Assessment of Explicit Representation of Dynamic Viral Processes in Regional Marine Ecological Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites
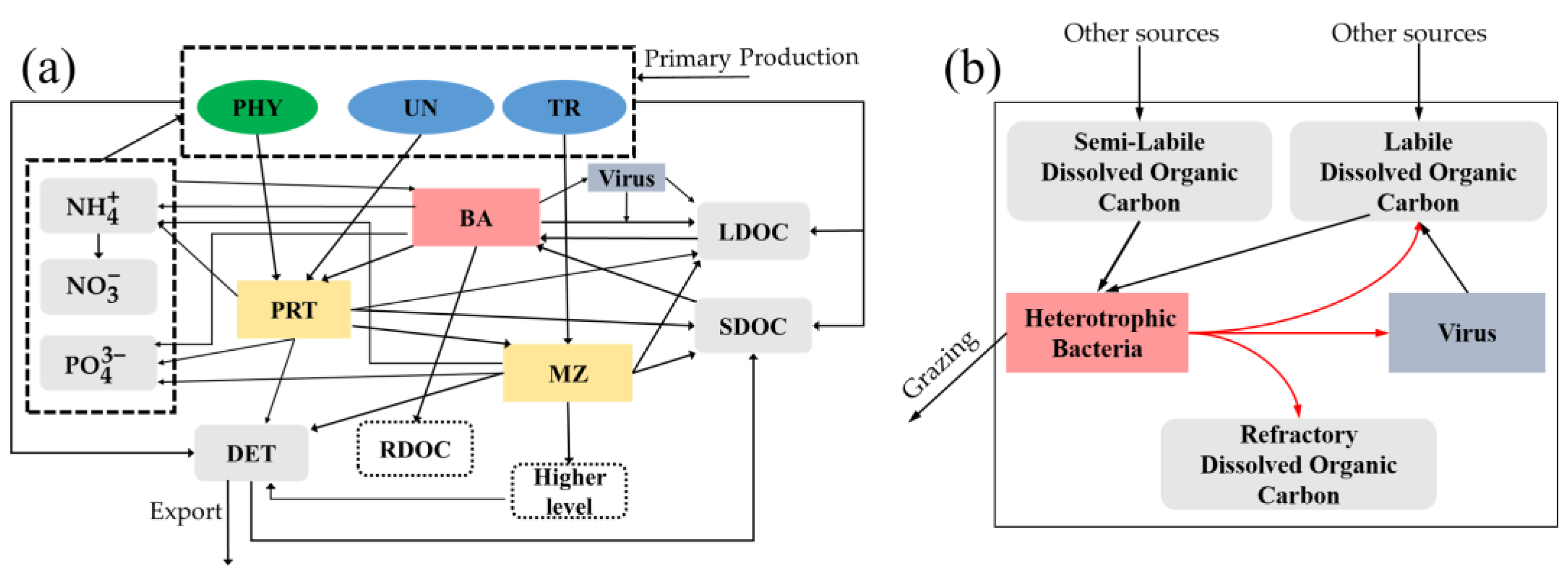
2.2. Standard Model
2.3. Viral Module
3. Results
3.1. Model Performance
3.2. Average Model Results
3.3. Spatiotemporally Variable Simulation of Viruses and Their Induced Bacterial Mortality
3.4. Cascading Effects of the Viral Module
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix B

References
- Xie, L.; Wei, W.; Cai, L.; Chen, X.; Huang, Y.; Jiao, N.; Zhang, R.; Luo, Y.-W. A global viral oceanography database (gVOD). Earth Syst. Sci. Data 2021, 13, 1251–1271. [Google Scholar] [CrossRef]
- Fuhrman, J.A. Marine viruses and their biogeochemical and ecological effects. Nature 1999, 399, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.M.; Milo, R. The biomass composition of the oceans: A blueprint of our blue planet. Cell 2019, 179, 1451–1454. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M. Marine viruses: Truth or dare. Ann. Rev. Mar. Sci. 2012, 4, 425–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinbauer, M.G. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 2004, 28, 127–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuhrman, J.A.; Noble, R.T. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol. Oceanogr. 1995, 40, 1236–1242. [Google Scholar] [CrossRef]
- Danovaro, R.; Corinaldesi, C.; Dell’anno, A.; Fuhrman, J.A.; Middelburg, J.J.; Noble, R.T.; Suttle, C.A. Marine viruses and global climate change. FEMS Microbiol. Rev. 2011, 35, 993–1034. [Google Scholar] [CrossRef]
- Fuhrman, J.A. Bacterioplankton roles in cycling of organic matter: The microbial food web. In Primary Productivity and Biogeochemical Cycles in the Sea; Falkowski, P.G., Woodhead, A.D., Vivirito, K., Eds.; Springer: Boston, MA, USA, 1992; pp. 361–383. [Google Scholar] [CrossRef]
- Suttle, C.A.; Chan, A.M. Marine cyanophages infecting oceanic and coastal strains of Synechococcus—Abundance, morphology, cross-infectivity and growth-characteristics. Mar. Ecol. Prog. Ser. 1993, 92, 99–109. [Google Scholar] [CrossRef]
- Suttle, C.A. Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 2007, 5, 801–812. [Google Scholar] [CrossRef]
- Winter, C.; Bouvier, T.; Weinbauer, M.G.; Thingstad, T.F. Trade-offs between competition and defense specialists among unicellular planktonic organisms: The “killing the winner” hypothesis revisited. Microbiol. Mol. Biol. Rev. 2010, 74, 42–57. [Google Scholar] [CrossRef] [Green Version]
- Jover, L.F.; Effler, T.C.; Buchan, A.; Wilhelm, S.W.; Weitz, J.S. The elemental composition of virus particles: Implications for marine biogeochemical cycles. Nat. Rev. Microbiol. 2014, 12, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Riemann, L.; Middelboe, M. Viral lysis of marine bacterioplankton: Implications for organic matter cycling and bacterial clonal composition. Ophelia 2002, 56, 57–68. [Google Scholar] [CrossRef]
- Suttle, C.A. Viruses in the sea. Nature 2005, 437, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Beltrami, E.; Carroll, T.O. Modeling the role of viral disease in recurrent phytoplankton blooms. J. Math. Biol. 1994, 32, 857–863. [Google Scholar] [CrossRef]
- Fuhrman, K.M.; Pinter, G.A.; Berges, J.A. Dynamics of a virus–host model with an intrinsic quota. Math. Comput. Model. 2011, 53, 716–730. [Google Scholar] [CrossRef]
- Singh, B.K.; Chattopadhyay, J.; Sinha, S. The role of virus infection in a simple phytoplankton zooplankton system. J. Biol. 2004, 231, 153–166. [Google Scholar] [CrossRef]
- Rhodes, C.J.; Martin, A.P. The influence of viral infection on a plankton ecosystem undergoing nutrient enrichment. J. Biol. 2010, 265, 225–237. [Google Scholar] [CrossRef] [Green Version]
- Richards, K.J. Viral infections of oceanic plankton blooms. J. Biol. 2017, 412, 27–35. [Google Scholar] [CrossRef] [Green Version]
- Beretta, E.; Kuang, Y. Modeling and analysis of a marine bacteriophage infection. Math. Biosci. 1998, 149, 57–76. [Google Scholar] [CrossRef]
- Thyrhaug, R.; Larsen, A.; Thingstad, T.F.; Bratbak, G. Stable coexistence in marine algal host-virus systems. Mar. Ecol. Prog. Ser. 2003, 254, 27–35. [Google Scholar] [CrossRef]
- Beretta, E.; Kuang, Y. Modeling and analysis of a marine bacteriophage infection with latency period. Nonlinear Anal-Real. 2001, 2, 35–74. [Google Scholar] [CrossRef]
- Carletti, M. On the stability properties of a stochastic model for phage-bacteria interaction in open marine environment. Math. Biosci. 2002, 175, 117–131. [Google Scholar] [CrossRef]
- Chattopadhyay, J.; Pal, S. Viral infection on phytoplankton-zooplankton system—A mathematical model. Ecol. Model. 2002, 151, 15–28. [Google Scholar] [CrossRef]
- Keller, D.P.; Hood, R.R. Comparative simulations of dissolved organic matter cycling in idealized oceanic, coastal, and estuarine surface waters. J. Mar. Syst. 2013, 109–110, 109–128. [Google Scholar] [CrossRef]
- Bongiorni, L.; Magagnini, M.; Armeni, M.; Noble, R.; Danovaro, R. Viral production, decay rates, and life strategies along a trophic gradient in the North Adriatic Sea. Appl. Environ. Microbiol. 2005, 71, 6644–6650. [Google Scholar] [CrossRef] [Green Version]
- Winter, C.; Smit, A.; Szoeke-Denes, T.; Herndl, G.J.; Weinbauer, M.G. Modelling viral impact on bacterioplankton in the North Sea using artificial neural networks. Environ. Microbiol. 2005, 7, 881–893. [Google Scholar] [CrossRef]
- Parada, V.; Baudoux, A.C.; Sintes, E.; Weinbauer, M.G.; Herndl, G.J. Dynamics and diversity of newly produced virioplankton in the North Sea. ISME J. 2008, 2, 924–936. [Google Scholar] [CrossRef] [Green Version]
- Rowe, J.M.; Saxton, M.A.; Cottrell, M.T.; DeBruyn, J.M.; Berg, G.M.; Kirchman, D.L.; Hutchins, D.A.; Wilhelm, S.W. Constraints on viral production in the Sargasso Sea and North Atlantic. Aquat. Microb. Ecol. 2008, 52, 233–244. [Google Scholar] [CrossRef]
- Boras, J.A.; Sala, M.M.; Vazquez-Dominguez, E.; Weinbauer, M.G.; Vaque, D. Annual changes of bacterial mortality due to viruses and protists in an oligotrophic coastal environment (NW Mediterranean). Environ. Microbiol. 2009, 11, 1181–1193. [Google Scholar] [CrossRef]
- Chen, X.; Liu, H.; Weinbauer, M.; Chen, B.; Jiao, N. Viral dynamics in the surface water of the western South China Sea in summer 2007. Aquat. Microb. Ecol. 2011, 63, 145–160. [Google Scholar] [CrossRef]
- Jasna, V.; Parvathi, A.; Pradeep Ram, A.S.; Balachandran, K.K.; Madhu, N.V.; Nair, M.; Jyothibabu, R.; Jayalakshmy, K.V.; Revichandran, C.; Sime-Ngando, T. Viral-induced mortality of prokaryotes in a tropical monsoonal estuary. Front. Microbiol. 2017, 8, 895. [Google Scholar] [CrossRef] [PubMed]
- Kostner, N.; Scharnreitner, L.; Jurgens, K.; Labrenz, M.; Herndl, G.J.; Winter, C. High viral abundance as a consequence of low viral decay in the Baltic Sea redoxcline. PLoS ONE 2017, 12, e0178467. [Google Scholar] [CrossRef] [PubMed]
- Ordulj, M.; Krstulović, N.; Šantić, D.; Jozić, S.; Šolić, M. Viral dynamics in two trophically different areas in the central Adriatic Sea. Helgol. Mar. Res. 2017, 71, 22. [Google Scholar] [CrossRef] [Green Version]
- Vaque, D.; Boras, J.A.; Torrent-Llagostera, F.; Agusti, S.; Arrieta, J.M.; Lara, E.; Castillo, Y.M.; Duarte, C.M.; Sala, M.M. Viruses and protists induced-mortality of prokaryotes around the Antarctic Peninsula during the austral summer. Front. Microbiol. 2017, 8, 241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mojica, K.D.; Brussaard, C.P. Factors affecting virus dynamics and microbial host-virus interactions in marine environments. FEMS Microbiol. Ecol. 2014, 89, 495–515. [Google Scholar] [CrossRef] [Green Version]
- Edwards, K.F.; Steward, G.F.; Schvarcz, C.R. Making sense of virus size and the tradeoffs shaping viral fitness. Ecol. Lett. 2021, 24, 363–373. [Google Scholar] [CrossRef]
- Suttle, C.A. The significance of viruses to mortality in aquatic microbial communities. Microb. Ecol. 1994, 28, 237–243. [Google Scholar] [CrossRef]
- Rhodes, C.J.; Truscott, J.E.; Martin, A.P. Viral infection as a regulator of oceanic phytoplankton populations. J. Mar. Syst. 2008, 74, 216–226. [Google Scholar] [CrossRef]
- Weitz, J.S.; Stock, C.A.; Wilhelm, S.W.; Bourouiba, L.; Coleman, M.L.; Buchan, A.; Follows, M.J.; Fuhrman, J.A.; Jover, L.F.; Lennon, J.T.; et al. A multitrophic model to quantify the effects of marine viruses on microbial food webs and ecosystem processes. ISME J. 2015, 9, 1352–1364. [Google Scholar] [CrossRef] [Green Version]
- Talmy, D.; Beckett, S.J.; Taniguchi, D.A.A.; Brussaard, C.P.D.; Weitz, J.S.; Follows, M.J. An empirical model of carbon flow through marine viruses and microzooplankton grazers. Environ. Microbiol. 2019, 21, 2171–2181. [Google Scholar] [CrossRef]
- Pourtois, J.; Tarnita, C.E.; Bonachela, J.A. Impact of lytic phages on phosphorus- vs. nitrogen-limited marine microbes. Front. Microbiol. 2020, 11, 221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.W.; Friedrichs, M.A.M.; Doney, S.C.; Church, M.J.; Ducklow, H.W. Oceanic heterotrophic bacterial nutrition by semilabile DOM as revealed by data assimilative modeling. Aquat. Microb. Ecol. 2010, 60, 273–287. [Google Scholar] [CrossRef] [Green Version]
- Karl, D.M.; Bjorkman, K.M.; Dore, J.E.; Fujieki, L.; Hebel, D.V.; Houlihan, T.; Letelier, R.M.; Tupas, L.M. Ecological nitrogen-to-phosphorus stoichiometry at station ALOHA. Deep-Sea Res. II 2001, 48, 1529–1566. [Google Scholar] [CrossRef] [Green Version]
- Lawson, L.M.; Spitz, Y.H.; Hofmann, E.E.; Long, R.B. A data assimilation technique applied to a predator-prey mode. Bull. Math. Biol. 1995, 57, 593–617. [Google Scholar] [CrossRef]
- Church, M.J.; Ducklow, H.W.; Letelier, R.M.; Karl, D.M. Temporal and vertical dynamics in picoplankton photoheterotrophic production in the subtropical North Pacific Ocean. Aquat. Microb. Ecol. 2006, 45, 41–53. [Google Scholar] [CrossRef] [Green Version]
- Ducklow, H. Bacterial production and biomass in the ocean. In Microbial Ecology of the Oceans, 2nd ed.; Kirchman, D., Ed.; Wiley: New York, NY, USA, 2000; pp. 85–120. [Google Scholar]
- Jasna, V.; Pradeep Ram, A.S.; Parvathi, A.; Sime-Ngando, T. Differential impact of lytic viruses on prokaryotic morphopopulations in a tropical estuarine system (Cochin estuary, India). PLoS ONE 2018, 13, e0194020. [Google Scholar] [CrossRef] [Green Version]
- Parada, V.; Herndl, G.J.; Weinbauer, M.G. Viral burst size of heterotrophic prokaryotes in aquatic systems. J. Mar. Biol. Assoc. UK 2006, 86, 613–621. [Google Scholar] [CrossRef]
- Motegi, C.; Nagata, T. Enhancement of viral production by addition of nitrogen or nitrogen plus carbon in subtropical surface waters of the South Pacific. Aquat. Microb. Ecol. 2007, 48, 27–34. [Google Scholar] [CrossRef]
- Hwang, C.Y.; Cho, B.C. Virus-infected bacteria in oligotrophic open waters of the East Sea, Korea. Aquat. Microb. Ecol. 2002, 30, 1–9. [Google Scholar] [CrossRef] [Green Version]
- González, J.M.; Suttle, C.A. Grazing by marine nanoflagellates on viruses and virus-sized particles: Ingestion and digestion. Mar. Ecol. Prog. Ser. 1993, 94, 1–10. [Google Scholar] [CrossRef]
- Weinbauer, M.G.; Bettarel, Y.; Cattaneo, R.; Luef, B.; Maier, C.; Motegi, C.; Peduzzi, P.; Mari, X. Viral ecology of organic and inorganic particles in aquatic systems: Avenues for further research. Aquat. Microb. Ecol. 2009, 57, 321–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brum, J.R.; Ignacio-Espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S.; Cruaud, C.; de Vargas, C.; Gasol, J.M.; et al. Patterns and ecological drivers of ocean viral communities. Science 2015, 348, 1261498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crameri, F.; Shephard, G.E.; Heron, P.J. The misuse of colour in science communication. Nat. Commun. 2020, 11, 5444. [Google Scholar] [CrossRef] [PubMed]
- Shelford, E.J.; Suttle, C.A. Virus-mediated transfer of nitrogen from heterotrophic bacteria to phytoplankton. Biogeosciences 2018, 15, 809–819. [Google Scholar] [CrossRef] [Green Version]
- Brum, J.R. Concentration, production and turnover of viruses and dissolved DNA pools at Stn ALOHA, North Pacific Subtropical Gyre. Aquat. Microb. Ecol. 2005, 41, 103–113. [Google Scholar] [CrossRef]
- Luo, Y.-W.; Ducklow, H.W.; Friedrichs, M.A.M.; Church, M.J.; Karl, D.M.; Doney, S.C. Interannual variability of primary production and dissolved organic nitrogen storage in the North Pacific Subtropical Gyre. J. Geophys. Res. Biogeosci. 2012, 117, G03019. [Google Scholar] [CrossRef] [Green Version]
- De Corte, D.; Sintes, E.; Yokokawa, T.; Lekunberri, I.; Herndl, G.J. Large-scale distribution of microbial and viral populations in the South Atlantic Ocean. Environ. Microbiol. Rep. 2016, 8, 305–315. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Zhang, Y.; Zhang, Y.; Luo, T.; Rivkin, R.B.; Jiao, N. Distributions and relationships of virio- and picoplankton in the epi-, meso- and bathypelagic zones of the Western Pacific Ocean. FEMS Microbiol. Ecol. 2017, 93, 105044. [Google Scholar] [CrossRef]
- Wigington, C.H.; Sonderegger, D.; Brussaard, C.P.D.; Buchan, A.; Finke, J.F.; Fuhrman, J.A.; Lennon, J.T.; Middelboe, M.; Suttle, C.A.; Stock, C.; et al. Re-examination of the relationship between marine virus and microbial cell abundances. Nat. Microbiol. 2016, 1, 15024. [Google Scholar] [CrossRef] [Green Version]
- Stock, C.A.; Dunne, J.P.; John, J.G. Global-scale carbon and energy flows through the marine planktonic food web: An analysis with a coupled physical–biological model. Prog. Oceanogr. 2014, 120, 1–28. [Google Scholar] [CrossRef]
- Wei, W.; Zhang, R.; Peng, L.; Liang, Y.; Jiao, N. Effects of temperature and photosynthetically active radiation on virioplankton decay in the western Pacific Ocean. Sci. Rep. 2018, 8, 1525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, C.; Herndl, G.J.; Weinbauer, M.G. Diel cycles in viral infection of bacterioplankton in the North Sea. Aquat. Microb. Ecol. 2004, 35, 207–216. [Google Scholar] [CrossRef]
- Murray, A.G.; Jackson, G.A. Viral dynamics: A model of the effects of size shape, motion and abundance of single-celled planktonic organisms and other particles. Mar. Ecol. Prog. Ser. 1992, 89, 103–116. [Google Scholar] [CrossRef]
- Weinbauer, M.G.; Wilhelm, S.W.; Suttle, C.A.; Pledger, R.J.; Mitchell, D.L. Sunlight-induced DNA damage and resistance in natural viral communities. Aquat. Microb. Ecol. 1999, 17, 111–120. [Google Scholar] [CrossRef]
- Bettarel, Y.; Bouvier, T.; Bouvier, C.; Carre, C.; Desnues, A.; Domaizon, I.; Jacquet, S.; Robin, A.; Sime-Ngando, T. Ecological traits of planktonic viruses and prokaryotes along a full-salinity gradient. FEMS Microbiol. Ecol. 2011, 76, 360–372. [Google Scholar] [CrossRef] [Green Version]
- Knowles, B.; Silveira, C.B.; Bailey, B.A.; Barott, K.; Cantu, V.A.; Cobián-Güemes, A.G.; Coutinho, F.H.; Dinsdale, E.A.; Felts, B.; Furby, K.A.; et al. Lytic to temperate switching of viral communities. Nature 2016, 531, 466–470. [Google Scholar] [CrossRef]
- Weitz, J.S.; Beckett, S.J.; Brum, J.R.; Cael, B.B.; Dushoff, J. Lysis, lysogeny and virus-microbe ratios. Nature 2017, 549, E1–E3. [Google Scholar] [CrossRef]
- Wang, I.N. Lysis timing and bacteriophage fitness. Genetics 2006, 172, 17–26. [Google Scholar] [CrossRef] [Green Version]
- Middelboe, M. Bacterial growth rate and marine virus–host dynamics. Microb. Ecol. 2000, 40, 114–124. [Google Scholar] [CrossRef]
- Maat, D.S.; Crawfurd, K.J.; Timmermans, K.R.; Brussaard, C.P. Elevated CO2 and phosphate limitation favor Micromonas pusilla through stimulated growth and reduced viral impact. Appl. Environ. Microbiol. 2014, 80, 3119–3127. [Google Scholar] [CrossRef] [Green Version]
- Choua, M.; Bonachela, J.A. Ecological and evolutionary consequences of viral plasticity. Am. Nat. 2019, 193, 346–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, K.F.; Steward, G.F. Host traits drive viral life histories across phytoplankton viruses. Am. Nat. 2018, 191, 566–581. [Google Scholar] [CrossRef] [PubMed]
- Noble, R.T.; Fuhrman, J.A. Virus decay and its causes in coastal waters. Appl. Environ. Microbiol. 1997, 63, 77–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, L.E.; Deming, J.W. Effects of temperature, salinity and clay particles on inactivation and decay of cold-active marine Bacteriophage 9A. Aquat. Microb. Ecol. 2006, 45, 31–39. [Google Scholar] [CrossRef]
- Wilhelm, S.W.; Jeffrey, W.H.; Suttle, C.A.; Mitchell, D.L. Estimation of biologically damaging UV levels in marine surface waters with DNA and viral dosimeters. Photochem. Photobiol. 2002, 76, 268–273. [Google Scholar] [CrossRef]
- Wommack, K.E.; Hill, R.T.; Muller, T.A.; Colwell, R.R. Effects of sunlight on bacteriophage viability and structure. Appl. Environ. Microbiol. 1996, 62, 1336–1341. [Google Scholar] [CrossRef] [Green Version]
- Carlson, M.C.G.; Ribalet, F.; Maidanik, I.; Durham, B.P.; Hulata, Y.; Ferron, S.; Weissenbach, J.; Shamir, N.; Goldin, S.; Baran, N.; et al. Viruses affect picocyanobacterial abundance and biogeography in the North Pacific Ocean. Nat. Microbiol. 2022, 7, 570–580. [Google Scholar] [CrossRef]
- Suttle, C.A.; Chan, A.M.; Cottrell, M.T. Infection of phytoplankton by viruses and reduction of primary productivity. Nature 1990, 347, 467–469. [Google Scholar] [CrossRef]
- Brussaard, C.P. Viral control of phytoplankton populations—A review. J. Eukaryot. Microbiol. 2004, 51, 125–138. [Google Scholar] [CrossRef]





| NO3− | PO43− | MZc | PHYn | CHL | PP | BAc | BP | sDOC | |
|---|---|---|---|---|---|---|---|---|---|
| HOT site | |||||||||
| Standard model | 1.2 | 0.4 | 1.2 | 1.4 | 9.1 | 24 | 11 | 2.3 | 1.8 |
| Viral model | 0.9 | 0.4 | 1.1 | 1.4 | 8.9 | 24 | 11 | 1.7 | 1.5 |
| AS site | |||||||||
| Standard model | 6.8 | 1.7 | 1.4 | 38 | 4.4 | 10.6 | 5.5 | 7.3 | 3.1 |
| Viral model | 5.9 | 1.4 | 1.3 | 38 | 4.3 | 10.4 | 6.3 | 6.9 | 3.3 |
| sDON | sDOP | POC | PON | POP | STc | STn | STp | Total | |
| HOT site | |||||||||
| Standard model | 1.5 | 1.9 | 3 | 1.7 | 1.7 | 0.8 | 0.9 | 0.56 | 64.9 |
| Viral model | 1.4 | 1.8 | 3 | 1.7 | 1.6 | 0.8 | 0.9 | 0.56 | 62.7 |
| AS site | |||||||||
| Standard model | 1.7 | 2.4 | 5.0 | 5.4 | 4.7 | 97.4 | |||
| Viral model | 1.5 | 2.4 | 5.0 | 4.9 | 4.3 | 95.7 | |||
| Symbol | Value at HOT | Value at AS | Unit | Description |
|---|---|---|---|---|
| 1.82 × 10−14 | 2.17 × 10−14 | m3 particle−1 d−1 | Viral infection rate on heterotrophic bacteria | |
| 15.7 | 23 | unitless | Burst size | |
| 1.84 × 10−14 | 5.11 × 10−14 | m3 particle−1 d−1 | Parameter for viral decay | |
| 0.05 | 0.05 | fg C particle−1 | Carbon content per virus | |
| 10 | 10 | fg C cell−1 | Carbon content per heterotrophic bacterial cell |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, L.; Zhang, R.; Luo, Y.-W. Assessment of Explicit Representation of Dynamic Viral Processes in Regional Marine Ecological Models. Viruses 2022, 14, 1448. https://doi.org/10.3390/v14071448
Xie L, Zhang R, Luo Y-W. Assessment of Explicit Representation of Dynamic Viral Processes in Regional Marine Ecological Models. Viruses. 2022; 14(7):1448. https://doi.org/10.3390/v14071448
Chicago/Turabian StyleXie, Le, Rui Zhang, and Ya-Wei Luo. 2022. "Assessment of Explicit Representation of Dynamic Viral Processes in Regional Marine Ecological Models" Viruses 14, no. 7: 1448. https://doi.org/10.3390/v14071448






