Identifying Structural Features of Nucleotide Analogues to Overcome SARS-CoV-2 Exonuclease Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Extension Reactions with Reverse Transcriptase to Produce Nucleotide Analogue-Extended RNA
2.2. Extension Reactions with RNA Polymerase to Produce Nucleotide Analogue-Extended RNA
2.3. Exonuclease Reactions with SARS-CoV-2 Nsp14/Nsp10 Complex
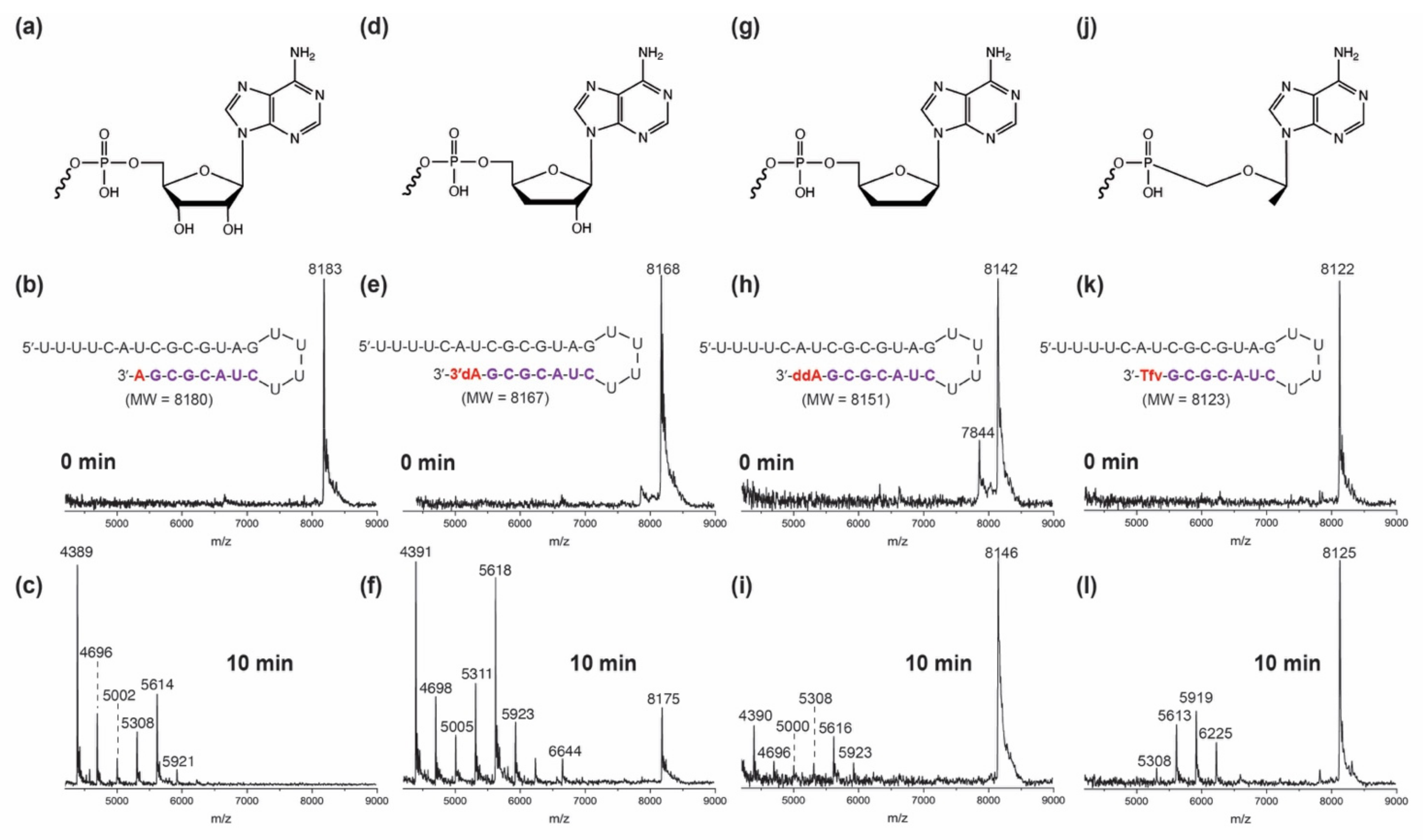
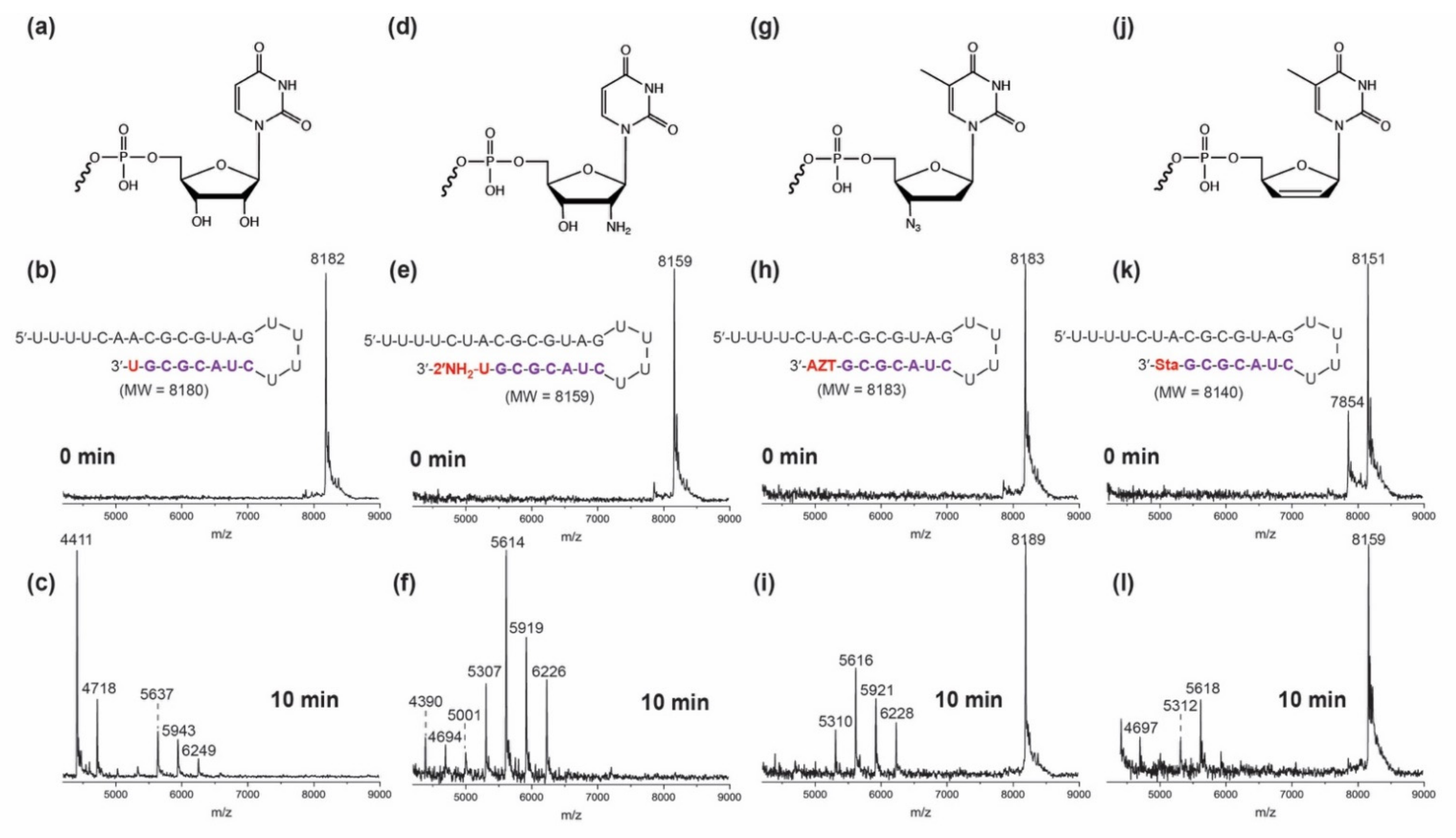
3. Results and Discussion
3.1. Sequence Conservation in Nsp14 and Nsp10
3.2. Excision of Nucleotide Analogues from RNA by SARS-CoV-2 ExoN
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A novel coronavirus from patients with pneumonia in China 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Chen, B.; Tian, E.-K.; He, B.; Tian, L.; Han, R.; Wang, S.; Xiang, Q.; Zhang, S.; El Arnaout, T.; Cheng, W. Overview of lethal human coronaviruses. Signal Transduct. Target. Ther. 2020, 5, 89. [Google Scholar] [CrossRef] [PubMed]
- Vakulenko, Y.; Deviatkin, A.; Drexler, J.F.; Lukashev, A. Modular evolution of coronavirus genomes. Viruses 2021, 13, 1270. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Román, E.; Gibbs, A.J. Ecology and evolution of betacoronaviruses. In Coronavirus Disease—COVID-19; Rezaie, N., Ed.; Springer: Cham, Switzerland, 2021; Volume 1318, pp. 41–60. [Google Scholar]
- Bai, C.; Zhong, Q.; Gao, G.F. Overview of SARS-CoV-2 genome-encoded proteins. Sci. China Life Sci. 2022, 65, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Rohaim, M.A.; El Naggar, R.F.; Clayton, E.; Munir, M. Structural and functional insights into non-structural proteins of coronaviruses. Microb. Pathog. 2021, 150, 104641. [Google Scholar] [CrossRef]
- Baddock, H.T.; Brolih, S.; Yosaatmadja, Y.; Ratnaweera, M.; Bielinski, M.; Swift, L.P.; Cruz-Migoni, A.; Fan, H.; Keown, J.R.; Walker, A.P.; et al. Characterization of the SARS-CoV-2 ExoN (nsp14ExoN-nsp10) complex: Implications for its role in viral genome stability and inhibitor identification. Nucleic Acids Res. 2022, 50, 1484–1500. [Google Scholar] [CrossRef]
- Saramago, M.; Bárria, C.; Costa, V.G.; Souza, C.S.; Viegas, S.C.; Domingues, S.; Lousa, D.; Soares, C.M.; Arraiano, C.M.; Matos, R.G. New targets for drug design: Importance of nsp14/nsp10 complex formation for the 3’–5’ exoribonucleolytic activity on SARS-CoV-2. FEBS J. 2021, 288, 5130–5147, Epub ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Bouvet, M.; Imbert, I.; Subissi, L.; Gluais, L.; Canard, B.; Decroly, E. RNA 3’-end mismatch excision by the severe acute respiratory syndrome coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. USA. 2012, 109, 9372–9377. [Google Scholar] [CrossRef] [Green Version]
- Gribble, J.; Stevens, L.J.; Agostini, M.L.; Anderson-Daniels, J.; Chappell, J.D.; Lu, X.; Pruijssers, A.J.; Routh, A.L.; Denison, M.R. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 2021, 17, e1009226. [Google Scholar] [CrossRef]
- Liu, C.; Shi, W.; Becker, S.T.; Schatz, D.G.; Liu, B.; Yang, Y. Structural basis of mismatch recognition by a SARS-CoV-2 proofreading enzyme. Science 2021, 373, 1142–1146. [Google Scholar] [CrossRef]
- Ma, Y.; Wu, L.; Shaw, N.; Gao, Y.; Wang, J.; Sun, Y.; Lou, Z.; Yan, L.; Zhang, R.; Rao, Z. Structural basis and functional analysis of the SARS coronavirus nsp14–nsp10 complex. Proc. Natl. Acad. Sci. USA 2015, 112, 9436–9441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moeller, N.H.; Shi, K.; Demir, Ö.; Belica, C.; Banerjee, S.; Yin, L.; Durfee, C.; Amaro, R.E.; Aihara, H. Structure and dynamics of SARS-CoV-2 proofreading exoribonuclease ExoN. Proc. Natl. Acad. Sci. USA 2022, 119, e2106379119. [Google Scholar] [CrossRef] [PubMed]
- Ogando, N.S.; Zevenhoven-Dobbe, J.; van der Meer, Y.; Bredenbeek, P.J.; Posthuma, C.C.; Snijder, E.J. The enzymatic activity of the nsp14 exoribonuclease is critical for replication of MERS-CoV and SARS-CoV-2. J. Virol. 2020, 94, e01246-20. [Google Scholar] [CrossRef]
- Rona, G.; Zeke, A.; Miwatani-Minter, B.; de Vries, M.; Kaur, R.; Schinlever, A.; Garcia, S.F.; Goldberg, H.V.; Wang, H.; Hinds, T.R.; et al. The NSP14/NSP10 RNA repair complex as a pan-coronavirus therapeutic target. Cell Death Differ. 2022, 29, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Tahir, M. Coronavirus genomic nsp14-ExoN, structure, role, mechanism, and potential application as a drug target. J. Med. Virol. 2021, 93, 4258–4264. [Google Scholar] [CrossRef]
- Robson, F.; Khan, K.S.; Le, T.K.; Paris, C.; Demirbag, S.; Barfuss, P.; Rocchi, P.; Ng, W.-L. Coronavirus RNA proofreading: Molecular basis and therapeutic targeting. Mol. Cell 2020, 79, 710–727. [Google Scholar] [CrossRef]
- Hsu, J.C.-C.; Laurent-Rolle, M.; Pawlak, J.B.; Wilen, C.B.; Cresswell, P. Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc. Natl. Acad. Sci. USA 2021, 118, e2101161118. [Google Scholar] [CrossRef]
- Gao, Y.; Yan, L.; Huang, Y.; Liu, F.; Zhao, Y.; Cao, L.; Wang, T.; Sun, Q.; Ming, Z.; Zhang, Y.; et al. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. [Google Scholar] [CrossRef] [Green Version]
- Hillen, H.S.; Kokic, G.; Farnung, L.; Dienemann, C.; Tegunov, D.; Cramer, P. Structure of replicating SARS-CoV-2 polymerase. Nature 2020, 584, 154–156. [Google Scholar] [CrossRef]
- Wang, Q.; Wu, J.; Wang, H.; Gao, Y.; Liu, Q.; Mu, A.; Ji, W.; Yan, L.; Zhu, Y.; Zhu, C.; et al. Structural basis for RNA replication by the SARS-CoV-2 polymerase. Cell 2020, 182, 417–428. [Google Scholar] [CrossRef]
- Yin, W.; Mao, C.; Luan, X.; Shen, D.-D.; Shen, Q.; Su, H.; Wang, X.; Zhou, F.; Zhao, W.; Gao, M.; et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science 2020, 368, 1499–1504. [Google Scholar] [CrossRef] [PubMed]
- Kokic, G.; Hillen, H.S.; Tegunov, D.; Dienemann, C.; Seitz, F.; Schmitzova, J.; Farnung, L.; Siewert, A.; Hobartner, C.; Cramer, P. Mechanism of SARS-CoV-2 polymerase stalling by remdesivir. Nat. Commun. 2021, 12, 279. [Google Scholar] [CrossRef] [PubMed]
- Naydenova, K.; Muir, K.W.; Wu, L.-F.; Zhang, Z.; Coscia, F.; Peet, M.J.; Castro-Hartmann, P.; Qian, P.; Sader, K.; Dent, K.; et al. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc. Natl. Acad. Sci. USA 2021, 118, e2021946118. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Chen, H.; Chen, Z.; Yang, F.; Ye, F.; Zheng, Y.; Yang, J.; Lin, X.; Sun, H.; Wang, L.; et al. Crystal structure of SARS-CoV-2 nsp10 bound to nsp14-ExoN domain reveals an exoribonuclease with both structural and functional integrity. Nucleic Acids Res. 2021, 49, 5382–5392. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, Y.; Ge, J.; Zheng, L.; Gao, Y.; Wang, T.; Jia, Z.; Wang, H.; Huang, Y.; Li, M.; et al. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020, 11, 5874. [Google Scholar] [CrossRef]
- Yan, L.; Ge, J.; Zheng, L.; Zhang, Y.; Gao, Y.; Wang, T.; Huang, Y.; Yang, Y.; Gao, S.; Li, M.; et al. Cryo-EM structure of an extended SARS-CoV-2 replication and transcription complex reveals an intermediate state in cap synthesis. Cell 2021, 184, 184–193. [Google Scholar] [CrossRef]
- Yan, L.; Yang, Y.; Li, M.; Zhang, Y.; Zheng, L.; Ge, J.; Huang, Y.C.; Liu, Z.; Wang, T.; Gao, S.; et al. Coupling of N7-methyltransferase and 30-50 exoribonuclease with SARS-CoV-2 polymerase reveals mechanisms for capping and proofreading. Cell 2021, 184, 3474–3485. [Google Scholar] [CrossRef]
- Chen, J.; Malone, B.; Llewellyn, E.; Grasso, M.; Shelton, P.M.M.; Olinares, P.D.B.; Maruthi, K.; Eng, E.T.; Vatandaslar, H.; Chait, B.T.; et al. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell 2020, 182, 1560–1573. [Google Scholar] [CrossRef]
- Malone, B.; Chen, J.; Wang, Q.; Llewellyn, E.; Choi, Y.J.; Olinares, P.D.B.; Cao, X.; Hernandez, C.; Eng, E.T.; Chait, B.T.; et al. Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex. Proc. Natl. Acad. Sci. USA 2021, 118, e2102516118. [Google Scholar] [CrossRef]
- Subissi, L.; Posthuma, C.C.; Collet, A.; Zevenhoven-Dobbe, J.C.; Gorbalenya, A.E.; Decroly, E.; Snijder, E.J.; Canard, B.; Imbert, I. One severe acute respiratory syndrome coronavirus protein complex integrates processive RNA polymerase and exonuclease activities. Proc. Natl. Acad. Sci. USA 2014, 111, E3900–E3909. [Google Scholar] [CrossRef] [Green Version]
- Ogando, N.S.; Ferron, F.; Decroly, E.; Canard, B.; Posthuma, C.C.; Snijder, E.J. The curious case of the Nidovirus exoribonuclease: Its role in RNA synthesis and replication fidelity. Front. Microbiol. 2019, 10, 813. [Google Scholar] [CrossRef] [PubMed]
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef]
- Eskier, D.; Suner, A.; Oktay, Y.; Karakülah, G. Mutations of SARS-CoV-2 nsp14 exhibit strong association with increased genome-wide mutation load. PeerJ 2020, 8, e10181. [Google Scholar] [CrossRef] [PubMed]
- Ogando, N.S.; El Kazzi, P.; Zevenhoven-Dobbe, J.C.; Bontes, B.W.; Decombe, A.; Posthuma, C.C.; Thiel, V.; Canard, B.; Ferron, F.; Decroly, E.; et al. Structure-function analysis of the nsp14 N7-guanine methyltransferase reveals an essential role in Betacoronavirus replication. Proc. Natl. Acad. Sci. USA 2021, 49, e2108709118. [Google Scholar] [CrossRef]
- Niu, X.; Kong, F.; Hou, Y.J.; Wang, Q. Crucial mutation in the exoribonuclease domain of nsp14 of PEDV leads to high genetic instability during viral replication. Cell Biosci. 2021, 11, 106. [Google Scholar] [CrossRef] [PubMed]
- Bouvet, M.; Lugari, A.; Posthuma, C.C.; Zevenhoven, J.C.; Bernard, S.; Betzi, S.; Imbert, I.; Canard, B.; Guillemot, J.-C.; Lécine, P.; et al. Coronavirus nsp10, a critical co-factor for activation of multiple replicative enzymes. J. Biol. Chem. 2014, 289, 25783–25796. [Google Scholar] [CrossRef] [Green Version]
- Keep, S.; Stevenson-Leggett, P.; Dowgier, G.; Everest, H.; Freimanis, G.; Oade, M.; Hammond, J.A.; Armesto, M.; Vila, R.; Bru, T.; et al. Identification of amino acids within nonstructural proteins 10 and 14 of the avian coronavirus infectious bronchitis virus that result in attenuation In Vivo and In Ovo. J. Virol. 2022, 96, e02059-21. [Google Scholar] [CrossRef]
- Jockusch, S.; Tao, C.; Li, X.; Chien, M.; Kumar, S.; Morozova, I.; Kalachikov, S.; Russo, J.J.; Ju, J. Sofosbuvir terminated RNA is more resistant to SARS-CoV-2 proofreader than RNA terminated by Remdesivir. Sci. Rep. 2020, 10, 16577. [Google Scholar] [CrossRef]
- Canal, B.; McClure, A.W.; Curran, J.F.; Wu, M.; Ulferts, R.; Weissman, F.; Zeng, J.; Bertolin, A.P.; Milligan, J.C.; Basu, S.; et al. Identifying SARS-CoV-2 antiviral compounds by screening for small molecule inhibitors of nsp14/nsp10 exoribonuclease. Bi-ohem. J. 2021, 478, 2445–2564. [Google Scholar] [CrossRef]
- Wang, X.; Sacramento, C.Q.; Jockusch, S.; Chaves, O.A.; Tao, C.; Fintelman-Rodrigues, N.; Chien, M.; Temerozo, J.R.; Li, X.; Kumar, S.; et al. Combination of antiviral drugs inhibits SARS-CoV-2 polymerase and exonuclease and demonstrates COVID-19 therapeutic potential in viral cell culture. Commun. Biol. 2022, 5, 154. [Google Scholar] [CrossRef]
- Gordon, C.J.; Tchesnokov, E.P.; Woolner, E.; Perry, J.K.; Feng, J.Y.; Porter, D.P.; Götte, M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bravo, J.P.K.; Dangerfield, T.L.; Taylor, D.W.; Johnson, K.A. Remdesivir is a delayed translocation inhibitor of SARS-CoV-2 replication. Mol. Cell 2021, 81, 1548–1552. [Google Scholar] [CrossRef] [PubMed]
- Vicenti, I.; Zazzi, M.; Saladini, F. SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19. Expert Opin. Ther. Pat. 2021, 31, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Seifert, M.; Bera, S.C.; van Nies, P.; Kirchdoerfer, R.N.; Shannon, A.; Le, T.-T.-N.; Meng, X.; Xia, H.; Wood, J.M.; Harris, L.D.; et al. Inhibition of SARS-CoV-2 polymerase by nucleotide analogs from a single-molecule perspective. eLife 2021, 10, e70968. [Google Scholar] [CrossRef]
- Chien, M.; Anderson, T.K.; Jockusch, S.; Tao, C.; Li, X.; Kumar, S.; Russo, J.J.; Kirchdoerfer, R.N.; Ju, J. Nucleotide analogues as inhibitors of SARS-CoV-2 polymerase, a key drug target for COVID-19. J. Proteome Res. 2020, 19, 4690–4697. [Google Scholar] [CrossRef]
- Jockusch, S.; Tao, C.; Li, X.; Anderson, T.K.; Chien, M.; Kumar, S.; Russo, J.J.; Kirchdoerfer, R.N.; Ju, J. A library of nucleotide analogues terminate RNA synthesis catalyzed by polymerases of coronaviruses that cause SARS and COVID-19. Antivir. Res. 2020, 180, 104857. [Google Scholar] [CrossRef]
- Blasiak, A.; Lim, J.J.; Seah, S.G.K.; Kee, T.; Remus, A.; Chye, D.H.; Wong, P.S.; Hooi, L.; Truong, A.T.L.; Le, N.; et al. IDentif.AI: Rapidly optimizing combination therapy design against severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) with digital drug development. Bioeng. Transl. Med. 2020, 6, e10196. [Google Scholar] [CrossRef]
- Dejmek, M.; Konkoľová, E.; Eyer, L.; Straková, P.; Svoboda, P.; Šála, M.; Krejčová, K.; Růžek, D.; Boura, E.; Nencka, R. Non-nucleotide RNA-dependent RNA polymerase inhibitor that blocks SARS-CoV-2 replication. Viruses 2021, 13, 1585. [Google Scholar] [CrossRef]
- Yang, H.; Rao, Z. Structural biology of SARS-CoV-2 and implications for therapeutic development. Nat. Rev. Microbiol. 2021, 19, 685–700. [Google Scholar] [CrossRef]
- Minskaia, E.; Hertzig, T.; Gorbalenya, A.E.; Campanacci, V.; Cambillau, C.; Canard, B.; Ziebhur, J. Discovery of an RNA virus 3’–>5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 5108–5113. [Google Scholar] [CrossRef] [Green Version]
- Gitto, S.; Gamal, N.; Andreone, P. NS5A inhibitors for the treatment of hepatitis C infection. J. Viral Hepat. 2017, 24, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Sacramento, C.Q.; Fintelman-Rodrigues, N.; Temerozo, J.R.; de Paul Dias Da Silva, A.; da Silva Gomes Dias, S.; dos Santos da Silva, C.; Ferreira, A.C.; Mattos, M.; Pão, C.R.R.; de Freitas, C.S.; et al. In vitro antiviral activity of the anti-HCV drugs daclatasvir and sofosbuvir against SARS-CoV-2, the aetiological agent of COVID-19. J. Antimicrob. Chemother. 2021, 76, 1874–1885. [Google Scholar] [CrossRef] [PubMed]
- Sanders, R. Can Hepatitis C Drugs Help Remdesivir Fight COVID-19? Available online: https://news.berkeley.edu/2021/02/03/can-hepatitis-c-drugs-help-remdesiver-fight-covid-19/ (accessed on 20 May 2022).
- Khater, S.; Kumar, P.; Dasgupta, N.; Das, G.; Ray, S.; Prakash, A. Combining SARS-CoV-2 proofreading exonuclease and RNA-dependent RNA polymerase inhibitors as a strategy to combat COVID-19: A high-throughput in silico screen. Front. Microbiol. 2021, 12, 647693. [Google Scholar] [CrossRef] [PubMed]
- Nguyenla, X.; Wehri, E.; Van Dis, E.; Biering, S.B.; Yamashiro, L.H.; Stroumza, J.; Dugast-Darzacq, C.; Graham, T.; Stanley, S.; Schaletzky, J. Discovery of SARS-CoV-2 antiviral synergy between remdesivir and approved drugs in human lung cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Chen, Z.; Azman, A.S.; Chen, X.; Zou, J.; Tian, Y.; Sun, R.; Xu, X.; Wu, Y.; Lu, W.; Ge, S.; et al. Global landscape of SARS-CoV-2 genomic surveillance and data sharing. Nat. Genet. 2022, 54, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Hu, X.; Chen, Y.; Peng, X.; Fu, Y.; Zheng, Y.; Yu, J.; He, J. Biological significance of the genome variation and structural dynamics of SARS-CoV-2 B.1.617. Front. Microbiol. 2021, 12, 750725. [Google Scholar] [CrossRef] [PubMed]
- Wiegand, T.; McVey, A.; Nemudraia, A.; Nemudryi, A.; Wiedenheft, A. The rise and fall of SARS-CoV-2 variants and the mutational profile of Omicron. bioRxiv 2021. [Google Scholar] [CrossRef]
- Jung, C.; Kmiec, D.; Koepke, L.; Zech, F.; Jacob, T.; Sparrer, K.M.J.; Kirchhoff, F. Omicron: What makes the latest SARS-CoV-2 variant of concern so concerning? J. Virol. 2022, 96, e02077-21. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022. online ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.P.; Leroy, C.; Sander, C. MView: A web-compatible database search or multiple alignment viewer. Bioinformatics 1998, 14, 380–381. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.C.; Case, J.B.; Blanc, H.; Isakov, O.; Shomron, N.; Vignuzzi, M.; Denisonon, M.R. Mutations in coronavirus nonstructural protein 10 decrease viral replication fidelity. J. Virol. 2015, 89, 6418–6426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabie, A.M. Potent inhibitory activities of the adenosine analogue cordycepin on SARS-CoV-2 replication. ACS Omega 2022, 7, 2960–2969. [Google Scholar] [CrossRef]
- Arnold, J.J.; Sharma, S.D.; Feng, J.Y.; Ray, A.S.; Smidansky, E.D.; Kireeva, M.L.; Cho, A.; Perry, J.; Vela, J.E.; Park, Y.; et al. Sensitivity of mitochondrial transcription and resistance of RNA polymerase II dependent nuclear transcription to antiviral ribonucleosides. PLoS Pathog. 2012, 8, e1003030. [Google Scholar] [CrossRef]
- Wu, Y.; Chang, K.Y.; Lou, L.; Edwards, L.G.; Doma, B.K.; Xie, Z.R. In silico identification of drug candidates against COVID-19. Inform. Med. Unlocked 2020, 21, 100461. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Tao, C.; Morozova, I.; Kalachikov, S.; Li, X.; Kumar, S.; Russo, J.J.; Ju, J. Identifying Structural Features of Nucleotide Analogues to Overcome SARS-CoV-2 Exonuclease Activity. Viruses 2022, 14, 1413. https://doi.org/10.3390/v14071413
Wang X, Tao C, Morozova I, Kalachikov S, Li X, Kumar S, Russo JJ, Ju J. Identifying Structural Features of Nucleotide Analogues to Overcome SARS-CoV-2 Exonuclease Activity. Viruses. 2022; 14(7):1413. https://doi.org/10.3390/v14071413
Chicago/Turabian StyleWang, Xuanting, Chuanjuan Tao, Irina Morozova, Sergey Kalachikov, Xiaoxu Li, Shiv Kumar, James J. Russo, and Jingyue Ju. 2022. "Identifying Structural Features of Nucleotide Analogues to Overcome SARS-CoV-2 Exonuclease Activity" Viruses 14, no. 7: 1413. https://doi.org/10.3390/v14071413
APA StyleWang, X., Tao, C., Morozova, I., Kalachikov, S., Li, X., Kumar, S., Russo, J. J., & Ju, J. (2022). Identifying Structural Features of Nucleotide Analogues to Overcome SARS-CoV-2 Exonuclease Activity. Viruses, 14(7), 1413. https://doi.org/10.3390/v14071413





