Sequence-Specific Features of Short Double-Strand, Blunt-End RNAs Have RIG-I- and Type 1 Interferon-Dependent or -Independent Anti-Viral Effects
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Viruses
2.3. dsRNA Oligos
2.4. Transfections and Infections
2.5. Imaging and Image Analysis
2.6. Gene Expression Profiling of miR29b-1*-Transfected A549 Cells
2.7. Measurement of IFN Production from dsRNA-Transfected Cells
2.8. siRNA Transfections
2.9. RT-qPCR
3. Results
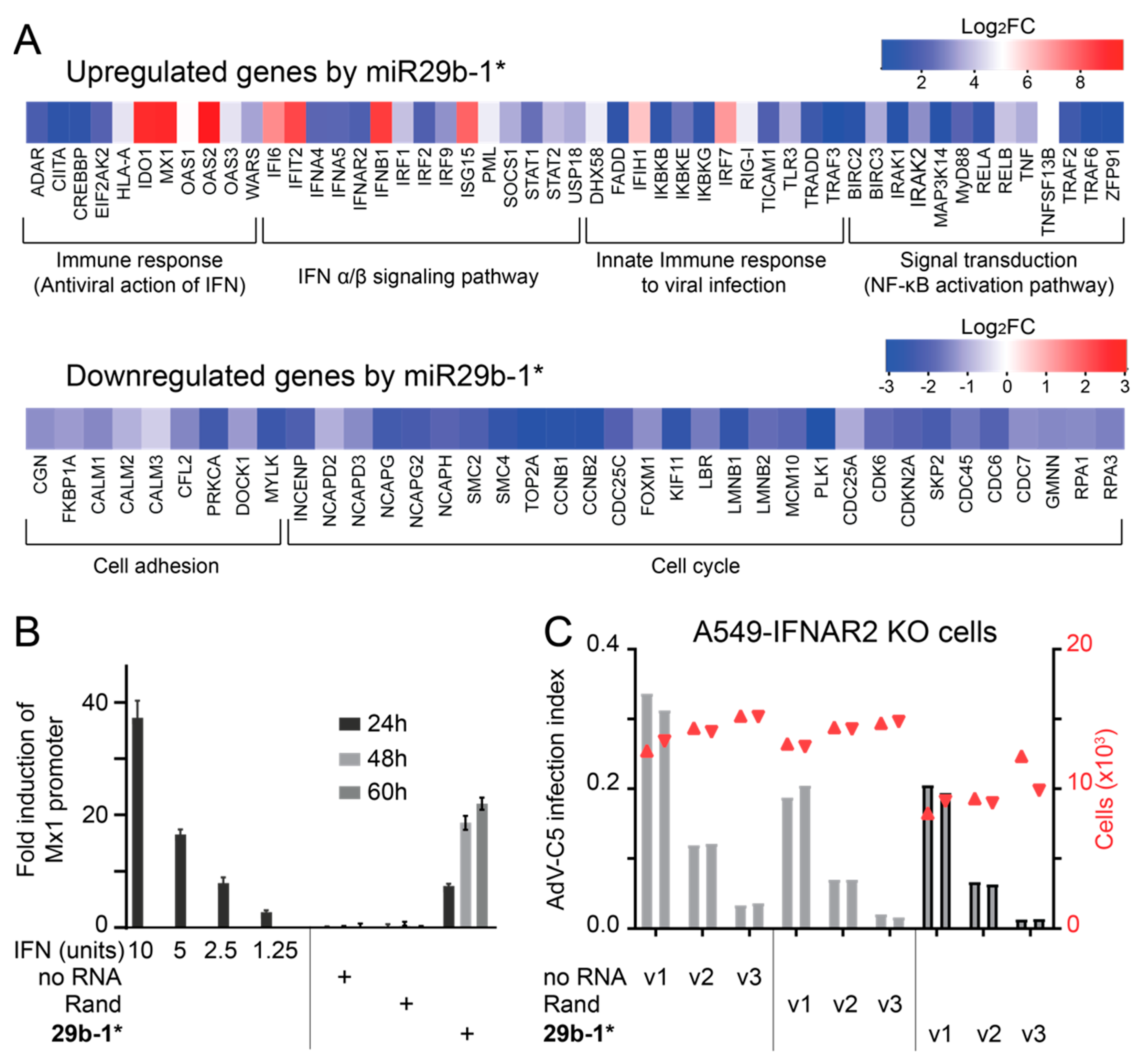
3.1. A Short Blunt-End dsRNA Oligo with hsa-miR-29b-1* Sequence Reduces AdV-C5 Infection
3.2. Blunt-End dsRNA miR29b-1* Mimic Induces Interferon Response When Transfected into A549 Cells
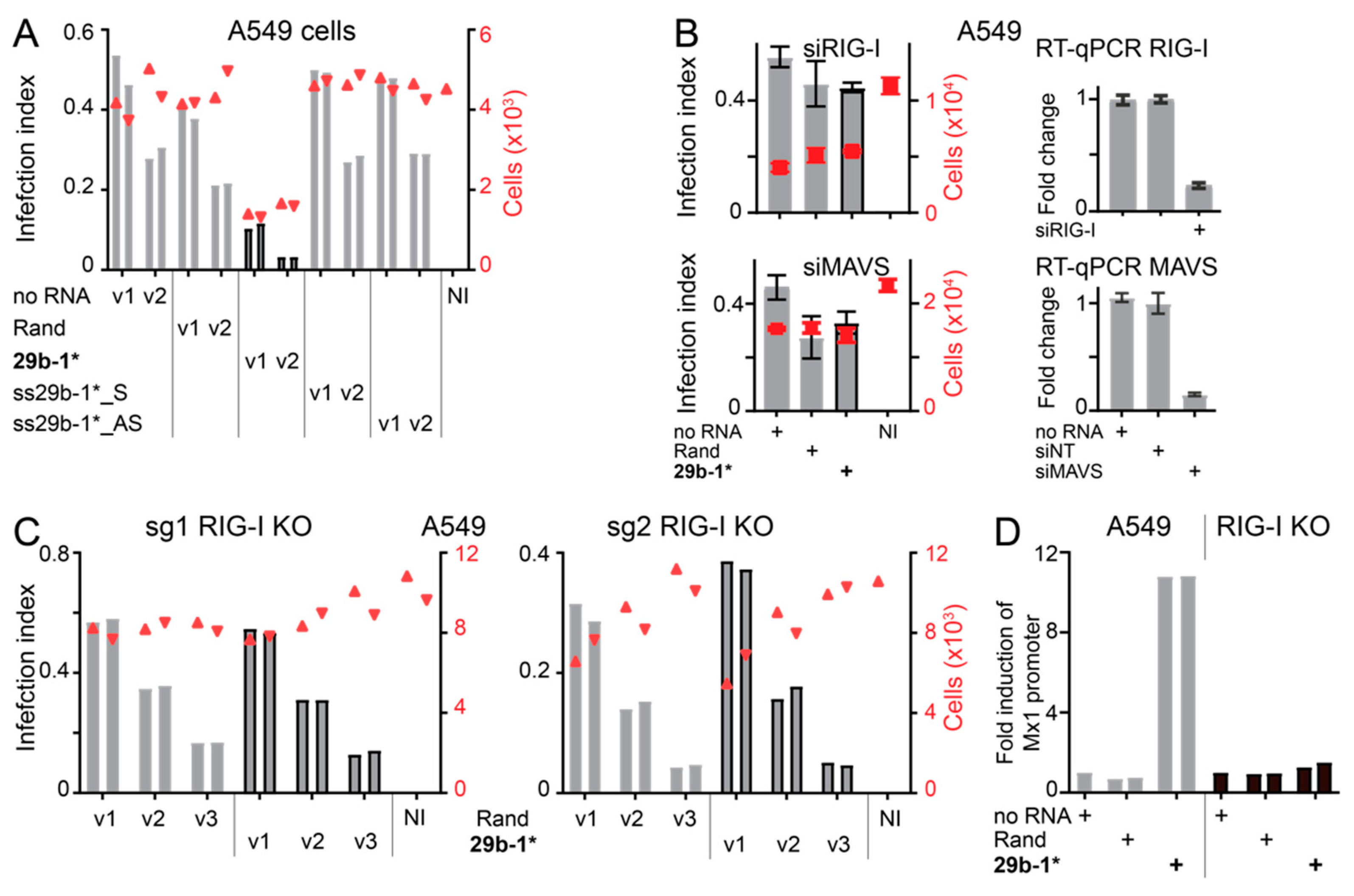
3.3. Anti-Viral Effects of Blunt-End miR29b-1* Mimic Are Mediated by RIG-I
3.4. Identification of miR-29b-1* Mimic-Related and Unrelated dsRNAs Inhibiting AdV-C5 Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iwakawa, H.O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.; Flatt, J.W.; Seiler, D.; Cardel, B.; Emmenlauer, M.; Boucke, K.; Suomalainen, M.; Hemmi, S.; Greber, U.F. The E3 Ubiquitin Ligase Mind Bomb 1 Controls Adenovirus Genome Release at the Nuclear Pore Complex. Cell Rep. 2019, 29, 3785–3795.e3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-Sancho, L.; Tripathi, S.; Rodriguez-Frandsen, A.; Pache, L.; Sanchez-Aparicio, M.; McGregor, M.J.; Haas, K.M.; Swaney, D.L.; Nguyen, T.T.; Mamede, J.I.; et al. Restriction factor compendium for influenza A virus reveals a mechanism for evasion of autophagy. Nat. Microbiol. 2021, 6, 1319–1333. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Kriegel, A.J.; Liu, Y.; Fang, Y.; Ding, X.; Liang, M. The miR-29 family: Genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol. Genom. 2012, 44, 237–244. [Google Scholar] [CrossRef]
- Watanabe, T.; Watanabe, S.; Kawaoka, Y. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 2010, 7, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Karlas, A.; Machuy, N.; Shin, Y.; Pleissner, K.P.; Artarini, A.; Heuer, D.; Becker, D.; Khalil, H.; Ogilvie, L.A.; Hess, S.; et al. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 2010, 463, 818–822. [Google Scholar] [CrossRef]
- Munk, C.; Sommer, A.F.; Konig, R. Systems-biology approaches to discover anti-viral effectors of the human innate immune response. Viruses 2011, 3, 1112–1130. [Google Scholar] [CrossRef] [Green Version]
- Stertz, S.; Shaw, M.L. Uncovering the global host cell requirements for influenza virus replication via RNAi screening. Microbes Infect. Inst. Pasteur 2011, 13, 516–525. [Google Scholar] [CrossRef] [Green Version]
- Snijder, B.; Sacher, R.; Ramo, P.; Liberali, P.; Mench, K.; Wolfrum, N.; Burleigh, L.; Scott, C.C.; Verheije, M.H.; Mercer, J.; et al. Single-cell analysis of population context advances RNAi screening at multiple levels. Mol. Syst. Biol. 2012, 8, 579. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, I.; Yamauchi, Y.; Helenius, A.; Horvath, P. High-content analysis of sequential events during the early phase of influenza A virus infection. PLoS ONE 2013, 8, e68450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, W.C.; Chen, Y.C.; Tseng, C.H.; Hsu, P.W.; Tung, K.F.; Jeng, K.S.; Lai, M.M. Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry. Proc. Natl. Acad. Sci. USA 2013, 110, 17516–17521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, S.; Pohl, M.O.; Zhou, Y.; Rodriguez-Frandsen, A.; Wang, G.; Stein, D.A.; Moulton, H.M.; DeJesus, P.; Che, J.; Mulder, L.C.; et al. Meta- and Orthogonal Integration of Influenza “OMICs” Data Defines a Role for UBR4 in Virus Budding. Cell Host Microbe 2015, 18, 723–735. [Google Scholar] [CrossRef] [Green Version]
- de Wilde, A.H.; Wannee, K.F.; Scholte, F.E.; Goeman, J.J.; Ten Dijke, P.; Snijder, E.J.; Kikkert, M.; van Hemert, M.J. A Kinome-Wide Small Interfering RNA Screen Identifies Proviral and Antiviral Host Factors in Severe Acute Respiratory Syndrome Coronavirus Replication, Including Double-Stranded RNA-Activated Protein Kinase and Early Secretory Pathway Proteins. J. Virol. 2015, 89, 8318–8333. [Google Scholar] [CrossRef] [Green Version]
- Ambike, S.; Cheng, C.C.; Feuerherd, M.; Velkov, S.; Baldassi, D.; Afridi, S.Q.; Porras-Gonzalez, D.; Wei, X.; Hagen, P.; Kneidinger, N.; et al. Targeting genomic SARS-CoV-2 RNA with siRNAs allows efficient inhibition of viral replication and spread. Nucleic Acids Res. 2022, 50, 333–349. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, M.; Pfeifer, G.; Binder, S.; Aigner, A.; Vollmer Barbosa, P.; Makert, G.R.; Fertey, J.; Ulbert, S.; Bodem, J.; Konig, E.M.; et al. Selection and Validation of siRNAs Preventing Uptake and Replication of SARS-CoV-2. Front. Bioeng. Biotechnol. 2022, 10, 801870. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Scherer, J.; Yi, J.; Vallee, R.B. Role of kinesins in directed adenovirus transport and cytoplasmic exploration. PLoS Pathog. 2018, 14, e1007055. [Google Scholar] [CrossRef]
- Hao, L.; He, Q.; Wang, Z.; Craven, M.; Newton, M.A.; Ahlquist, P. Limited agreement of independent RNAi screens for virus-required host genes owes more to false-negative than false-positive factors. PLoS Comput. Biol. 2013, 9, e1003235. [Google Scholar] [CrossRef] [Green Version]
- Ramo, P.; Drewek, A.; Arrieumerlou, C.; Beerenwinkel, N.; Ben-Tekaya, H.; Cardel, B.; Casanova, A.; Conde-Alvarez, R.; Cossart, P.; Csucs, G.; et al. Simultaneous analysis of large-scale RNAi screens for pathogen entry. BMC Genom. 2014, 15, 1162. [Google Scholar] [CrossRef] [Green Version]
- Greber, U.F.; Suomalainen, M. Adenovirus Entry—Stability, Uncoating and Nuclear Import. Mol. Microbiol. 2022. [CrossRef]
- Grundhoff, A.; Sullivan, C.S. Virus-encoded microRNAs. Virology 2011, 411, 325–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makkoch, J.; Poomipak, W.; Saengchoowong, S.; Khongnomnan, K.; Praianantathavorn, K.; Jinato, T.; Poovorawan, Y.; Payungporn, S. Human microRNAs profiling in response to influenza A viruses (subtypes pH1N1, H3N2, and H5N1). Exp. Biol. Med. 2016, 241, 409–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, L.; Zhang, X.; Wu, Z.; Huang, K.; Sun, X.; Chen, H.; Jin, M. The Downregulation of MicroRNA hsa-miR-340-5p in IAV-Infected A549 Cells Suppresses Viral Replication by Targeting RIG-I and OAS2. Mol. Ther. Nucleic Acids 2019, 14, 509–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, S.; Cullen, B.R. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J. Virol. 2004, 78, 12868–12876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersson, M.G.; Haasnoot, P.C.; Xu, N.; Berenjian, S.; Berkhout, B.; Akusjarvi, G. Suppression of RNA interference by adenovirus virus-associated RNA. J. Virol. 2005, 79, 9556–9565. [Google Scholar] [CrossRef] [Green Version]
- Aparicio, O.; Carnero, E.; Abad, X.; Razquin, N.; Guruceaga, E.; Segura, V.; Fortes, P. Adenovirus VA RNA-derived miRNAs target cellular genes involved in cell growth, gene expression and DNA repair. Nucleic Acids Res. 2010, 38, 750–763. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.; Segerman, B.; Zhou, X.; Akusjarvi, G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes. J. Virol. 2007, 81, 10540–10549. [Google Scholar] [CrossRef] [Green Version]
- Bellutti, F.; Kauer, M.; Kneidinger, D.; Lion, T.; Klein, R. Identification of RISC-associated adenoviral microRNAs, a subset of their direct targets, and global changes in the targetome upon lytic adenovirus 5 infection. J. Virol. 2015, 89, 1608–1627. [Google Scholar] [CrossRef] [Green Version]
- Pawlica, P.; Yario, T.A.; White, S.; Wang, J.; Moss, W.N.; Hui, P.; Vinetz, J.M.; Steitz, J.A. SARS-CoV-2 expresses a microRNA-like small RNA able to selectively repress host genes. Proc. Natl. Acad. Sci. USA 2021, 118, e2116668118. [Google Scholar] [CrossRef]
- Singh, M.; Chazal, M.; Quarato, P.; Bourdon, L.; Malabat, C.; Vallet, T.; Vignuzzi, M.; van der Werf, S.; Behillil, S.; Donati, F.; et al. A virus-derived microRNA targets immune response genes during SARS-CoV-2 infection. EMBO Rep. 2022, 23, e54341. [Google Scholar] [CrossRef]
- Farr, R.J.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.H.O.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.A.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009759. [Google Scholar] [CrossRef] [PubMed]
- Siniscalchi, C.; Di Palo, A.; Russo, A.; Potenza, N. Human MicroRNAs Interacting With SARS-CoV-2 RNA Sequences: Computational Analysis and Experimental Target Validation. Front. Genet. 2021, 12, 678994. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Lowey, B.; Sodroski, C.; Krishnamurthy, S.; Alao, H.; Cha, H.; Chiu, S.; El-Diwany, R.; Ghany, M.G.; Liang, T.J. Cellular microRNA networks regulate host dependency of hepatitis C virus infection. Nat. Commun. 2017, 8, 1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birmingham, A.; Anderson, E.M.; Reynolds, A.; Ilsley-Tyree, D.; Leake, D.; Fedorov, Y.; Baskerville, S.; Maksimova, E.; Robinson, K.; Karpilow, J.; et al. 3’ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods 2006, 3, 199–204. [Google Scholar] [CrossRef]
- Jackson, A.L.; Burchard, J.; Schelter, J.; Chau, B.N.; Cleary, M.; Lim, L.; Linsley, P.S. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 2006, 12, 1179–1187. [Google Scholar] [CrossRef] [Green Version]
- Franceschini, A.; Meier, R.; Casanova, A.; Kreibich, S.; Daga, N.; Andritschke, D.; Dilling, S.; Ramo, P.; Emmenlauer, M.; Kaufmann, A.; et al. Specific inhibition of diverse pathogens in human cells by synthetic microRNA-like oligonucleotides inferred from RNAi screens. Proc. Natl. Acad. Sci. USA 2014, 111, 4548–4553. [Google Scholar] [CrossRef] [Green Version]
- Goodchild, A.; Nopper, N.; King, A.; Doan, T.; Tanudji, M.; Arndt, G.M.; Poidinger, M.; Rivory, L.P.; Passioura, T. Sequence determinants of innate immune activation by short interfering RNAs. BMC Immunol. 2009, 10, 40. [Google Scholar] [CrossRef] [Green Version]
- Hornung, V.; Guenthner-Biller, M.; Bourquin, C.; Ablasser, A.; Schlee, M.; Uematsu, S.; Noronha, A.; Manoharan, M.; Akira, S.; de Fougerolles, A.; et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005, 11, 263–270. [Google Scholar] [CrossRef]
- Judge, A.D.; Sood, V.; Shaw, J.R.; Fang, D.; McClintock, K.; MacLachlan, I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005, 23, 457–462. [Google Scholar] [CrossRef]
- Marques, J.T.; Devosse, T.; Wang, D.; Zamanian-Daryoush, M.; Serbinowski, P.; Hartmann, R.; Fujita, T.; Behlke, M.A.; Williams, B.R. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006, 24, 559–565. [Google Scholar] [CrossRef]
- Barreau, C.; Dutertre, S.; Paillard, L.; Osborne, H.B. Liposome-mediated RNA transfection should be used with caution. RNA 2006, 12, 1790–1793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomson, D.W.; Bracken, C.P.; Szubert, J.M.; Goodall, G.J. On measuring miRNAs after transient transfection of mimics or antisense inhibitors. PLoS ONE 2013, 8, e55214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sioud, M. Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: A central role for 2’-hydroxyl uridines in immune responses. Eur. J. Immunol. 2006, 36, 1222–1230. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W. Interferon-Stimulated Genes: What Do They All Do? Annu. Rev. Virol. 2019, 6, 567–584. [Google Scholar] [CrossRef] [PubMed]
- Carty, M.; Guy, C.; Bowie, A.G. Detection of Viral Infections by Innate Immunity. Biochem. Pharmacol. 2021, 183, 114316. [Google Scholar] [CrossRef] [PubMed]
- Rehwinkel, J.; Gack, M.U. RIG-I-like receptors: Their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020, 20, 537–551. [Google Scholar] [CrossRef]
- Weber, M.; Gawanbacht, A.; Habjan, M.; Rang, A.; Borner, C.; Schmidt, A.M.; Veitinger, S.; Jacob, R.; Devignot, S.; Kochs, G.; et al. Incoming RNA Virus Nucleocapsids Containing a 5’-Triphosphorylated Genome Activate RIG-I and Antiviral Signaling. Cell Host Microbe 2013, 13, 336–346. [Google Scholar] [CrossRef] [Green Version]
- Hur, S. Double-Stranded RNA Sensors and Modulators in Innate Immunity. Annu. Rev. Immunol. 2019, 37, 349–375. [Google Scholar] [CrossRef]
- Vignuzzi, M.; Lopez, C.B. Defective viral genomes are key drivers of the virus-host interaction. Nat. Microbiol. 2019, 4, 1075–1087. [Google Scholar] [CrossRef]
- Suomalainen, M.; Greber, U.F. Virus Infection Variability by Single-Cell Profiling. Viruses 2021, 13, 1568. [Google Scholar] [CrossRef]
- Feng, Q.; Hato, S.V.; Langereis, M.A.; Zoll, J.; Virgen-Slane, R.; Peisley, A.; Hur, S.; Semler, B.L.; van Rij, R.P.; van Kuppeveld, F.J. MDA5 detects the double-stranded RNA replicative form in picornavirus-infected cells. Cell Rep. 2012, 2, 1187–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, X.; Riva, L.; Pu, Y.; Martin-Sancho, L.; Kanamune, J.; Yamamoto, Y.; Sakai, K.; Gotoh, S.; Miorin, L.; De Jesus, P.D.; et al. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021, 34, 108628. [Google Scholar] [CrossRef] [PubMed]
- Onomoto, K.; Onoguchi, K.; Yoneyama, M. Regulation of RIG-I-like receptor-mediated signaling: Interaction between host and viral factors. Cell. Mol. Immunol. 2021, 18, 539–555. [Google Scholar] [CrossRef] [PubMed]
- Vavassori, S.; Chou, J.; Faletti, L.E.; Haunerdinger, V.; Opitz, L.; Joset, P.; Fraser, C.J.; Prader, S.; Gao, X.; Schuch, L.A.; et al. Multisystem inflammation and susceptibility to viral infections in human ZNFX1 deficiency. J. Allergy Clin. Immunol. 2021, 148, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Volkmer, B.; Planas, R.; Gossweiler, E.; Lunemann, A.; Opitz, L.; Mauracher, A.; Nuesch, U.; Gayden, T.; Kaiser, D.; Drexel, B.; et al. Recurrent inflammatory disease caused by a heterozygous mutation in CD48. J. Allergy Clin. Immunol. 2019, 144, 1441–1445.e1417. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murer, L.; Volle, R.; Andriasyan, V.; Petkidis, A.; Gomez-Gonzalez, A.; Yang, L.; Meili, N.; Suomalainen, M.; Bauer, M.; Policarpo Sequeira, D.; et al. Identification of broad anti-coronavirus chemical agents for repurposing against SARS-CoV-2 and variants of concern. Curr. Res. Virol. Sci. 2022, 3, 100019. [Google Scholar] [CrossRef]
- Perez, A.R.; Pritykin, Y.; Vidigal, J.A.; Chhangawala, S.; Zamparo, L.; Leslie, C.S.; Ventura, A. GuideScan software for improved single and paired CRISPR guide RNA design. Nat. Biotechnol. 2017, 35, 347–349. [Google Scholar] [CrossRef] [Green Version]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef] [Green Version]
- Brinkman, E.K.; Chen, T.; Amendola, M.; van Steensel, B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014, 42, e168. [Google Scholar] [CrossRef]
- Greber, U.F.; Willetts, M.; Webster, P.; Helenius, A. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 1993, 75, 477–486. [Google Scholar] [CrossRef] [Green Version]
- Thao, T.T.N.; Labroussaa, F.; Ebert, N.; V’Kovski, P.; Stalder, H.; Portmann, J.; Kelly, J.; Steiner, S.; Holwerda, M.; Kratzel, A.; et al. Rapid reconstruction of SARS-CoV-2 using a synthetic genomics platform. Nature 2020, 582, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Marin-Gonzalez, A.; Aicart-Ramos, C.; Marin-Baquero, M.; Martin-Gonzalez, A.; Suomalainen, M.; Kannan, A.; Vilhena, J.G.; Greber, U.F.; Moreno-Herrero, F.; Perez, R. Double-stranded RNA bending by AU-tract sequences. Nucleic Acids Res. 2020, 48, 12917–12928. [Google Scholar] [CrossRef] [PubMed]
- Burckhardt, C.J.; Suomalainen, M.; Schoenenberger, P.; Boucke, K.; Hemmi, S.; Greber, U.F. Drifting motions of the adenovirus receptor CAR and immobile integrins initiate virus uncoating and membrane lytic protein exposure. Cell Host Microbe 2011, 10, 105–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suomalainen, M.; Luisoni, S.; Boucke, K.; Bianchi, S.; Engel, D.A.; Greber, U.F. A direct and versatile assay measuring membrane penetration of adenovirus in single cells. J. Virol. 2013, 87, 12367–12379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [Green Version]
- Suomalainen, M.; Prasad, V.; Kannan, A.; Greber, U.F. Cell-to-cell and genome-to-genome variability of adenovirus transcription tuned by the cell cycle. J. Cell Sci. 2020, 134, jcs.252544. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Meth. 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Ekins, S.; Nikolsky, Y.; Bugrim, A.; Kirillov, E.; Nikolskaya, T. Pathway mapping tools for analysis of high content data. Methods Mol. Biol. 2007, 356, 319–350. [Google Scholar] [CrossRef]
- Jorns, C.; Holzinger, D.; Thimme, R.; Spangenberg, H.C.; Weidmann, M.; Rasenack, J.; Blum, H.E.; Haller, O.; Kochs, G. Rapid and simple detection of IFN-neutralizing antibodies in chronic hepatitis C non-responsive to IFN-alpha. J. Med. Virol. 2006, 78, 74–82. [Google Scholar] [CrossRef]
- Santhakumar, D.; Forster, T.; Laqtom, N.N.; Fragkoudis, R.; Dickinson, P.; Abreu-Goodger, C.; Manakov, S.A.; Choudhury, N.R.; Griffiths, S.J.; Vermeulen, A.; et al. Combined agonist-antagonist genome-wide functional screening identifies broadly active antiviral microRNAs. Proc. Natl. Acad. Sci. USA 2010, 107, 13830–13835. [Google Scholar] [CrossRef] [Green Version]
- Ho, V.; Yong, H.Y.; Chevrier, M.; Narang, V.; Lum, J.; Toh, Y.X.; Lee, B.; Chen, J.; Tan, E.Y.; Luo, D.; et al. RIG-I Activation by a Designer Short RNA Ligand Protects Human Immune Cells against Dengue Virus Infection without Causing Cytotoxicity. J. Virol. 2019, 93, e00102-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohlway, A.; Luo, D.; Rawling, D.C.; Ding, S.C.; Pyle, A.M. Defining the functional determinants for RNA surveillance by RIG-I. EMBO Rep. 2013, 14, 772–779. [Google Scholar] [CrossRef] [PubMed]
- Lotzerich, M.; Roulin, P.S.; Boucke, K.; Witte, R.; Georgiev, O.; Greber, U.F. Rhinovirus 3C protease suppresses apoptosis and triggers caspase-independent cell death. Cell Death Dis. 2018, 9, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Yuan, S.; Jia, X.; Ge, Y.; Ling, T.; Nie, M.; Lan, X.; Chen, S.; Xu, A. Mitochondria-localised ZNFX1 functions as a dsRNA sensor to initiate antiviral responses through MAVS. Nat. Cell Biol. 2019, 21, 1346–1356. [Google Scholar] [CrossRef] [PubMed]
- Marin-Gonzalez, A.; Vilhena, J.G.; Perez, R.; Moreno-Herrero, F. A molecular view of DNA flexibility. Q. Rev. Biophys. 2021, 54, e8. [Google Scholar] [CrossRef]
- Szabo, G.T.; Mahiny, A.J.; Vlatkovic, I. COVID-19 mRNA vaccines: Platforms and current developments. Mol. Ther. 2022, 30, 1850–1868. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Kensch, O.; Connolly, B.A.; Steinhoff, H.J.; McGregor, A.; Goody, R.S.; Restle, T. HIV-1 reverse transcriptase-pseudoknot RNA aptamer interaction has a binding affinity in the low picomolar range coupled with high specificity. J. Biol. Chem. 2000, 275, 18271–18278. [Google Scholar] [CrossRef] [Green Version]
- Gopinath, S.C. Antiviral aptamers. Arch. Virol. 2007, 152, 2137–2157. [Google Scholar] [CrossRef]
- Torres-Vazquez, B.; de Lucas, A.M.; Garcia-Crespo, C.; Garcia-Martin, J.A.; Fragoso, A.; Fernandez-Algar, M.; Perales, C.; Domingo, E.; Moreno, M.; Briones, C. In vitro Selection of High Affinity DNA and RNA Aptamers that Detect Hepatitis C Virus Core Protein of Genotypes 1 to 4 and Inhibit Virus Production in Cell Culture. J. Mol. Biol. 2022, 434, 167501. [Google Scholar] [CrossRef]
- Filipowicz, W.; Bhattacharyya, S.N.; Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008, 9, 102–114. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.K.; Chow, M.Y.; Zhang, Y.; Leung, S.W. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids 2015, 4, e252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malathi, K.; Dong, B.; Gale, M., Jr.; Silverman, R.H. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 2007, 448, 816–819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kariko, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Kariko, K.; Muramatsu, H.; Keller, J.M.; Weissman, D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol. Ther. 2012, 20, 948–953. [Google Scholar] [CrossRef] [Green Version]
- Kwon, J.J.; Factora, T.D.; Dey, S.; Kota, J. A Systematic Review of miR-29 in Cancer. Mol. Ther. Oncolytics 2019, 12, 173–194. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Carmichael, G.G. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 2004, 68, 432–452, table of contents. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.Y.; Gonzalez-Martin, A.; Miletic, A.V.; Lai, M.; Knight, S.; Sabouri-Ghomi, M.; Head, S.R.; Macauley, M.S.; Rickert, R.C.; Xiao, C. Transfection of microRNA Mimics Should Be Used with Caution. Front. Genet. 2015, 6, 340. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, T.; Ogasawara, K.; Takaoka, A.; Tanaka, N. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 2001, 19, 623–655. [Google Scholar] [CrossRef]
- Borden, E.C.; Sen, G.C.; Uze, G.; Silverman, R.H.; Ransohoff, R.M.; Foster, G.R.; Stark, G.R. Interferons at age 50: Past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007, 6, 975–990. [Google Scholar] [CrossRef] [PubMed]
- Woeckel, V.J.; Eijken, M.; van de Peppel, J.; Chiba, H.; van der Eerden, B.C.; van Leeuwen, J.P. IFNbeta impairs extracellular matrix formation leading to inhibition of mineralization by effects in the early stage of human osteoblast differentiation. J. Cell Physiol. 2012, 227, 2668–2676. [Google Scholar] [CrossRef] [PubMed]
- Goubau, D.; Deddouche, S.; Reis, E.S.C. Cytosolic sensing of viruses. Immunity 2013, 38, 855–869. [Google Scholar] [CrossRef] [Green Version]
- Schlee, M.; Roth, A.; Hornung, V.; Hagmann, C.A.; Wimmenauer, V.; Barchet, W.; Coch, C.; Janke, M.; Mihailovic, A.; Wardle, G.; et al. Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 2009, 31, 25–34. [Google Scholar] [CrossRef] [Green Version]
- Linehan, M.M.; Dickey, T.H.; Molinari, E.S.; Fitzgerald, M.E.; Potapova, O.; Iwasaki, A.; Pyle, A.M. A minimal RNA ligand for potent RIG-I activation in living mice. Sci. Adv. 2018, 4, e1701854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marq, J.B.; Hausmann, S.; Veillard, N.; Kolakofsky, D.; Garcin, D. Short double-stranded RNAs with an overhanging 5’ ppp-nucleotide, as found in arenavirus genomes, act as RIG-I decoys. J. Biol. Chem. 2011, 286, 6108–6116. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Linehan, M.M.; Iwasaki, A.; Pyle, A.M. RIG-I Selectively Discriminates against 5’-Monophosphate RNA. Cell Rep. 2019, 26, 2019–2027.e2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahasi, K.; Yoneyama, M.; Nishihori, T.; Hirai, R.; Kumeta, H.; Narita, R.; Gale, M., Jr.; Inagaki, F.; Fujita, T. Nonself RNA-sensing mechanism of RIG-I helicase and activation of antiviral immune responses. Mol. Cell 2008, 29, 428–440. [Google Scholar] [CrossRef]
- Weitzer, S.; Martinez, J. The human RNA kinase hClp1 is active on 3’ transfer RNA exons and short interfering RNAs. Nature 2007, 447, 222–226. [Google Scholar] [CrossRef]
- Taghavi, A.; Yildirim, I. Computational Investigation of Bending Properties of RNA AUUCU, CCUG, CAG, and CUG Repeat Expansions Associated With Neuromuscular Disorders. Front. Mol. Biosci. 2022, 9, 830161. [Google Scholar] [CrossRef]
- Nabet, B.Y.; Qiu, Y.; Shabason, J.E.; Wu, T.J.; Yoon, T.; Kim, B.C.; Benci, J.L.; DeMichele, A.M.; Tchou, J.; Marcotrigiano, J.; et al. Exosome RNA Unshielding Couples Stromal Activation to Pattern Recognition Receptor Signaling in Cancer. Cell 2017, 170, 352–366.e313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Liu, C.X.; Xue, W.; Zhang, Y.; Jiang, S.; Yin, Q.F.; Wei, J.; Yao, R.W.; Yang, L.; Chen, L.L. Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell 2017, 67, 214–227.e217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.G.; Chen, R.; Ahmad, S.; Verma, R.; Kasturi, S.P.; Amaya, L.; Broughton, J.P.; Kim, J.; Cadena, C.; Pulendran, B.; et al. N6-Methyladenosine Modification Controls Circular RNA Immunity. Mol. Cell 2019, 76, 96–109.e9. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kannan, A.; Suomalainen, M.; Volle, R.; Bauer, M.; Amsler, M.; Trinh, H.V.; Vavassori, S.; Schmid, J.P.; Vilhena, G.; Marín-González, A.; et al. Sequence-Specific Features of Short Double-Strand, Blunt-End RNAs Have RIG-I- and Type 1 Interferon-Dependent or -Independent Anti-Viral Effects. Viruses 2022, 14, 1407. https://doi.org/10.3390/v14071407
Kannan A, Suomalainen M, Volle R, Bauer M, Amsler M, Trinh HV, Vavassori S, Schmid JP, Vilhena G, Marín-González A, et al. Sequence-Specific Features of Short Double-Strand, Blunt-End RNAs Have RIG-I- and Type 1 Interferon-Dependent or -Independent Anti-Viral Effects. Viruses. 2022; 14(7):1407. https://doi.org/10.3390/v14071407
Chicago/Turabian StyleKannan, Abhilash, Maarit Suomalainen, Romain Volle, Michael Bauer, Marco Amsler, Hung V. Trinh, Stefano Vavassori, Jana Pachlopnik Schmid, Guilherme Vilhena, Alberto Marín-González, and et al. 2022. "Sequence-Specific Features of Short Double-Strand, Blunt-End RNAs Have RIG-I- and Type 1 Interferon-Dependent or -Independent Anti-Viral Effects" Viruses 14, no. 7: 1407. https://doi.org/10.3390/v14071407
APA StyleKannan, A., Suomalainen, M., Volle, R., Bauer, M., Amsler, M., Trinh, H. V., Vavassori, S., Schmid, J. P., Vilhena, G., Marín-González, A., Perez, R., Franceschini, A., Mering, C. v., Hemmi, S., & Greber, U. F. (2022). Sequence-Specific Features of Short Double-Strand, Blunt-End RNAs Have RIG-I- and Type 1 Interferon-Dependent or -Independent Anti-Viral Effects. Viruses, 14(7), 1407. https://doi.org/10.3390/v14071407






