Combination of E- and NS1-Derived DNA Vaccines: The Immune Response and Protection Elicited in Mice against DENV2
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Dengue Virus
2.2. Previous DNA Vaccines
2.3. Construction of the Plasmid pNS1/E/D2
2.4. DNA Vaccine Purification
2.5. Transfection of BHK Cells and Detection of the Recombinant Proteins
2.5.1. Immunofluorescence Assay
2.5.2. Flow Cytometry
2.5.3. Mass Spectrometry
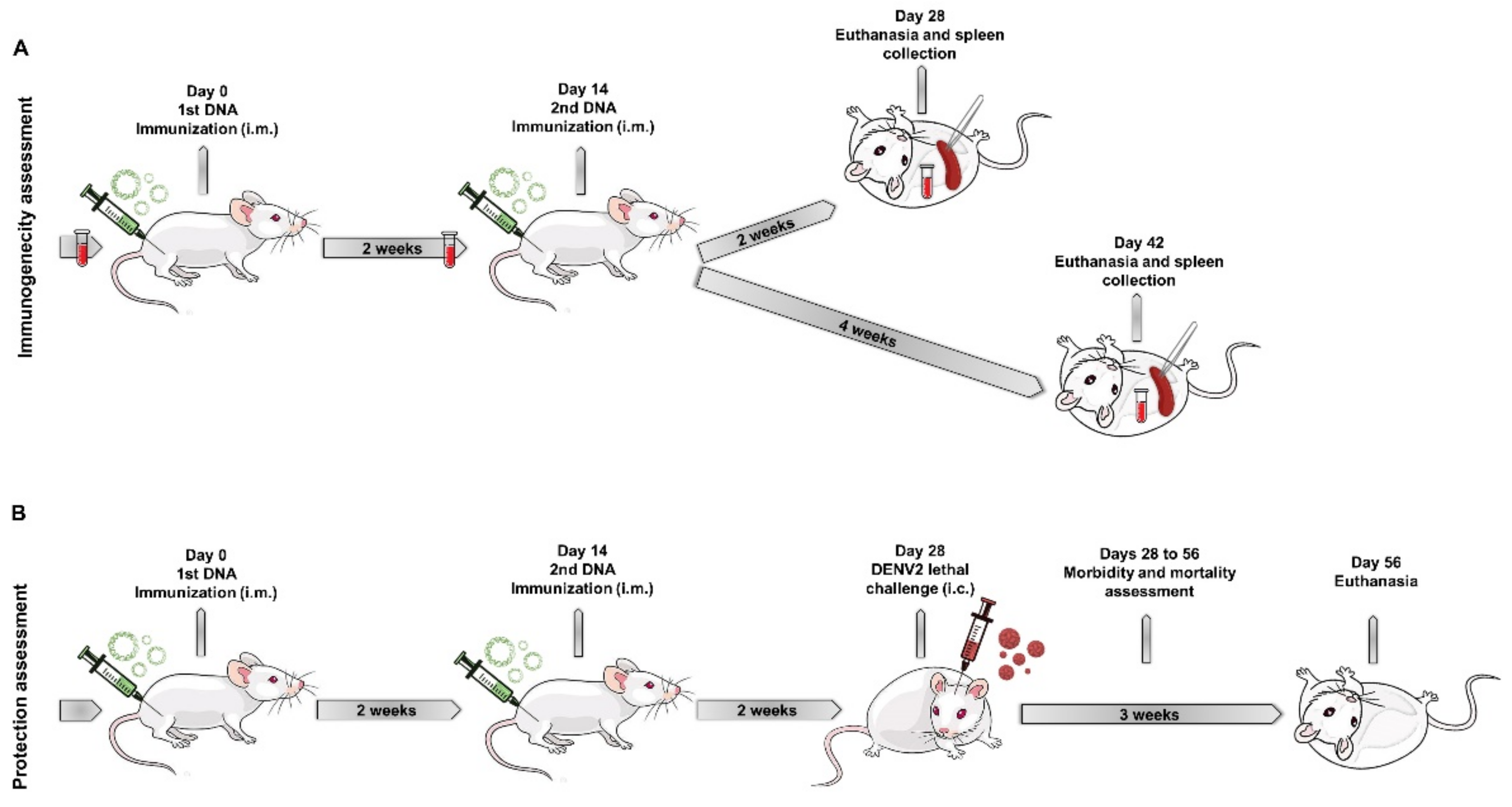
2.6. Mice Immunization and Challenge with DENV2
2.7. Detection of Antibodies against E and NS1 Proteins
2.8. Plaque Reduction Neutralization Test (PRNT50)
2.9. Interferon Gamma ELISPOT Assay
2.10. Statistics
3. Results
3.1. Detection of the Recombinant E and NS1 Proteins in Cells Transfected with the DNA Vaccines
3.2. Identification and Quantification of Secreted E and NS1 Proteins in the Supernatant of Cells Transfected with the DNA Vaccines
3.3. Antibody Response against the E Protein Generated by the DNA Vaccines
3.4. Antibody Response against the NS1 Protein Induced by the DNA Vaccines
3.5. Activation of the Cellular Immune Response by the DNA Vaccines
3.6. Protection Elicited in BALB/c Mice Immunized with the Different DNA Vaccines
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Special Programme for Research and Training in Tropical Diseases. In Dengue: Guidelines for Diagnosis, Treatment, Prevention and Control, New ed.; World Health Organization: Geneva, Switzerland, 2009; ISBN 978-92-4-154787-1. [Google Scholar]
- Brady, O.J.; Gething, P.W.; Bhatt, S.; Messina, J.P.; Brownstein, J.S.; Hoen, A.G.; Moyes, C.L.; Farlow, A.W.; Scott, T.W.; Hay, S.I. Refining the Global Spatial Limits of Dengue Virus Transmission by Evidence-Based Consensus. PLoS Negl. Trop. Dis. 2012, 6, e1760. [Google Scholar] [CrossRef]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef]
- Pang, E.L.; Loh, H.-S. Towards development of a universal dengue vaccine—How close are we? Asian Pac. J. Trop. Med. 2017, 10, 220–228. [Google Scholar] [CrossRef]
- Guy, B.; Guirakhoo, F.; Barban, V.; Higgs, S.; Monath, T.P.; Lang, J. Preclinical and clinical development of YFV 17D-based chimeric vaccines against dengue, West Nile and Japanese encephalitis viruses. Vaccine 2010, 28, 632–649. [Google Scholar] [CrossRef]
- WHO. Dengue vaccine: WHO position paper—July 2016. Wkly. Epidemiol. Rec. 2016, 91, 349–364. [Google Scholar]
- Hadinegoro, S.R.; Arredondo-García, J.L.; Capeding, M.R.; Deseda, C.; Chotpitayasunondh, T.; Dietze, R.; Ismail, H.H.M.; Reynales, H.; Limkittikul, K.; Rivera-Medina, D.M.; et al. Efficacy and Long-Term Safety of a Dengue Vaccine in Regions of Endemic Disease. N. Engl. J. Med. 2015, 373, 1195–1206. [Google Scholar] [CrossRef] [Green Version]
- Aguiar, M.; Stollenwerk, N. Dengvaxia Efficacy Dependency on Serostatus: A Closer Look at More Recent Data. Clin. Infect. Dis. 2017, 66, 641–642. [Google Scholar] [CrossRef] [Green Version]
- Halstead, S.B. Dengvaxia sensitizes seronegatives to vaccine enhanced disease regardless of age. Vaccine 2017, 35, 6355–6358. [Google Scholar] [CrossRef]
- WHO. Dengue vaccine: WHO position paper—September 2018. Wkly. Epidemiol. Rec. 2018, 93, 457–476. [Google Scholar]
- A Henchal, E.; Putnak, J.R. The dengue viruses. Clin. Microbiol. Rev. 1990, 3, 376–396. [Google Scholar] [CrossRef]
- Gubler, D.J. Dengue and Dengue Hemorrhagic Fever. Clin. Microbiol. Rev. 1998, 11, 480–496. [Google Scholar] [CrossRef] [Green Version]
- Modis, Y.; Ogata, S.; Clements, D.; Harrison, S.C. Structure of the dengue virus envelope protein after membrane fusion. Nature 2004, 427, 313–319. [Google Scholar] [CrossRef]
- Zhang, W.; Chipman, P.R.; Corver, J.; Johnson, P.R.; Zhang, Y.; Mukhopadhyay, S.; Baker, T.S.; Strauss, J.H.; Rossmann, M.G.; Kuhn, R.J. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Mol. Biol. 2003, 10, 907–912. [Google Scholar] [CrossRef] [Green Version]
- Lindenbach, B.D.; Thiel, H.-J.; Rice, C.M. Flaviviridae: The Viruses and Their Replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Eds.; Lippincott-Raven Publishers: Philadelphia, PA, USA, 2007; pp. 1101–1152. [Google Scholar]
- Rouvinski, A.; Guardado-Calvo, P.; Barba-Spaeth, G.; Duquerroy, S.; Vaney, M.-C.; Kikuti, C.M.; Sanchez, M.E.N.; Dejnirattisai, W.; Wongwiwat, W.; Haouz, A.; et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 2015, 520, 109–113. [Google Scholar] [CrossRef]
- Gan, E.S.; Ting, D.H.R.; Chan, K.R. The mechanistic role of antibodies to dengue virus in protection and disease pathogenesis. Expert Rev. Anti. Infect. Ther. 2017, 15, 111–119. [Google Scholar] [CrossRef] [Green Version]
- Halstead, S.B. Neutralization and Antibody-Dependent Enhancement of Dengue Viruses. In Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2003; Volume 60, pp. 421–467. ISBN 978-0-12-039860-7. [Google Scholar]
- Weiskopf, D.; Cerpas, C.; Angelo, M.A.; Bangs, D.J.; Sidney, J.; Paul, S.; Peters, B.; Sanches, F.P.; Silvera, C.G.T.; Costa, P.R.; et al. Human CD8+T-Cell Responses Against the 4 Dengue Virus Serotypes Are Associated With Distinct Patterns of Protein Targets. J. Infect. Dis. 2015, 212, 1743–1751. [Google Scholar] [CrossRef] [Green Version]
- Weiskopf, D.; Bangs, D.J.; Sidney, J.; Kolla, R.V.; de Silva, A.D.; de Silva, A.M.; Crotty, S.; Peters, B.; Sette, A. Dengue virus infection elicits highly polarized CX3CR1 + cytotoxic CD4 + T cells associated with protective immunity. Proc. Natl. Acad. Sci. USA 2015, 112, E4256–E4263. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-W.; Hu, H.-M.; Wu, S.-H.; Chiang, C.-Y.; Hsiao, Y.-J.; Wu, C.-K.; Hsieh, C.-H.; Chung, H.-H.; Chong, P.; Leng, C.-H.; et al. The Immunodominance Change and Protection of CD4+ T-Cell Responses Elicited by an Envelope Protein Domain III-Based Tetravalent Dengue Vaccine in Mice. PLoS ONE 2015, 10, e0145717. [Google Scholar] [CrossRef]
- Pryor, M.J.; Wright, P.J. Glycosylation Mutants of Dengue Virus NS1 Protein. J. Gen. Virol. 1994, 75, 1183–1187. [Google Scholar] [CrossRef]
- Jacobs, M.G.; Robinson, P.J.; Bletchly, C.; Mackenzie, J.M.; Young, P.R. Dengue virus nonstructural protein 1 is expressed in a glycosyl-phosphatidylinositol-linked form that is capable of signal transduction. FASEB J. 2000, 14, 1603–1610. [Google Scholar] [CrossRef]
- Flamand, M.; Megret, F.; Mathieu, M.; Lepault, J.; Rey, F.A.; Deubel, V. Dengue Virus Type 1 Nonstructural Glycoprotein NS1 Is Secreted from Mammalian Cells as a Soluble Hexamer in a Glycosylation-Dependent Fashion. J. Virol. 1999, 73, 6104–6110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlesinger, J.J.; Brandriss, M.W.; Walsh, E.E. Protection of Mice Against Dengue 2 Virus Encephalitis by Immunization with the Dengue 2 Virus Non-structural Glycoprotein NS1. J. Gen. Virol. 1987, 68, 853–857. [Google Scholar] [CrossRef] [PubMed]
- Henchal, E.A.; Henchal, L.S.; Schlesinger, J.J. Synergistic Interactions of Anti-NS1 Monoclonal Antibodies Protect Passively Immunized Mice from Lethal Challenge with Dengue 2 Virus. J. Gen. Virol. 1988, 69, 2101–2107. [Google Scholar] [CrossRef] [PubMed]
- Kao, Y.-S.; Yu, C.-Y.; Huang, H.-J.; Tien, S.-M.; Wang, W.-Y.; Yang, M.; Anderson, R.; Yeh, T.-M.; Lin, Y.-S.; Wan, S.-W. Combination of Modified NS1 and NS3 as a Novel Vaccine Strategy against Dengue Virus Infection. J. Immunol. 2019, 203, 1909–1917. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, D.A.; Beatty, P.R.; Reiner, G.L.; Sivick, K.E.; Glickman, L.H.; Dubensky, T.W.; Harris, E. Cyclic Dinucleotide–Adjuvanted Dengue Virus Nonstructural Protein 1 Induces Protective Antibody and T Cell Responses. J. Immunol. 2019, 202, 1153–1162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, S.M.; Paes, M.; Barreto, D.F.; Pinhão, A.T.; Barth, O.M.; Queiroz, J.L.; Armôa, G.R.; Freire, M.S.; Alves, A.M. Protection against dengue type 2 virus induced in mice immunized with a DNA plasmid encoding the non-structural 1 (NS1) gene fused to the tissue plasminogen activator signal sequence. Vaccine 2006, 24, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Amorim, J.H.; Diniz, M.O.; Cariri, F.A.; Rodrigues, J.F.; Bizerra, R.S.P.; Gonçalves, A.J.; de Barcelos Alves, A.M.; de Souza Ferreira, L.C. Protective immunity to DENV2 after immunization with a recombinant NS1 protein using a genetically detoxified heat-labile toxin as an adjuvant. Vaccine 2012, 30, 837–845. [Google Scholar] [CrossRef] [Green Version]
- Weiskopf, D.; Angelo, M.A.; de Azeredo, E.L.; Sidney, J.; Greenbaum, J.A.; Fernando, A.N.; Broadwater, A.; Kolla, R.V.; de Silva, A.D.; de Silva, A.M.; et al. Comprehensive analysis of dengue virus-specific responses supports an HLA-linked protective role for CD8+ T cells. Proc. Natl. Acad. Sci. USA 2013, 110, E2046–E2053. [Google Scholar] [CrossRef] [Green Version]
- Gonçalves, A.J.S.; Oliveira, E.R.; Costa, S.M.; Paes, M.; Silva, J.F.A.; Azevedo, A.S.; Mantuano-Barradas, M.; Nogueira, A.C.M.A.; Almeida, C.J.; Alves, A.M.B. Cooperation between CD4+ T Cells and Humoral Immunity Is Critical for Protection against Dengue Using a DNA Vaccine Based on the NS1 Antigen. PLoS Negl. Trop. Dis. 2015, 9, e0004277. [Google Scholar] [CrossRef] [Green Version]
- Azevedo, A.S.; Yamamura, A.M.Y.; Freire, M.S.; Trindade, G.; Bonaldo, M.; Galler, R.; Alves, A.M.B. DNA Vaccines against Dengue Virus Type 2 Based on Truncate Envelope Protein or Its Domain III. PLoS ONE 2011, 6, e20528. [Google Scholar] [CrossRef]
- Costa, S.; Azevedo, A.; Paes, M.; Sarges, F.; Freire, M.; Alves, A. DNA vaccines against dengue virus based on the ns1 gene: The influence of different signal sequences on the protein expression and its correlation to the immune response elicited in mice. Virology 2007, 358, 413–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azevedo, A.S.; Gonçalves, A.J.S.; Archer, M.; Freire, M.S.; Galler, R.; Alves, A.M.B. The Synergistic Effect of Combined Immunization with a DNA Vaccine and Chimeric Yellow Fever/Dengue Virus Leads to Strong Protection against Dengue. PLoS ONE 2013, 8, e58357. [Google Scholar] [CrossRef] [PubMed]
- Pinto, P.B.A.; de Assis, M.; Vallochi, A.L.; Pacheco, A.R.; Lima, L.M.; Quaresma, K.R.L.; Pereira, B.; Costa, S.M.; Alves, A.M.B. T Cell Responses Induced by DNA Vaccines Based on the DENV2 E and NS1 Proteins in Mice: Importance in Protection and Immunodominant Epitope Identification. Front. Immunol. 2019, 10, 1522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caufour, P.; Motta, M.; Yamamura, A.; Vazquez, S.; Ferreira, I.; Jabor, A.; Bonaldo, M.; Freire, M.; Galler, R. Construction, characterization and immunogenicity of recombinant yellow fever 17D-dengue type 2 viruses. Virus Res. 2001, 79, 1–14. [Google Scholar] [CrossRef]
- Angel-Camilo, K.L.; Guerrero-Vargas, J.A.; Carvalho, E.F.; Lima-Silva, K.; de Siqueira, R.J.B.; Freitas, L.B.N.; Sousa, J.a.C.; Mota, M.R.L.; Santos, A.A.D.; Neves-Ferreira, A.; et al. Disorders on cardiovascular parameters in rats and in human blood cells caused by Lachesis acrochorda snake venom. Toxicon 2020, 184, 180–191. [Google Scholar] [CrossRef]
- Camacho, E.; Sanz, L.; Escalante, T.; Perez, A.; Villalta, F.; Lomonte, B.; Neves-Ferreira, A.G.; Feoli, A.; Calvete, J.J.; Gutierrez, J.M.; et al. Novel Catalytically-Inactive PII Metalloproteinases from a Viperid Snake Venom with Substitutions in the Canonical Zinc-Binding Motif. Toxins 2016, 8, 292. [Google Scholar] [CrossRef] [Green Version]
- Santos, M.D.M.; Lima, D.B.; Fischer, J.S.G.; Clasen, M.A.; Kurt, L.U.; Camillo-Andrade, A.C.; Monteiro, L.C.; Aquino, P.F.; Neves-Ferreira, A.G.C.; Valente, R.H.; et al. Simple, efficient and thorough shotgun proteomic analysis with PatternLab V. Nat. Protoc. 2022. [Google Scholar] [CrossRef]
- Rothman, A.L.; Kurane, I.; Ennis, F.A. Multiple Specificities in the Murine CD4+ and CD8+ T-Cell Response to Dengue Virus. J. Virol. 1996, 70, 7. [Google Scholar] [CrossRef] [Green Version]
- Gao, G.; Wang, Q.; Dai, Z.; Calcedo, R.; Sun, X.; Li, G.; Wilson, J.M. Adenovirus-Based Vaccines Generate Cytotoxic T Lymphocytes to Epitopes of NS1 from Dengue Virus That Are Present in All Major Serotypes. Hum. Gene Ther. 2008, 19, 927–936. [Google Scholar] [CrossRef]
- Deng, S.-Q.; Yang, X.; Wei, Y.; Chen, J.-T.; Wang, X.-J.; Peng, H.-J. A Review on Dengue Vaccine Development. Vaccines 2020, 8, 63. [Google Scholar] [CrossRef] [Green Version]
- Izmirly, A.M.; Alturki, S.O.; Alturki, S.O.; Connors, J.; Haddad, E.K. Challenges in Dengue Vaccines Development: Pre-existing Infections and Cross-Reactivity. Front. Immunol. 2020, 11, 1055. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Dar, L. Dengue vaccines: Challenges, development, current status and prospects. Indian J. Med. Microbiol. 2015, 33, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.K.; Putnak, J.R.; Warren, R.L.; Hoke, C.H. Mice immunized with a dengue type 2 virus E and NS1 fusion protein made in Escherichia coli are protected against lethal dengue virus infection. Vaccine 1995, 13, 1251–1258. [Google Scholar] [CrossRef]
- Mellado-Sánchez, G.; García-Machorro, J.; Sandoval-Montes, C.; Gutiérrez-Castañeda, B.; Rojo-Domínguez, A.; García-Cordero, J.; Santos-Argumedo, L.; Cedillo-Barrón, L. A plasmid encoding parts of the dengue virus E and NS1 proteins induces an immune response in a mouse model. Arch. Virol. 2010, 155, 847–856. [Google Scholar] [CrossRef]
- Zheng, Q.; Fan, D.; Gao, N.; Chen, H.; Wang, J.; Ming, Y.; Li, J.; An, J. Evaluation of a DNA vaccine candidate expressing prM-E-NS1 antigens of dengue virus serotype 1 with or without granulocyte-macrophage colony-stimulating factor (GM-CSF) in immunogenicity and protection. Vaccine 2010, 29, 763–771. [Google Scholar] [CrossRef]
- Lu, H.; Xu, X.-F.; Gao, N.; Fan, D.-Y.; Wang, J.; An, J. Preliminary evaluation of DNA vaccine candidates encoding dengue-2 prM/E and NS1: Their immunity and protective efficacy in mice. Mol. Immunol. 2012, 54, 109–114. [Google Scholar] [CrossRef]
- Dey, A.; Rajanathan, T.C.; Chandra, H.; Pericherla, H.P.; Kumar, S.; Choonia, H.S.; Bajpai, M.; Singh, A.K.; Sinha, A.; Saini, G.; et al. Immunogenic potential of DNA vaccine candidate, ZyCoV-D against SARS-CoV-2 in animal models. Vaccine 2021, 39, 4108–4116. [Google Scholar] [CrossRef]
- Eflingai, S.; Eczerwonko, M.; Egoodman, J.; Kudchodkar, S.B.; Emuthumani, K.; Weiner, D.B. Synthetic DNA Vaccines: Improved Vaccine Potency by Electroporation and Co-Delivered Genetic Adjuvants. Front. Immunol. 2013, 4, 354. [Google Scholar] [CrossRef] [Green Version]
- Schlesinger, J.J.; Brandriss, M.W.; Cropp, C.B.; Monath, T.P. Protection against yellow fever in monkeys by immunization with yellow fever virus non-structural protein. J. Virol. 1986, 60, 1153–1155. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.-F.; Liao, C.-L.; Lin, Y.-L.; Yeh, C.-T.; Chen, L.-K.; Huang, Y.-F.; Chou, H.-Y.; Huang, J.-L.; Shaio, M.-F.; Sytwu, H.-K. Evaluation of protective efficacy and immune mechanisms of using a non-structural protein NS1 in DNA vaccine against dengue 2 virus in mice. Vaccine 2003, 21, 3919–3929. [Google Scholar] [CrossRef]
- Whiteman, M.C.; Li, L.; Wicker, J.A.; Kinney, R.M.; Huang, C.; Beasley, D.W.; Chung, K.M.; Diamond, M.S.; Solomon, T.; Barrett, A.D. Development and characterization of non-glycosylated E and NS1 mutant viruses as a potential candidate vaccine for West Nile virus. Vaccine 2009, 28, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.-C.; Chuang, Y.-C.; Liu, C.-C.; Ho, T.-S.; Lin, Y.-S.; Anderson, R.; Yeh, T.-M. Antibodies Against Modified NS1 Wing Domain Peptide Protect Against Dengue Virus Infection. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.J.; Duehr, J.; Dulin, H.; Broecker, F.; Brown, J.A.; Arumemi, F.O.; González, M.C.B.; Leyva-Grado, V.H.; Evans, M.J.; Simon, V.; et al. Human antibodies targeting Zika virus NS1 provide protection against disease in a mouse model. Nat. Commun. 2018, 9, 4560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Alwis, R.; Smith, S.A.; Olivarez, N.P.; Messer, W.B.; Huynh, J.P.; Wahala, W.M.P.B.; White, L.J.; Diamond, M.S.; Baric, R.S.; Crowe, J.E.; et al. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc. Natl. Acad. Sci. USA 2012, 109, 7439–7444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, W.-Y.; Chen, H.-L.; Tsai, J.-J.; Dejnirattisai, W.; Jumnainsong, A.; Mongkolsapaya, J.; Screaton, G.; Crowe, J.E.; Wang, W.-K. Potent Neutralizing Human Monoclonal Antibodies Preferentially Target Mature Dengue Virus Particles: Implication for Novel Strategy for Dengue Vaccine. J. Virol. 2018, 92, e00556-18. [Google Scholar] [CrossRef] [Green Version]
- Modhiran, N.; Song, H.; Liu, L.; Bletchly, C.; Brillault, L.; Amarilla, A.A.; Xu, X.; Qi, J.; Chai, Y.; Cheung, S.T.M.; et al. A broadly protective antibody that targets the flavivirus NS1 protein. Science 2021, 371, 190–194. [Google Scholar] [CrossRef] [PubMed]
- Yauch, L.E.; Zellweger, R.M.; Kotturi, M.F.; Qutubuddin, A.; Sidney, J.; Peters, B.; Prestwood, T.R.; Sette, A.; Shresta, S. A Protective Role for Dengue Virus-Specific CD8+T Cells. J. Immunol. 2009, 182, 4865–4873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zompi, S.; Santich, B.; Beatty, P.R.; Harris, E. Protection from Secondary Dengue Virus Infection in a Mouse Model Reveals the Role of Serotype Cross-Reactive B and T Cells. J. Immunol. 2011, 188, 404–416. [Google Scholar] [CrossRef] [Green Version]
- Zellweger, R.M.; Tang, W.W.; Eddy, W.E.; King, K.; Sanchez, M.C.; Shresta, S. CD8 + T Cells Can Mediate Short-Term Protection against Heterotypic Dengue Virus Reinfection in Mice. J. Virol. 2015, 89, 6494–6505. [Google Scholar] [CrossRef] [Green Version]
- Ngono, A.E.; Chen, H.-W.; Tang, W.; Joo, Y.; King, K.; Weiskopf, D.; Sidney, J.; Sette, A.; Shresta, S. Protective Role of Cross-Reactive CD8 T Cells Against Dengue Virus Infection. eBioMedicine 2016, 13, 284–293. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Grifoni, A.; Sette, A.; Weiskopf, D. Human T Cell Response to Dengue Virus Infection. Front. Immunol. 2019, 10, 2125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivino, L.; Kumaran, E.A.P.; Jovanovic, V.; Nadua, K.; Teo, E.W.; Pang, S.W.; Teo, G.H.; Gan, V.C.H.; Lye, D.C.; Leo, Y.S.; et al. Differential Targeting of Viral Components by CD4 + versus CD8 + T Lymphocytes in Dengue Virus Infection. J. Virol. 2013, 87, 2693–2706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zellweger, R.M.; Shresta, S. Mouse models to study dengue virus immunology and pathogenesis. Front. Immunol. 2014, 5, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guabiraba, R.; Ryffel, B. Dengue virus infection: Current concepts in immune mechanisms and lessons from murine models. Immunology 2014, 141, 143–156. [Google Scholar] [CrossRef]
- Porter, K.R.; Kochel, T.J.; Wu, S.-J.; Raviprakash, K.; Phillips, I.; Hayes, C.G. Protective efficacy of a dengue 2 DNA vaccine in mice and the effect of CpG immuno-stimulatory motifs on antibody responses. Arch. Virol. 1998, 143, 997–1003. [Google Scholar] [CrossRef]
- Clements, D.E.; Coller, B.-A.G.; Lieberman, M.M.; Ogata, S.; Wang, G.; Harada, K.E.; Putnak, J.R.; Ivy, J.M.; McDonell, M.; Bignami, G.S.; et al. Development of a recombinant tetravalent dengue virus vaccine: Immunogenicity and efficacy studies in mice and monkeys. Vaccine 2010, 28, 2705–2715. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, E.; Amorim, J.; Paes, M.; Azevedo, A.; Gonçalves, A.; Costa, S.; Mantuano-Barradas, M.; Póvoa, T.; de Meis, J.; Basílio-De-Oliveira, C.; et al. Peripheral effects induced in BALB/c mice infected with DENV by the intracerebral route. Virology 2016, 489, 95–107. [Google Scholar] [CrossRef] [Green Version]






| Gel Fraction (a) | DNA Vaccine (b) | Protein (c) | Mass (d) | UniquePep (e) | UniqueSC (f) | NSAF (g) | %Coverage (h) | PtnScore (i) |
|---|---|---|---|---|---|---|---|---|
| F3-F4 | pcTPANS1 | NS1 | 42,525 | 87 | 627 | 0.2637 | 84 | 206.65 |
| F7-F8 | pE1D2 | E | 46,881 | 31 | 85 | 0.0379 | 67 | 82.73 |
| F9-F10 | pE1D2 + pcTPANS1 | NS1 | 42,525 | 66 | 484 | 0.1838 | 80 | 168.49 |
| E | 46,881 | 31 | 71 | 0.0239 | 65 | 80.26 | ||
| F11-F12 | pNS1/E/D2 | NS1 | 42,525 | 25 | 58 | 0.0293 | 55 | 53.17 |
| E | 46,881 | 30 | 62 | 0.0278 | 61 | 80.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto, P.B.A.; Barros, T.A.C.; Lima, L.M.; Pacheco, A.R.; Assis, M.L.; Pereira, B.A.S.; Gonçalves, A.J.S.; Azevedo, A.S.; Neves-Ferreira, A.G.C.; Costa, S.M.; et al. Combination of E- and NS1-Derived DNA Vaccines: The Immune Response and Protection Elicited in Mice against DENV2. Viruses 2022, 14, 1452. https://doi.org/10.3390/v14071452
Pinto PBA, Barros TAC, Lima LM, Pacheco AR, Assis ML, Pereira BAS, Gonçalves AJS, Azevedo AS, Neves-Ferreira AGC, Costa SM, et al. Combination of E- and NS1-Derived DNA Vaccines: The Immune Response and Protection Elicited in Mice against DENV2. Viruses. 2022; 14(7):1452. https://doi.org/10.3390/v14071452
Chicago/Turabian StylePinto, Paolla Beatriz A., Tamiris A. C. Barros, Lauro M. Lima, Agatha R. Pacheco, Maysa L. Assis, Bernardo A. S. Pereira, Antônio J. S. Gonçalves, Adriana S. Azevedo, Ana Gisele C. Neves-Ferreira, Simone M. Costa, and et al. 2022. "Combination of E- and NS1-Derived DNA Vaccines: The Immune Response and Protection Elicited in Mice against DENV2" Viruses 14, no. 7: 1452. https://doi.org/10.3390/v14071452






