeng, Z.; Liang, H.; Wang, B.; Huang, T.; Tang, D. Diagnosis of a goat intranasal tumor. China Herbiv. Sci. 2018, 2, 75–77. (In Chinese) [Google Scholar]
- De las Heras, M.; Garcia de Jalon, J.A.; Minguijon, E.; Gray, E.W.; Dewar, P.; Sharp, J.M. Experimental transmission of enzootic intranasal tumors of goats. Vet. Pathol. 1995, 32, 19–23. [Google Scholar] [CrossRef] [Green Version]
- Walsh, S.R.; Stinson, K.J.; Wootton, S.K. Seroconversion of sheep experimentally infected with enzootic nasal tumor virus. BMC Res. Notes 2016, 9, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wootton, S.K.; Metzger, M.J.; Hudkins, K.L.; Alpers, C.E.; York, D.; DeMartini, J.C.; Miller, A.D. Lung cancer induced in mice by the envelope protein of jaagsiekte sheep retrovirus (JSRV) closely resembles lung cancer in sheep infected with JSRV. Retrovirology 2006, 3, 94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Q.; Ye, C.; Chen, T.; Jiang, J.; Peng, Y.; Chen, J.; Fang, R. EvaGreen-based real-time PCR assay for sensitive detection of enzootic nasal tumor virus 2. Mol. Cell. Probes 2019, 44, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.; Xie, Z.; Liao, H.; Liu, D.; Guo, L.; Liu, J.; Yang, S.; Shu, L.; Yan, Q. Estabilshment of an RT-PCR method for detection of enzootic nasal tumor virus in goat. Chin. Vet. Sci. 2014, 44, 933–938. (In Chinese) [Google Scholar]
- He, R.; Du, Y.; Gan, L.; Mohsin, M.A.; He, B.X. Development of a SYBR Green-based real-time quantitative polymerase chain reaction assay to detect enzootic nasal tumor virus in goats. Can. J. Vet. Res. Rev. Can. De Rech. Vet. 2021, 85, 145–150. [Google Scholar]
- Zhang, J.P.; H, J.J.; Lin, Y.S.; You, W.; Liu, D.Q.; Mao, K.M.; Jiang, B.; Hu, Q.L. A SYBR-Green I RT-qPCR assay for detecting enzootic nasal tumor virus in goats. Fujian J. Agric. Sci. 2021, 36, 779–784. (In Chinese) [Google Scholar]
- Xiao, S.; Zhai, S.; Chen, X.; Xie, Y.; Zhou, X.; Lyv, D.; Wen, X.; Zhai, Q.; Jia, C.; Wei, W.; et al. Development and Application of a Fluorescent PCR Method for Detection of Enzootic Nasal Tumor Virus. Chin. J. Anim. Infect. Dis. 2021, 29, 9–14. (In Chinese) [Google Scholar]
- Davison, A.J. Herpesvirus systematics. Vet. Microbiol. 2010, 143, 52–69. [Google Scholar] [CrossRef] [Green Version]
- Thiry, J.; Saegerman, C.; Chartier, C.; Mercier, P.; Keuser, V.; Thiry, E. Serological evidence of caprine herpesvirus 1 infection in Mediterranean France. Vet. Microbiol. 2008, 128, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Berrios, P.E.; McKercher, D.G.; Knight, H.D. Pathogenicity of a caprine herpesvirus. Am. J. Vet. Res. 1975, 36, 1763–1769. [Google Scholar]
- Tempesta, M.; Camero, M.; Greco, G.; Pratelli, A.; Martella, V.; Buonavoglia, C. A classical inactivated vaccine induces protection against caprine herpesvirus 1 infection in goats. Vaccine 2001, 19, 3860–3864. [Google Scholar] [CrossRef]
- Gonzalez, J.; Passantino, G.; Esnal, A.; Cuesta, N.; Garcia Vera, J.A.; Mechelli, L.; Saez, A.; Garcia Marin, J.F.; Tempesta, M. Abortion in goats by Caprine alphaherpesvirus 1 in Spain. Reprod. Domest. Anim. Zuchthyg. 2017, 52, 1093–1096. [Google Scholar] [CrossRef]
- Suavet, F.; Champion, J.L.; Bartolini, L.; Bernou, M.; Alzieu, J.P.; Brugidou, R.; Darnatigues, S.; Reynaud, G.; Perrin, C.; Adam, G.; et al. First Description of Infection of Caprine Herpesvirus 1 (CpHV-1) in Goats in Mainland France. Pathogens 2016, 5, 17. [Google Scholar] [CrossRef] [Green Version]
- Tarigan, S.; Webb, R.F.; Kirkland, D. Caprine herpesvirus from balanoposthitis. Aust. Vet. J. 1987, 64, 321. [Google Scholar] [CrossRef]
- Chenier, S.; Montpetit, C.; Helie, P. Caprine herpesvirus- 1 abortion storm in a goat herd in Quebec. Can. Vet. J. La Rev. Vet. Can. 2004, 45, 241–243. [Google Scholar]
- Camero, M.; Lanave, G.; Lucente, M.S.; Losurdo, M.; Di Paola, G.; Lorusso, E.; Martella, V.; Buonavoglia, C.; Tempesta, M. Bubaline alphaherpesvirus 1 induces a latent/reactivable infection in goats. Comp. Immunol. Microbiol. Infect. Dis. 2019, 62, 54–57. [Google Scholar] [CrossRef]
- Zell, R.; Delwart, E.; Gorbalenya, A.E.; Hovi, T.; King, A.M.Q.; Knowles, N.J.; Lindberg, A.M.; Pallansch, M.A.; Palmenberg, A.C.; Reuter, G.; et al. ICTV Virus Taxonomy Profile: Picornaviridae. J. Gen. Virol. 2017, 98, 2421–2422. [Google Scholar] [CrossRef]
- Anbalagan, S.; Hesse, R.A.; Hause, B.M. First identification and characterization of porcine enterovirus G in the United States. PLoS ONE 2014, 9, e97517. [Google Scholar] [CrossRef]
- Wang, M.; He, J.; Lu, H.; Liu, Y.; Deng, Y.; Zhu, L.; Guo, C.; Tu, C.; Wang, X. A novel enterovirus species identified from severe diarrheal goats. PLoS ONE 2017, 12, e0174600. [Google Scholar] [CrossRef] [Green Version]
- Bunke, J.; Receveur, K.; Oeser, A.C.; Fickenscher, H.; Zell, R.; Krumbholz, A. High genetic diversity of porcine enterovirus G in Schleswig-Holstein, Germany. Arch. Virol. 2018, 163, 489–493. [Google Scholar] [CrossRef]
- Boros, A.; Pankovics, P.; Knowles, N.J.; Reuter, G. Natural interspecies recombinant bovine/porcine enterovirus in sheep. J. Gen. Virol. 2012, 93, 1941–1951. [Google Scholar] [CrossRef]
- Dong, K.; Hu, J.Y.; Li, Z.C.; Cai, M.L.; Zhang, F.; Wang, W.Y.; Wang, Y.G.; Wang, X.P. Establishment and preliminary application of RT-PCR method for the detection of caprine/ovine enterovirus. Chin. J. Vet. Sci. 2021, 41, 1949–1952. (In Chinese) [Google Scholar]
- Lu, G. Epidemiological investigation and analysis of sheep enterovirus infection in some areas of Hebei province. Chin. J. Anim. Infect. Dis. 2021, 29, 88–92. (In Chinese) [Google Scholar]
- Lu, H.B.; Hu, J.Y.; Zhang, Q.; Wang, Y.G.; Guo, C.M.; Zhu, L.S.; Tu, C.C.; Wang, X.P. Development of a sandwich ELISA for detection of CEV antigen and epidemiological investigation. Chin. J. Vet. Sci. 2017, 37, 653–657. (In Chinese) [Google Scholar]
- Wang, R.D.; Lin, Q.; Wang, W.Y.; Hu, J.Y.; Zhang, Z.C.; Zhu, J.F.; Wang, X.P. Epidemiological investigarion and analysis of sheep enterovirus infection in Henan province. Chin. J. Vet. Sci. 2020, 40, 902–906. (In Chinese) [Google Scholar]
- Li, X.; Hu, J.Y.; Lin, Q.; Wang, X.; Zheng, B.W.; Zhang, Z.C.; Hu, L.P.; Wang, X.P. Epidemiological investigarion on enterovirus infection in cattle/sheep/goat herds in Shandong province. Chin. J. Vet. Sci. 2019, 39, 223–227. (In Chinese) [Google Scholar]

| Interferon Type | Key Points or Pathways | Related Viral Proteins | References |
|---|---|---|---|
| type I IFN pathways | MAVS | / | [20] |
| type I interferon | IRF3, TBK1–IRF3 complex | N | [24] |
| IFN-λ3, IFN-β, and IFN-λ2 | RIG-I-like receptor, IRF3 | P | [25] |
| IFN-β | JAK–STAT, MAVS, and RIG-I signaling pathway | C | [26] |
| IFN-β, IFN-γ | STAT1 | N and P | [23] |
| IFN | ATG13 | / | [21] |
| type I IFN pathway | miR-3 | / | [27] |
| Region | Province | Positive No./Total No. Positive Rate (%) | Region | Province | Positive No./Total No. Positive Rate (%) | ||
|---|---|---|---|---|---|---|---|
| Goat | Sheep | Goat | Sheep | ||||
| East China | Jiangsu | 1137/2842 40.01 | 356/1078 33.02 | Northwest China | Xinjiang | / | 408/568 71.83 |
| Anhui | 273/658 41.49 | 24/219 10.96 | Qinghai | / | 73/118 61.86 | ||
| Shandong | 162/855 b 18.95 | 342/728 46.98 | Gansu | / | 592/848 69.81 | ||
| Zhejiang | 31/98 31.63 | / | Shannxi | 66/254 25.98 | / | ||
| North China | Inner Mongolia | / | 89/105 84.76 | Southwest China | Tibet | / | 12/104 12.54 |
| Hebei | / | 150/332 45.18 | Guizhou | 46/158 29.11 | / | ||
| Shanxi | 39/93 41.94 | / | Northeast China | Jilin | 173/480 36.04 | / | |
| South China | Guangdong | 0/200 0 | / | Liaoning | 38/167 22.75 | / | |
| Guangxi | 171/634 a 26.97 | / | |||||
| Methods | Primer | Sequence (5′-3′) | Size | Reference |
|---|---|---|---|---|
| RT-PCR | F | AGTGATCTAGATGATGATCCA | 329bp | [79] |
| R | GTTATTGATCCAATTGCTGT | |||
| RT-PCR | F | GCAATCCACCAAAGCATGGGGT | 346bp | [80] |
| R | GGGGCAAGTGCTACTTTTTGAGCA | |||
| RT-qPCR | qF | GCTTGGCTTCTTTGAAATGG | 150bp | [67] |
| qR | GCCTGCAGAAGTTCCTTGTC | |||
| Probe | FAM-CAATCGGACTAGCCAAGTATGGTGGGA-TAMRA |
| Methods | Primers | Sequences (5′-3′) | Target | Size (bp) | Reference |
|---|---|---|---|---|---|
| RT-PCR | 324 | ATGCCCWTAGTAGGACTAGCA | 5′-UTR | 288 | [109] |
| 326 | TCAACTCCATGTGCCATGTAC | ||||
| RT-PCR | PBD1 | TCGTGGTGAGATCCCTGAG | 5′-UTR | 225 | [110] |
| PBD2 | GCAGAGATTTTTTATACTAGCCTATRC | ||||
| RT-PCR | 320F | GCCTGATAGGGTGYWGCAGAG | Npro-C | 740 | [111] |
| 1040R | TTYCCTTTCTTCTTYACCTGGTA | ||||
| RT-PCR | BD1 | TCTCTGCTGTACATGGCACATG | Npro-C | 738 | [112] |
| BD2 | TTGTTRTGGTACARRCCGTC | ||||
| RT-qPCR (Probe) | BDV87F | CCGTGTTAACCATACACGTAGTAGGA | 5′-UTR | 155 | [113] |
| BDV237 | GCCCTCGTCCACGTAGCA | ||||
| BDV136T (probe) | VIC-CTCAGGGATCTCACCACGA-NFQ-MGB |
| Method | Targets | Primer | Sequence (5′-3′) | References |
|---|---|---|---|---|
| RT-PCR | ENTV-1U5-gag | F | GATGCTCCGTTCTCTCCTTATA | [120] |
| R | GGGACGCGACGAATGTAGG | |||
| PCR | ENTV-1LTR | F | AAGCAAGTTAAGTAACTTGAGATC | [117] |
| R | GCTTAGCCGTCCTAAAAGAG | |||
| PCR | ENTV-2env | F | AGCTGCTCATACTGTGGATC | |
| R | GATCTTATCTGCTTATTTTCAG | |||
| RT-PCR | ENTV-2gag | F | AAATGCGACCTTCCGATAATGATGA | [135] |
| R | CTTCTGTAGCGGGGACATATTCTCA | |||
| PCR | ENTV-2gag | F | GTCCCTAAAAATGCGACCTT | [136] |
| R | GCGACTCCTGAGTTCTGTAAAACCAC | |||
| qPCR/RT-qPCR (Probe) | ENTV-2env-U3 | F | CCTAACCTTCAT TCRTTATGGCARAGT | [115] |
| R | CACCGGATCCTTAYGTAATCRGATTTCCTG | |||
| Probe | FAM-TGTTTAGTTCCTTGCCTCCTTCGTGG-IBFQ | |||
| qPCR (SYBR Green) | ENTV-2LTR | F | GAGATTTCTTACACATGAGAGC | [137] |
| R | TCCCAGGACTTAACCATTC | |||
| qPCR (Eva Green) | ENTV-2env | F | GAGGCAAATTGAGGCGTTGAT | [134] |
| R | CCCGTTCTGCATTCGCTGTAG | |||
| qPCR (Probe) | ENTV-2env | F | ATGGCAATAGTTTATATCTGCAAT | [138] |
| R | GATGGCCTTGTATCAACATAAATGG | |||
| Probe | FAM-ATATAAGAATCCCGTAACACCTACATCTC-BHQ1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, L.; Li, W.; Hao, F.; Yang, L.; Li, J.; Sun, M.; Zhang, W.; Liu, M.; Luo, X.; Cheng, Z. Research Progress on Emerging Viral Pathogens of Small Ruminants in China during the Last Decade. Viruses 2022, 14, 1288. https://doi.org/10.3390/v14061288
Mao L, Li W, Hao F, Yang L, Li J, Sun M, Zhang W, Liu M, Luo X, Cheng Z. Research Progress on Emerging Viral Pathogens of Small Ruminants in China during the Last Decade. Viruses. 2022; 14(6):1288. https://doi.org/10.3390/v14061288
Chicago/Turabian StyleMao, Li, Wenliang Li, Fei Hao, Leilei Yang, Jizong Li, Min Sun, Wenwen Zhang, Maojun Liu, Xuenong Luo, and Zilong Cheng. 2022. "Research Progress on Emerging Viral Pathogens of Small Ruminants in China during the Last Decade" Viruses 14, no. 6: 1288. https://doi.org/10.3390/v14061288
APA StyleMao, L., Li, W., Hao, F., Yang, L., Li, J., Sun, M., Zhang, W., Liu, M., Luo, X., & Cheng, Z. (2022). Research Progress on Emerging Viral Pathogens of Small Ruminants in China during the Last Decade. Viruses, 14(6), 1288. https://doi.org/10.3390/v14061288







