Serotype I and II Feline Coronavirus Replication and Gene Expression Patterns of Feline Cells—Building a Better Understanding of Serotype I FIPV Biology
Abstract
1. Introduction
2. Materials and Methods
2.1. FIPV Inoculum for In Vitro Experiments
2.2. RNA Processing
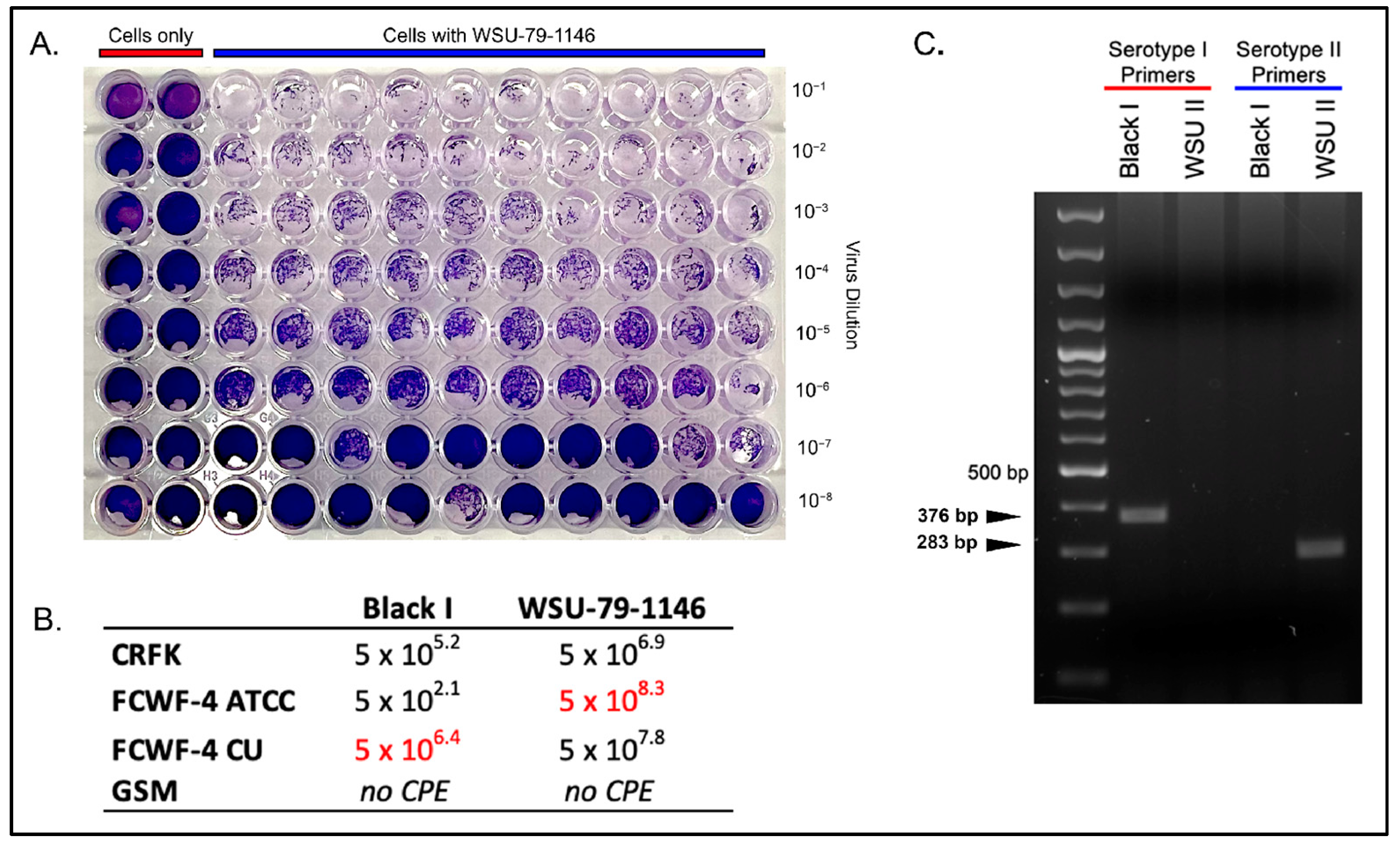
2.3. Black I and WSU-79-1146 Titers across Cell Types
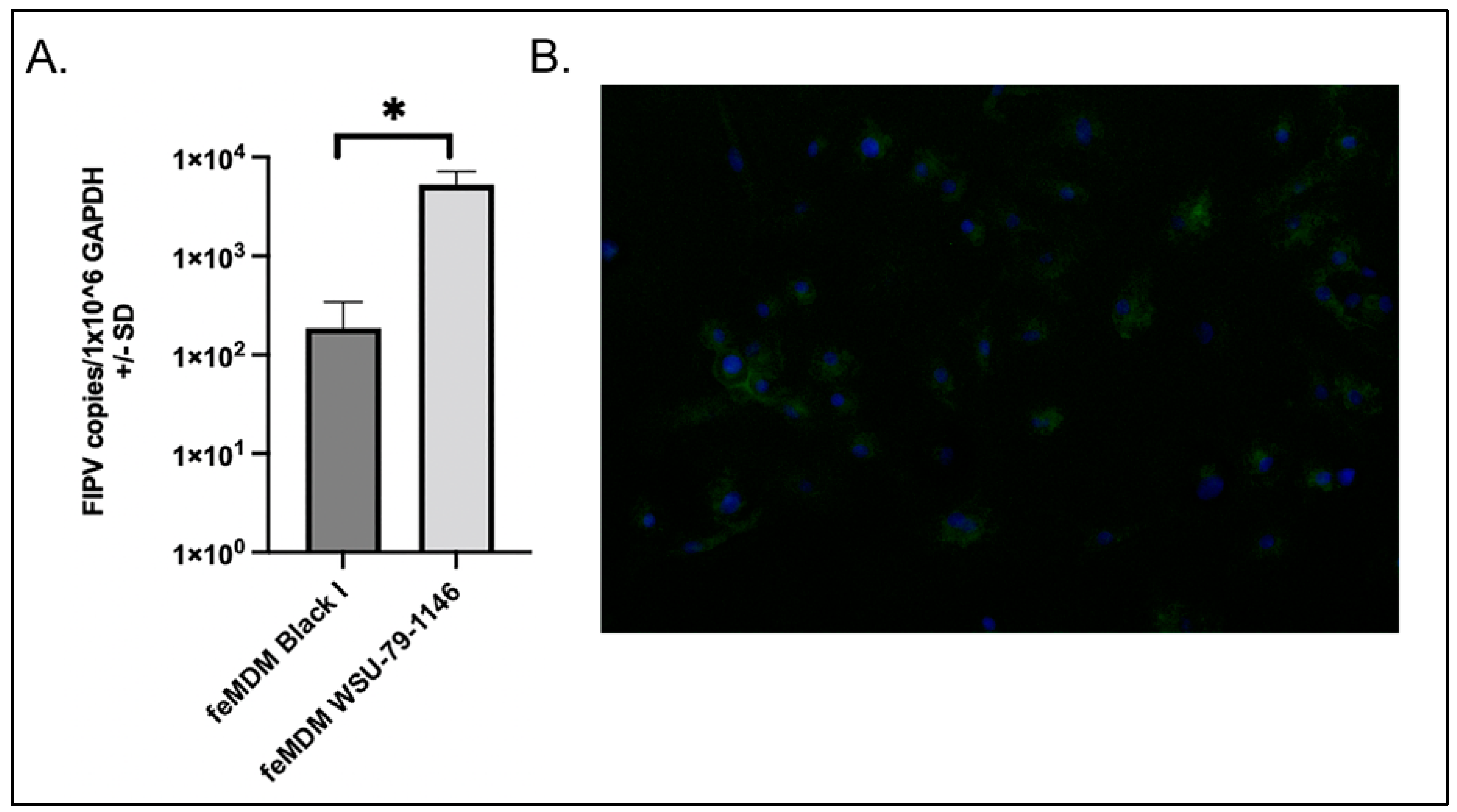
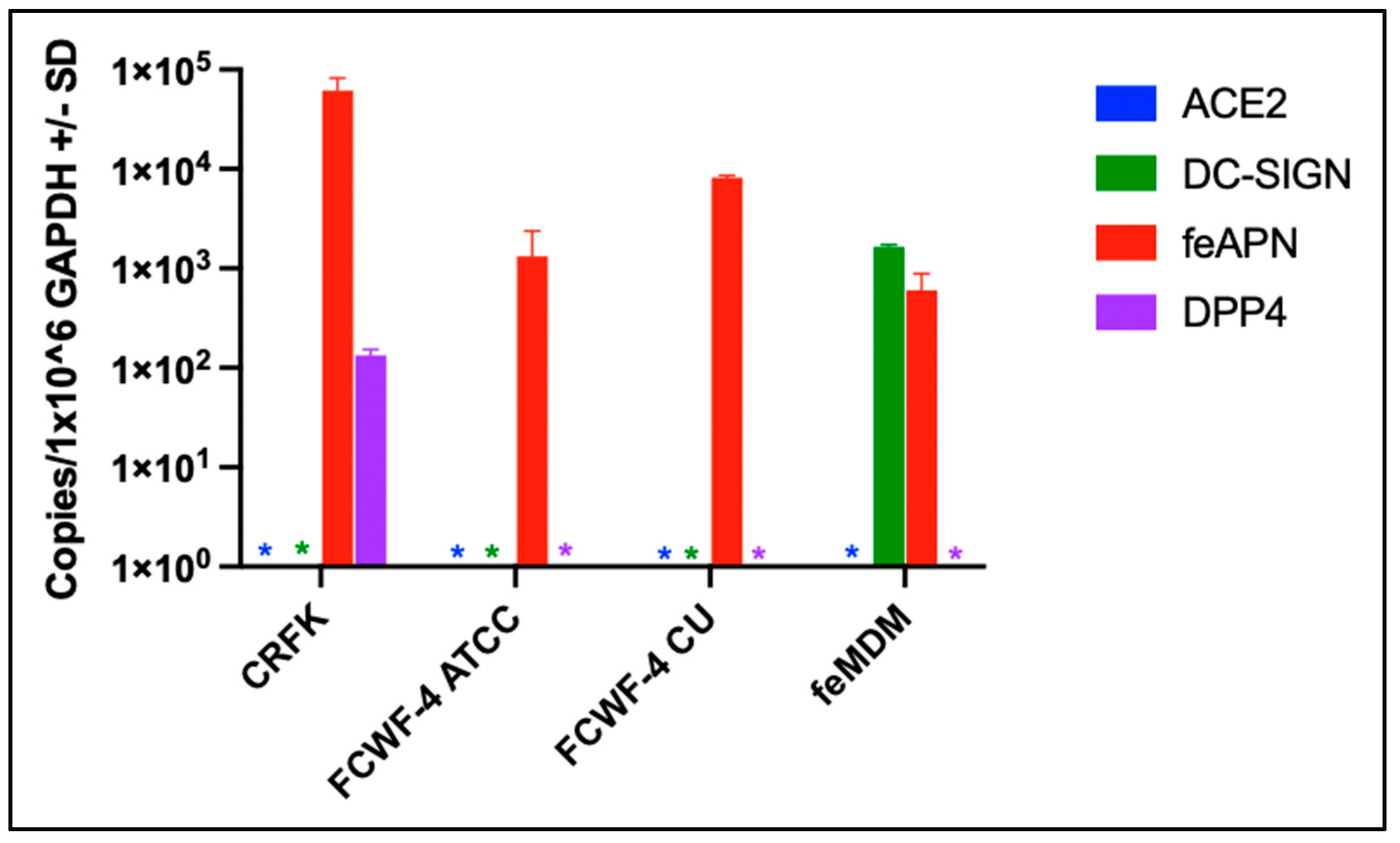
2.4. Quantification of FIPV and Cellular Gene Expression by RT-qPCR
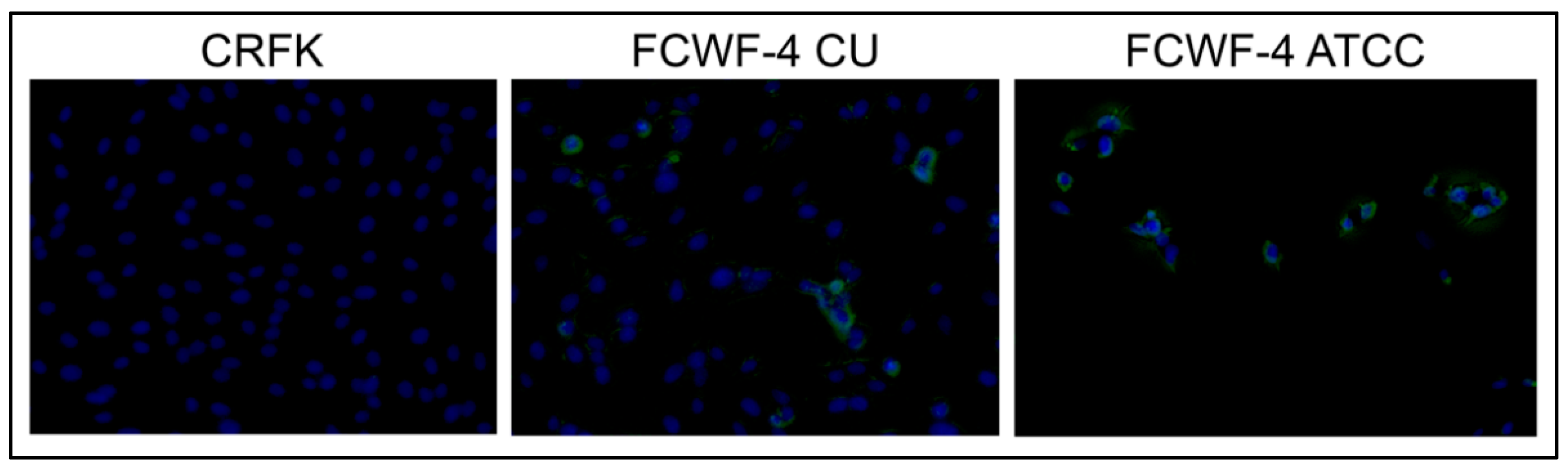
2.5. CD14 Immunofluorescence
2.6. Determining the Systemic Distribution of ACE2 and DC-SIGN in Feline Tissues
2.7. Gene Expression Profiles of FCWF-4 ATCC and FCWF-4 CU Cell Lines Using RNAseq
2.8. RNAseq
3. Results
3.1. Comparison of Serotype I and II Titers Produced in Each Cell Line
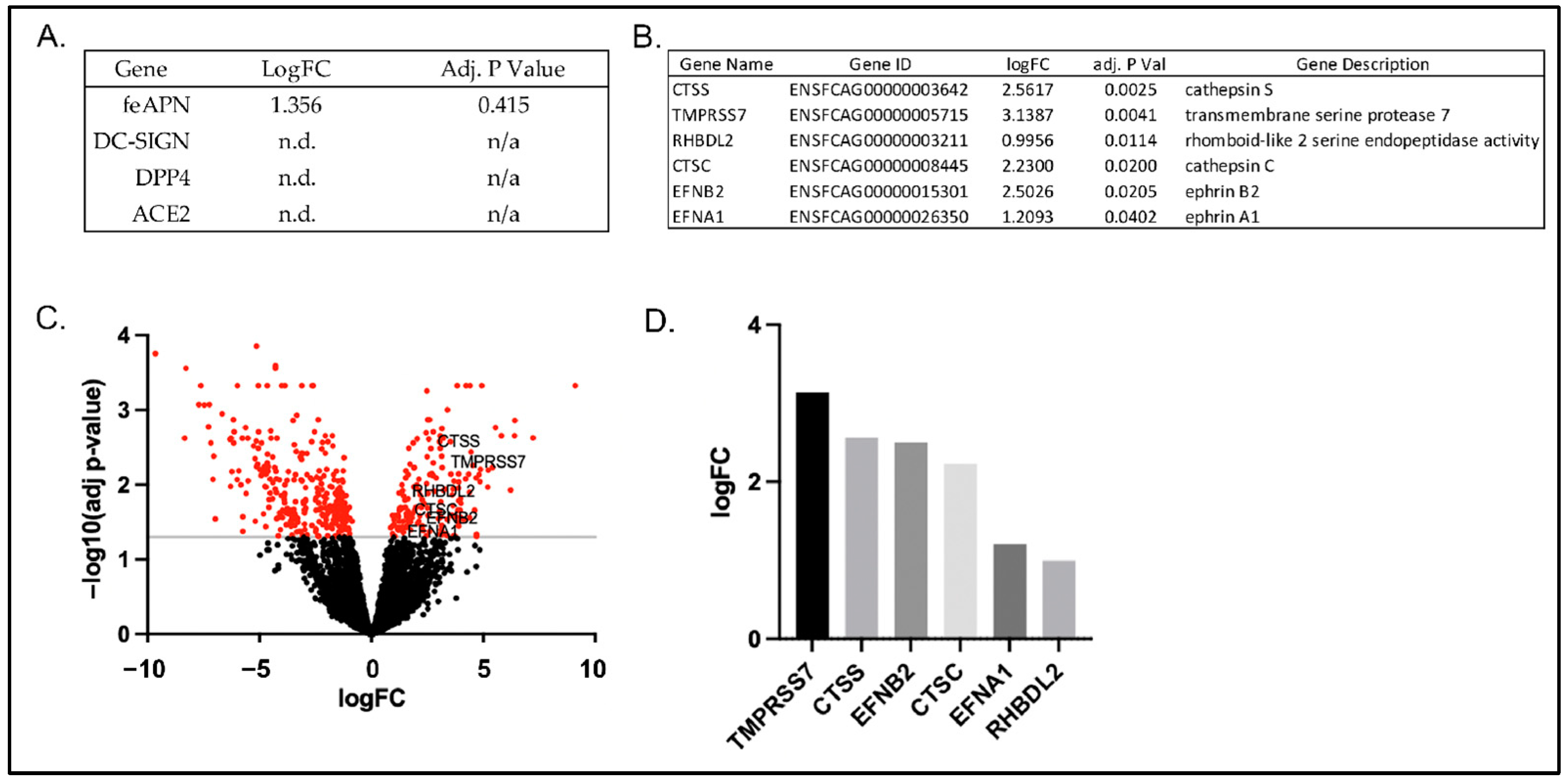
3.2. Cellular Expression of Known Coronavirus Receptors
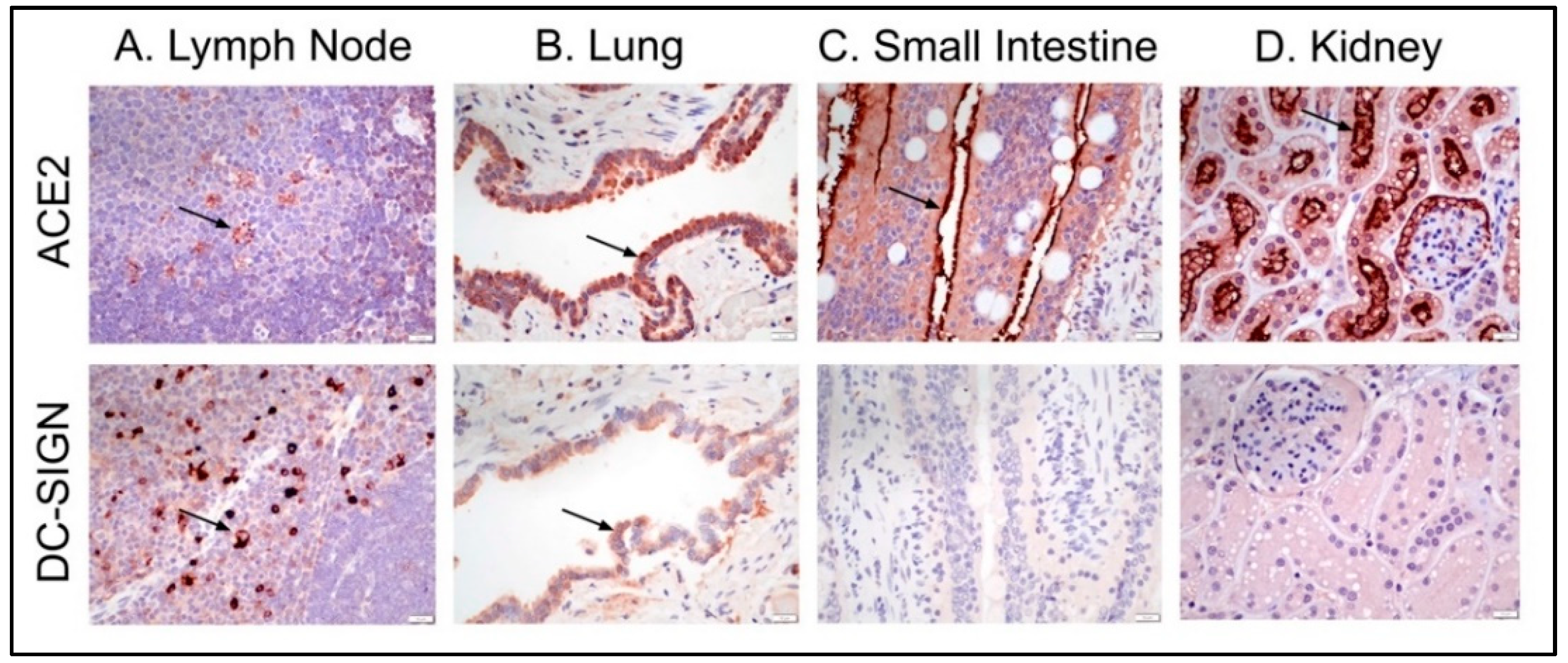
3.3. Feline Tissues and Cellular Distribution of ACE2 and DC-SIGN Proteins via IHC
3.4. RNAseq Gene Expression Profiles of FCWF-4 ATCC and FCWF-4 CU Cell Lines
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pedersen, N.C.; Liu, H.; Dodd, K.A.; Pesavento, P.A. Significance of coronavirus mutants in feces and diseased tissues of cats suffering from feline infectious peritonitis. Viruses 2009, 1, 166–184. [Google Scholar] [CrossRef] [PubMed]
- Vennema, H.; Poland, A.; Foley, J.; Pedersen, N.C. Feline infectious peritonitis viruses arise by mutation from endemic feline enteric coronaviruses. Virology 1998, 243, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, L.G.; Griesemer, R.A. Feline infectious peritonitis. Vet. Pathol. 1966, 3, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Montali, R.J.; Strandberg, J.D. Extraperitoneal lesions in feline infectious peritonitis. Vet. Pathol 1972, 9, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Kipar, A.; May, H.; Menger, S.; Weber, M.; Leukert, W.; Reinacher, M. Morphologic features and development of granulomatous vasculitis in feline infectious peritonitis. Vet. Pathol. 2005, 42, 321–330. [Google Scholar] [CrossRef] [PubMed]
- King, A.; Adams, M.; Carstens, E.; Lefkowitz, E. Nidovirales. Virus Taxon. 2012, 784–794. Available online: https://www.sciencedirect.com/science/article/pii/B9780123846846000665 (accessed on 29 May 2022).
- Fiscus, S.A.; Teramoto, Y.A. Antigenic comparison of feline coronavirus isolates: Evidence for markedly different peplomer glycoproteins. J. Virol. 1987, 61, 2607–2613. [Google Scholar] [CrossRef] [PubMed]
- Rottier, P.J.M.; Nakamura, K.; Schellen, P.; Volders, H.; Haijema, B.J. Acquisition of macrophage tropism during the pathogenesis of feline infectious peritonitis is determined by mutations in the feline coronavirus spike protein. J. Virol. 2005, 79, 14122–14130. [Google Scholar] [CrossRef]
- Haijema, B.J.; Volders, H.; Rottier, P.J.M. Switching species tropism: An effective way to manipulate the feline coronavirus genome. J. Virol. 2003, 77, 4528–4538. [Google Scholar] [CrossRef] [PubMed]
- Terada, Y.; Matsui, N.; Noguchi, K.; Kuwata, R.; Shimoda, H.; Soma, T.; Mochizuki, M.; Maeda, K. Emergence of pathogenic coronaviruses in cats by homologous recombination between feline and canine coronaviruses. PLoS ONE 2014, 9, e106534. [Google Scholar] [CrossRef]
- Herrewegh, A.A.P.M.; Smeenk, I.; Horzinek, M.C.; Rottier, P.J.M.; de Groot, R.J. Feline Coronavirus Type II Strains 79-1683 and 79-1146 originate from a double recombination between feline coronavirus type I and canine coronavirus. J. Virol. 1998, 72, 4508–4514. [Google Scholar] [CrossRef] [PubMed]
- Jaimes, J.A.; Millet, J.K.; Stout, A.E.; André, N.M.; Whittaker, G.R. A tale of two viruses: The distinct spike glycoproteins of feline coronaviruses. Viruses 2020, 12, 83. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Liu, Q.; Kong, F.; Guo, D.; Zhai, J.; Su, M.; Sun, D. Circulation and genetic diversity of Feline coronavirus type I and II from clinically healthy and FIP-suspected cats in China. Transbound. Emerg. Dis. 2019, 66, 763–775. [Google Scholar] [CrossRef] [PubMed]
- Kummrow, M.; Meli, M.L.; Haessig, M.; Goenczi, E.; Poland, A.; Pedersen, N.C.; Hofmann-Lehmann, R.; Lutz, H. Feline coronavirus serotypes 1 and 2: Seroprevalence and association with disease in Switzerland. Clin. Diagn. Lab. Immunol. 2005, 12, 1209–1215. [Google Scholar] [CrossRef] [PubMed]
- Benetka, V.; Kübber-Heiss, A.; Kolodziejek, J.; Nowotny, N.; Hofmann-Parisot, M.; Möstl, K. Prevalence of feline coronavirus types I and II in cats with histopathologically verified feline infectious peritonitis. Vet. Microbiol. 2004, 99, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Tresnan, D.B.; Levis, R.; Holmes, K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996, 70, 8669–8674. [Google Scholar] [CrossRef]
- Cham, T.-C.; Chang, Y.-C.; Tsai, P.-S.; Wu, C.-H.; Chen, H.-W.; Jeng, C.-R.; Pang, V.F.; Chang, H.-W. Determination of the cell tropism of serotype 1 feline infectious peritonitis virus using the spike affinity histochemistry in paraffin-embedded tissues. Microbiol. Immunol. 2017, 61, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Dye, C.; Temperton, N.; Siddell, S.G. Type I feline coronavirus spike glycoprotien fails to recognize aminopeptidase N as a functional receptor on feline cell lines. J. Gen. Virol. 2007, 88, 1753–1760. [Google Scholar] [CrossRef]
- Hohdatsu, T.; Izumiya, Y.; Yokoyama, Y.; Kida, K.; Koyama, H. Differences in virus receptor for type I and type II feline infectious peritonitis virus. Arch. Virol. 1998, 143, 839–850. [Google Scholar] [CrossRef]
- Pedersen, N.C.; Black, J.W.; Boyle, J.F.; Evermann, J.F.; McKeirnan, A.J.; Ott, R.L. Pathogenic differences between various feline coronavirus isolates. In Molecular Biology and Pathogenesis of Coronaviruses; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1984; pp. 365–380. [Google Scholar]
- Black, J. Recovery and in vitro cultivation of a coronavirus from laboratory-induced cases of feline infectious peritonitis (FIP). Vet. Med. Small Anim. Clin. 1980, 75, 811–814. [Google Scholar] [PubMed]
- O’Brien, A.; Mettelman, R.C.; Volk, A.; André, N.M.; Whittaker, G.R.; Baker, S.C. Characterizing replication kinetics and plaque production of type I feline infectious peritonitis virus in three feline cell lines. Virology 2018, 525, 1–9. [Google Scholar] [CrossRef]
- Regan, A.D.; Millet, J.K.; Tse, L.V.; Chillag, Z.; Rinaldi, V.; Licitra, B.N.; Dubovi, E.J.; Town, C.D.; Whittaker, G.R. Characterization of a recombinant canine coronavirus with a distinct receptor-binding (S1) domain. Virology 2012, 430, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Regan, A.D.; Ousterout, D.G.; Whittaker, G.R. Feline lectin activity is critical for the cellular entry of feline infectious peritonitis virus. J. Virol. 2010, 84, 7917–7921. [Google Scholar] [CrossRef]
- Hay, R.J.; Williams, C.D.; Macy, M.L.; Lavappa, K.S. Cultured cell lines for research on pulmonary physiology available through the American type culture collection. Am. Rev. Respir. Dis. 1982, 125, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Jacobse-Geels, H.E.L.; Horzinek, M.C. Expression of Feline Infectious Peritonitis Coronavirus Antigens on the Surface of Feline Macrophage-like Cells. J. Gen. Virol. 1983, 64, 1859–1866. [Google Scholar] [CrossRef] [PubMed]
- American Type Culture Collection, 2013. Fcwf4 [Fcwf] (ATCC CRL2787). Available online: https://www.atcc.org/products/crl-2787 (accessed on 1 May 2022).
- Tekes, G.; Spies, D.; Bank-Wolf, B.; Thiel, V.; Thiel, H.-J. A Reverse genetics approach to study feline infectious peritonitis. J. Virol. 2012, 86, 6994–6998. [Google Scholar] [CrossRef]
- Bálint, A.; Farsang, A.; Zádori, Z.; Hornyák, Á.; Dencső, L.; Almazán, F.; Enjuanes, L.; Belák, S. Molecular characterization of feline infectious peritonitis virus strain DF-2 and studies of the role of ORF3abc in viral cell tropism. J. Virol. 2012, 86, 6258–6267. [Google Scholar] [CrossRef]
- Wang, N.; Shi, X.; Jiang, L.; Zhang, S.; Wang, D.; Tong, P.; Guo, D.; Fu, L.; Cui, Y.; Liu, X.; et al. Complex structure of MERS-CoV RBD with DPP4 986 Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013, 23, 986–993. [Google Scholar]
- Hofmann, H.; Pyrc, K.; van der Hoek, L.; Geier, M.; Berkhout, B.; Pöhlmann, S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA 2005, 102, 7988–7993. [Google Scholar] [CrossRef]
- Yeager, C. Identification of Aminopeptidase N as a Cellular Receptor for Human Coronavirus-229E. Ph.D. Thesis, Uniformed Servies University of the Health Sciences, Bethesda, MD, USA, 12 May 1992. [Google Scholar]
- Li, W.; Moore, M.J.; Vasilieva, N.; Sui, J.; Wong, S.K.; Berne, M.A.; Somasundaran, M.; Sullivan, J.L.; Luzuriaga, K.; Greenough, T.C.; et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 2003, 426, 450–454. Available online: www.nature.com/nature (accessed on 4 April 2022).
- Regan, A.D.; Whittaker, G.R. Utilization of DC-SIGN for entry of feline coronaviruses into host cells. J. Virol. 2008, 82, 11992–11996. [Google Scholar] [CrossRef]
- Cook, S.E.; Vogel, H.; Castillo, D.; Olsen, M.; Pedersen, N.; Murphy, B.G. Investigation of monotherapy and combined anticoronaviral therapies against feline coronavirus serotype II in vitro. J. Feline Med. Surg. 2021, 1098612X211048647. [Google Scholar] [CrossRef] [PubMed]
- Addie, D.D.; Schaap, I.; Nicolson, L.; Jarrett, O. Persistence and transmission of natural type I feline coronavirus infection. J. Gen. Virol. 2003, 84, 2735–2744. [Google Scholar] [CrossRef]
- Klevjer-Anderson, P.; Cheevers, W. Characterization of the infection of caprine synovial membrane cells by the retrovirus caprine arthritis-encephalitis virus. Virology 1981, 110, 113–119. [Google Scholar] [CrossRef]
- Landmann, R.; Müller, B.; Zimmerli, W. CD14, new aspects of ligand and signal diversity. Microbes Infect. 2000, 2, 295–304. [Google Scholar] [CrossRef]
- Mitani, S.; Yabuki, A.; Sawa, M.; Chang, H.-S.; Yamato, O. Intrarenal distributions and changes of angiotensin-converting enzyme and angiotensin-converting enzyme 2 in feline and canine chronic kidney disease. J. Vet. Med. Sci. 2014, 76, 45–50. [Google Scholar] [CrossRef]
- Brand, J.M.A.V.D.; Haagmans, B.L.; Leijten, L.; van Riel, D.; Martina, B.E.E.; Osterhaus, A.D.M.E.; Kuiken, T. Pathology of experimental SARS coronavirus infection in cats and ferrets. Vet. Pathol. 2008, 45, 551–562. [Google Scholar] [CrossRef]
- HTStream. Available online: https://github.com/s4hts/HTStream (accessed on 22 April 2022).
- Smith, T.; Heger, A.; Sudbery, I. UMI-tools: Modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Res. 2017, 27, 491–499. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. Sequence analysis STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. Sequence analysis featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Ritchie, M.E.; Belinda, P.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Dodge, Y. Kolmogorov-Smirnov Test. In The Concise Encyclopedia of Statistics; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2008. [Google Scholar] [CrossRef]
- topGO: Enrichment Analysis for Gene Ontology Version from Bioconductor. Available online: https://rdrr.io/bioc/topGO/ (accessed on 5 May 2022).
- Haynes, W. Wilcoxon rank sum test BT—Encyclopedia of systems biology. In Encyclopedia of Systems Biology; Dubitzky, W., Wolkenhauer, O., Cho, K.-H., and Yokota, H., Eds.; Springer: New York, NY, USA, 2013; pp. 2354–2355. [Google Scholar]
- KEGGREST: Client-Side REST Access to the Kyoto Encyclopedia of Genes and Genomes (KEGG) Version 1.30.1 from Bioconductor. Available online: https://rdrr.io/bioc/KEGGREST/ (accessed on 5 May 2022).
- Shirato, K.; Chang, H.-W.; Rottier, P.J. Differential susceptibility of macrophages to serotype II feline coronaviruses correlates with differences in the viral spike protein. Virus Res. 2018, 255, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Van Hamme, E.; Desmarets, L.; Dewerchin, H.L.; Nauwynck, H.J. Intriguing interplay between feline infectious peritonitis virus and its receptors during entry in primary feline monocytes. Virus Res. 2011, 160, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, G. Development of Improved Cell Culture Systems for Feline Coronavirus and FIP Vaccine Development. Available online: https://www.vet.cornell.edu/research/awards/202011/development-improved-cell-culture-systems-feline-coronavirus-and-fip-vaccine-development (accessed on 26 May 2022).
- Castell-Rodríguez, A. Dendritic cells: Location, function, and clinical implications. In Biology of Myelomonocytic Cells; Piñón-Zárate, G., Ed.; IntechOpen: Rijeka, Croatia, 2017. [Google Scholar]
- Xiang, Q.; Feng, Z.; Diao, B.; Tu, C.; Qiao, Q.; Yang, H.; Zhang, Y.; Wang, G.; Wang, H.; Wang, C.; et al. SARS-CoV-2 Induces Lymphocytopenia by Promoting Inflammation and Decimates Secondary Lymphoid Organs. Front. Immunol. 2021, 12, 661052. Available online: https://www.frontiersin.org/article/10.3389/fimmu.2021.661052 (accessed on 29 May 2022). [CrossRef]
- Bosch, B.J.; Bartelink, W.; Rottier, P.J.M. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 2008, 82, 8887–8890. [Google Scholar] [CrossRef]
- Qiu, Z.; Hingley, S.T.; Simmons, G.; Yu, C.; Das Sarma, J.; Bates, P.; Weiss, S.R. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J. Virol. 2006, 80, 5768–5776. [Google Scholar] [CrossRef]
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134. [Google Scholar] [CrossRef]
- Peacock, T.P.; Goldhill, D.H.; Zhou, J.; Baillon, L.; Frise, R.; Swann, O.C.; Kugathasan, R.; Penn, R.; Brown, J.C.; David, R.Y.S.; et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat. Microbiol. 2021, 6, 899–909. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven article SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef]
- Zhou, Y.; Vedantham, P.; Lu, K.; Agudelo, J.; Carrion, R., Jr.; Nunneley, J.W.; Barnard, D.; Pöhlmann, S.; McKerrow, J.H.; Renslo, A.R.; et al. Protease inhibitors targeting coronavirus and filovirus entry. Antivir. Res. 2015, 116, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, M.; Itakura, Y.; Kishimoto, M.; Tabata, K.; Uemura, K.; Ito, N.; Sugiyama, M.; Wastika, C.E.; Orba, Y.; Sawa, H. Host serine proteases TMPRSS2 and TMPRSS11D mediate proteolytic activation and trypsin-independent infection in group A rotaviruses. J. Virol. 2021, 95, e00398-21. [Google Scholar] [CrossRef]
- Bonaparte, M.I.; Dimitrov, A.S.; Bossart, K.N.; Crameri, G.; Mungall, B.A.; Bishop, K.A.; Choudhry, V.; Dimitrov, D.S.; Wang, L.-F.; Eaton, B.T.; et al. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. USA 2005, 102, 10652–10657. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zheng, X.; Peng, Q.; Zhang, X.; Qin, Z. Eph receptors: The bridge linking host and virus. Cell Mol. Life Sci. 2020, 77, 2355–2365. [Google Scholar] [CrossRef] [PubMed]
- de Boer, E.C.; van Gils, J.M.; van Gils, M.J. Ephrin-Eph signaling usage by a variety of viruses. Pharmacol. Res. 2020, 159, 105038. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Wang, H.-B.; Zhang, A.; Chen, M.-L.; Fang, Z.-X.; Dong, X.-D.; Li, S.-B.; Du, Y.; Xiong, D.; et al. Ephrin receptor A2 is an epithelial cell receptor for Epstein–Barr virus entry. Nat. Microbiol. 2018, 3, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Zalpoor, H.; Akbari, A.; Samei, A.; Forghaniesfidvajani, R.; Kamali, M.; Afzalnia, A.; Manshouri, S.; Heidari, F.; Pornour, M.; Khoshmirsafa, M.; et al. The roles of Eph receptors, neuropilin-1, P2X7, and CD147 in COVID-19-associated neurodegenerative diseases: Inflammasome and JaK inhibitors as potential promising therapies. Cell Mol. Biol. Lett. 2022, 27, 10. [Google Scholar] [CrossRef]
- Beaudoin, C.A.; Jamasb, A.R.; Alsulami, A.F.; Copoiu, L.; van Tonder, A.J.; Hala, S.; Bannerman, B.P.; Thomas, S.E.; Vedithi, S.C.; Torres, P.H.M.; et al. Predicted structural mimicry of spike receptor-binding motifs from highly pathogenic human coronaviruses. Comput. Struct. Biotechnol. J. 2021, 19, 3938–3953. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Keidar, S.; Gamliel-Lazarovich, A.; Kaplan, M.; Pavlotzky, E.; Hamoud, S.; Hayek, T.; Karry, R.; Abassi, Z. Mineralocorticoid receptor blocker increases angiotensin-converting Enzyme 2 activity in congestive heart failure patients. Circ. Res. 2005, 97, 946–953. [Google Scholar] [CrossRef]
- Song, X.; Hu, W.; Yu, H.; Zhao, L.; Zhao, Y.; Zhao, Y. High expression of angiotensin-converting enzyme-2 (ACE2) on tissue macrophages that may be targeted by virus SARS-CoV-2 in COVID-19 patients. bioRxiv 2020, 1–27. [Google Scholar] [CrossRef]
- Song, X.; Hu, W.; Yu, H.; Zhao, L.; Zhao, Y.; Zhao, X.; Xue, H.; Zhao, Y. Little to no expression of angiotensin-converting enzyme-2 on most human peripheral blood immune cells but highly expressed on tissue macrophages. Cytom. Part A J. Int. Soc. Anal. Cytol. 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Hikmet, F.; Méar, L.; Edvinsson, Å.; Micke, P.; Uhlén, M.; Lindskog, C. The protein expression profile of ACE2 in human tissues. Mol. Syst. Biol. 2020, 16, e9610. [Google Scholar] [CrossRef] [PubMed]
- Hamming, I.; Timens, W.; Bulthuis, M.L.C.; Lely, A.T.; Navis, G.J.; van Goor, H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004, 203, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Lean, F.Z.X.; Núñez, A.; Spiro, S.; Priestnall, S.L.; Vreman, S.; Bailey, D.; James, J.; Wrigglesworth, E.; Suarez-Bonnet, A.; Conceicao, C.; et al. Differential susceptibility of SARS-CoV-2 in animals: Evidence of ACE2 host receptor distribution in companion animals, livestock and wildlife by immunohistochemical characterisation. Transbound. Emerg. Dis. 2021, 1–12. [Google Scholar] [CrossRef]
- Chiocchetti, R.; Galiazzo, G.; Fracassi, F.; Giancola, F.; Pietra, M. ACE2 expression in the cat and the tiger gastrointestinal tracts. Front. Vet. Sci. 2020, 7, 514. [Google Scholar] [CrossRef]
- Van Hamme, E.; Dewerchin, H.L.; Cornelissen, E.; Nauwynck, H.J. Attachment and internalization of feline infectious peritonitis virus in feline blood monocytes and Crandell feline kidney cells. J. Gen. Virol. 2007, 88, 2527–2532. [Google Scholar] [CrossRef]
- Granelli-Piperno, A.; Pritsker, A.; Pack, M.; Shimeliovich, I.; Arrighi, J.-F.; Park, C.G.; Trumpfheller, C.; Piguet, V.; Moran, T.M.; Steinman, R.M. Dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin/CD209 is abundant on macrophages in the normal human lymph node and is not required for dendritic cell stimulation of the mixed leukocyte reaction. J. Immunol. 2005, 175, 4265–4273. [Google Scholar] [CrossRef]
- Gioia, M.; Ciaccio, C.; Calligari, P.; De Simone, G.; Sbardella, D.; Tundo, G.; Fasciglione, G.F.; Di Masi, A.; Di Pierro, D.; Bocedi, A.; et al. Role of proteolytic enzymes in the COVID-19 infection and promising therapeutic approaches. Biochem. Pharmacol. 2020, 182, 114225. [Google Scholar]
- Kishimoto, M.; Uemura, K.; Sanaki, T.; Sato, A.; Hall, W.; Kariwa, H.; Orba, Y.; Sawa, H.; Sasaki, M. TMPRSS11D and TMPRSS13 Activate the SARS-CoV-2 Spike Protein. Viruses 2021, 13, 384. [Google Scholar] [CrossRef]
- Yadati, T.; Houben, T.; Bitorina, A.; Shiri-Sverdlov, R. The ins and outs of cathepsins: Physiological function and role in disease management. Cells 2020, 9, 1679. [Google Scholar] [CrossRef] [PubMed]
- Kawase, M.; Shirato, K.; Matsuyama, S.; Taguchi, F. Protease-mediated entry via the endosome of human coronavirus 229E. J. Virol. 2009, 83, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.; Gosalia, D.N.; Rennekamp, A.J.; Reeves, J.D.; Diamond, S.L.; Bates, P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA 2005, 102, 11876–11881. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.M.; Yang, W.L.; Yang, F.Y.; Zhang, L.; Huang, W.J.; Hou, W.; Fan, C.F.; Jin, R.H.; Feng, Y.M.; Wang, Y.C.; et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct. Target Ther. 2021, 6, 134. [Google Scholar] [CrossRef] [PubMed]
- Golden, J.; Bahe, J.A.; Lucas, W.T.; Nibert, M.L.; Schiff, L.A. Cathepsin S supports acid-independent infection by some reoviruses. J. Biol. Chem. 2004, 279, 8547–8557. [Google Scholar] [CrossRef]
- Tuzi, N.L.; Gullick, W.J. eph, the largest known family of putative growth factor receptors. Br. J. Cancer 1994, 69, 417–421. [Google Scholar] [CrossRef][Green Version]
- Bowden, T.; Aricescu, A.R.; Gilbert, R.; Grimes, J.M.; Jones, Y.; Stuart, D. Structural basis of Nipah and Hendra virus attachment to their cell-surface receptor ephrin-B2. Nat. Struct. Mol. Biol. 2008, 15, 567–572. [Google Scholar] [CrossRef]
- Lupberger, J.; Zeisel, M.B.; Xiao, F.; Thumann, C.; Fofana, I.; Zona, L.; Davis, C.; Mee, C.; Turek, M.; Gorke, S.; et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 2011, 17, 589–595. [Google Scholar] [CrossRef]
- Egal, E.; Helms, M.N.; Liou, T.G.; Mimche, P. Ephrin ligands are upregulated in the saliva of SARS-CoV-2 infected patients. FASEB J. 2022, 36. [Google Scholar] [CrossRef]

| Genus | Virus | Species | Receptor |
|---|---|---|---|
| Alpha | HCoV-NL63 | Human | ACE2 |
| 229E | Human | APN | |
| TGEV | Pig | APN | |
| PRCoV | Pig | APN | |
| FCoV Serotype I | Cat | ND | |
| FCoV Serotype II | Cat | APN, DC-SIGN | |
| CCoV | Dog | APN | |
| Beta | SARS-CoV-1 | Human | ACE2 |
| SARS-CoV-2 | Human | ACE2, DC-SIGN | |
| MERS-CoV | Human | DPP4 |
| Target | Sequence (5′-3′) | Annealing Temperature (°C) | Size (bp) | |
|---|---|---|---|---|
| FIPV | Forward | GGAAGTTTAGATTTGATTTGGCAATGCTAG | 58 | 112 |
| Reverse | AACAATCACTAGATCCAGACGTTAGCT | |||
| GAPDH | Forward | AAATTCCACGGCACAGTCAAG | 58 | 61 |
| Reverse | TGATGGGCTTTCCATTGATGA | |||
| CD16 | Forward | AACCAGCTTTCTGCTTCAGTATC | 51 | 98 |
| Reverse | AGACCTGAGCGCATAGAGTATC | |||
| ACE2 | Forward | GACAACAGCCTGGAGTTTCT | 51 | 134 |
| Reverse | GAGACGATGAGCAGGACAATAC | |||
| DC-SIGN | Forward | GGCCTTTAAGATCCTGAGAGAGA | 51 | 102 |
| Reverse | GCTGAGACCATCAACAGAAGAAG | |||
| feAPN | Forward | GGTGTTTGACTCCATCTCCTAC | 60 | 134 |
| Reverse | GGTAGATGGTGTTCCCGTATTT | |||
| DPP4 | Forward | ATGCAAGCACCGACTTCTAA | 55 | 117 |
| Reverse | CGTAGCACCTCTAGCCATAAC | |||
| FCoV I S For (lffs type I) | Forward | GTTTCAACCTAGAAAGCCTCAGAT | 50 | 376 |
| FCoV II S For (lcfs type II) | Forward | GCCTAGTATTATACCTGACTA | 283 | |
| FCoV I and II S Rev (lubs) | Reverse | CCACACATACCAAGGCC | ||
| nFCoV I S For (nlffles type I) | Forward | CCTAGAAAGCCTCAGATGAGTG | 47 | 360 |
| nFCoV II S For (nlcfs type II) | Forward | CAGACCAAACTGGACTGTAC | 218 | |
| nFCoV I and II S Rev (nlubs) | Reverse | CCAAGGCCATTTTACATA |
| Tissue | Cell | ACE2 | DC-SIGN |
|---|---|---|---|
| Cerebrum | Neurons | 0 | 0 |
| Glial cells | +0-2 (C) | 0 | |
| Cerebellum | Purkinje cells | 0 | 0 |
| Granular cells | 0 | 0 | |
| Choroid Plexus | Ependymal cells | +1 (C) | 0 |
| Meninges | Fibroblasts | +1 (C) | 0 |
| Eye | Ganglion cells | +2 (C) | 0 |
| Nuclear layer of retina | +0-1 (C) | 0 | |
| Fibroblasts | +2 (C) | 0 | |
| Pigmented epithelium | U | U | |
| Melanocytes | U | U | |
| Corneal epithelium | +0-1 (C) | 0 | |
| Keratocytes | 0 | 0 | |
| Ciliary body epithelium | +0-1 (C) | 0 | |
| Lens epithelium | +1 (C) | 0 | |
| Optic nerve | Glial cells | +2 (C) | 0 |
| Thyroid | Follicular cells | +2-3 (C) | +2 (C) |
| Parafollicular cells | +1 (C) | +0-1 (C) | |
| Parathyroid | Chief and oxyphil cells | 0 | 0 |
| Adrenal gland | Cortex | +0-2 (C) | +1 (C; reticularis) |
| Medulla | +1 (C) | 0 | |
| Lymph node | Paracortex and follicles (scattered cells) | +2 (C) | +3 (C, M) |
| Adipose tissue | Adipocyte | +2 (C) | +1 (C) |
| Skeletal muscle | Myocyte | +1 (C) | 0 |
| Smooth muscle | Myocyte | +0-1 (C) | 0 |
| Tongue | Epithelium | +2-3 (C, M) | 0 |
| Esophagus | Epithelium | +2 (M) | 0 |
| Trachea | Airway epithelium | +2 (C) | 0 |
| Submucosal gland epithelium | +2 (C) | 0 | |
| Lung | Bronchial/bronchiolar epithelium | +2-3 (C) | +1-2 (C) |
| Type I pneumocyte | +1-2 (U) | +1-2 (U) | |
| Type II pneumocyte | +2 (C) | +1 (C) | |
| Submucosal gland epithelium | +1-2 (C) | 0 | |
| Heart | Myocyte | +0-1 (C) | +0-1 (C) |
| Vessels | Endothelium | +1-2 (C) | 0 |
| Mural smooth muscle | +2 (C) | 0 | |
| Liver | Hepatocyte | +1 (C) | 0 |
| Biliary epithelium | 0 | 0 | |
| Kidney | Cortical tubular epithelium | +3 (M, apical); +1 (C) | 0 |
| Medullary tubular epithelium | +1 (C) | 0 | |
| Spleen | Red pulp, scattered cells | +1-2 (C) | +2-3 (C) |
| White pulp (rare) | 0 | +1 (C) | |
| Pancreas | Acinar cells | +0-1 (C) | 0 |
| Islet cells | 0 | 0 | |
| Stomach | Mucosal epithelium | +2 (C) | 0 |
| Small Intestine | Mucosal epithelium, apical | +3 (M); +1 (C) | 0 |
| Ileum | Mucosal epithelium, apical | +3 (M); +1 (C) | 0 |
| Colon | Mucosal epithelium | +3 (C) | 0 |
| Intestine as a whole | Lamina propria, scattered cells | +3 (C) | +3 (C) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cook, S.; Castillo, D.; Williams, S.; Haake, C.; Murphy, B. Serotype I and II Feline Coronavirus Replication and Gene Expression Patterns of Feline Cells—Building a Better Understanding of Serotype I FIPV Biology. Viruses 2022, 14, 1356. https://doi.org/10.3390/v14071356
Cook S, Castillo D, Williams S, Haake C, Murphy B. Serotype I and II Feline Coronavirus Replication and Gene Expression Patterns of Feline Cells—Building a Better Understanding of Serotype I FIPV Biology. Viruses. 2022; 14(7):1356. https://doi.org/10.3390/v14071356
Chicago/Turabian StyleCook, Sarah, Diego Castillo, Sonyia Williams, Christine Haake, and Brian Murphy. 2022. "Serotype I and II Feline Coronavirus Replication and Gene Expression Patterns of Feline Cells—Building a Better Understanding of Serotype I FIPV Biology" Viruses 14, no. 7: 1356. https://doi.org/10.3390/v14071356
APA StyleCook, S., Castillo, D., Williams, S., Haake, C., & Murphy, B. (2022). Serotype I and II Feline Coronavirus Replication and Gene Expression Patterns of Feline Cells—Building a Better Understanding of Serotype I FIPV Biology. Viruses, 14(7), 1356. https://doi.org/10.3390/v14071356







