Abstract
In an era of antibiotic therapy crisis caused by spreading antimicrobial resistance, and when recurrent urinary tract infections constitute a serious social and medical problem, the isolation and complex characterization of phages with a potential therapeutic application represents a promising solution. It is an inevitable, and even a necessary direction in the development of current phage research. In this paper, we present two newly isolated myoviruses that show lytic activity against multidrug-resistant clinical isolates of Enterobacter spp. (E. cloacae, E. hormaechei, and E. kobei), the genomes of which belong to a poorly represented phage group. Both phages were classified as part of the Tevenvirinae subfamily (Entb_43 was recognized as Karamvirus and Entb_45 as Kanagawavirus). Phage lytic spectra ranging from 40 to 60% were obtained. The most effective phage-to-bacteria ratios (MOI = 0.01 and MOI = 0.001) for both the phage amplification and their lytic activity against planktonic bacteria were also estimated. Complete adsorption to host cells were obtained after about 20 min for Entb_43 and 10 min for Entb_45. The phage lysates retained their initial titers even during six months of storage at both −70 °C and 4 °C, whereas storage at 37 °C caused a complete loss in their activity. We showed that phages retained their activity after incubation with solutions of silver and copper nanoparticles, which may indicate possible synergistic antibacterial activity. Moreover, a significant reduction in phage titers was observed after incubation with a disinfectant containing octenidinum dihydrochloridum and phenoxyethanol, as well as with 70% ethanol. The observed maintenance of phage activity during incubation in a urine sample, along with other described properties, may suggest a therapeutic potential of phages at the infection site after intravesical administration.
1. Introduction
Bacteria of the Enterobacter species were first described in 1960 by Hormaeche and Edwards as Gram-negative, facultative anaerobic bacilli with the ability to move [1]. They belong to the ESKAPE (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) group of pathogens, which include bacteria that are characterized by a constantly increasing resistance to antibiotics—this is a huge problem, particularly for hospital environments [2,3]. The name of this group of microorganisms is related to their “escape” from the effects of antibiotics and other antimicrobial agents [4,5]. According to a report by the World Health Organization (WHO) published in 2017 on the need to search for new antibiotics and ways to combat multidrug-resistant pathogens, bacteria from the Enterobacteriaceae family (including Enterobacter spp.) belong to the highest priority group [6]. Infections with these pathogens are especially dangerous in immunocompromised patients, e.g., in solid organ transplant recipients on chronic immunosuppressive therapy [7] or in hemodialysis patients [8]. Enterobacter cloacae, the main representative of Enterobacter spp., colonizes the mucosa of the human intestine in a physiological state; however, it is also described as the source of many different disorders, including bacteremia [9,10], urinary tract infections (UTIs) [11,12], wound infections [13,14,15], osteoarticular infections [16,17,18], and pneumonia/lower respiratory tract infections [19,20,21]. These bacteria also commonly colonize in various hospital wards, including neonatal intensive care units, which may result in the development of life-threatening infections such as meningitidis [22]. Moreover, it has been suggested that the presence of E. cloacae in the colon environment may be associated with the initiation and progression of colorectal cancer [23].
A significant proportion of clinical bacterial isolates are extended-spectrum β-lactamase (ESBL)-producing strains. The principal described genes responsible for this mechanism of resistance to β-lactam antibiotics (penicillins, cephalosporins, and monobactams) include blaTEM, blaCTX-M, and blaSHV [20,24,25]. Broad-spectrum antibiotic resistance also includes resistance to carbapenems, known as last-resort antibiotics, and to colistin, which often causes a wide range of side effects [26,27]. Related genes that have increasingly been found in clinical strains are, among others, blaKPC−2, as well as blaNDM, and the presence of efflux pumps is also important [25,27,28]. Recent studies describing Enterobacter-induced UTIs indicate the involvement of extensive drug resistant (XDR), multidrug resistant (MDR), as well as carbapenem-resistant Enterobacterales (CRE) strains in the pathogenesis of infection in hospitalized patients in Eastern Europe [29].
In an era of increasing antibiotic resistance, the search for new alternative infection treatment strategies is a major medical challenge. Although antibiotics are losing their effectiveness, one way to combat bacterial pathogens is bacteriophage therapy. Bacteriophages (phages)—viruses specific to their bacterial host—after penetrating into the host pathogen, amplify and destroy them, and should not negatively impact the physiological microbiota [30]. Various environmental samples are used to search and isolate new therapeutic bacteriophages [31]. Despite the ubiquitous presence of phages in the environment, there may be many obstacles in their isolation, as well as the possibility of using them as therapeutic agents. There is a constant need to obtain preclinical data on new phages in order to assess their efficacy and safety, both in vitro and in vivo [32], especially in the case of urinary tract infections in which Enterobacter strains are etiological agents. In our study, we have isolated and characterized two new bacteriophages which are specific for multidrug-resistant (MDR) Enterobacter spp. strains, and which are meant to be applicable in phage therapy.
2. Materials and Methods
2.1. Bacterial Strains
In the study, we used twenty clinical strains of antibiotic-resistant Enterobacter, which were isolated from patients with infections of the genitourinary system (n = 7); ears (n = 2); and wounds, ulcers, or purulent lesions (n = 11); who applied to qualify for experimental bacteriophage therapy at the Ludwik Hirszfeld Institute of Immunology and Experimental Therapy, Polish Academy of Sciences (HIIET PAS) in the years 2017–2021. The species of bacterial strains were determined using matrix-assisted laser desorption/ionization—time of flight (MALDI-TOF/TOF) mass spectrometry (Bruker Daltonics, Billerica, MA, USA). Antibiotic sensitivity was determined by the microdilution method using the MicroScan WalkAway analyzer (Beckman Coulter, Brea, CA, USA). All bacteria were grown on MacConkey agar (incubation at 37 °C, approximately 18 h), but were stored frozen in a 20% glycerol (Difco) solution at −70 °C prior to the experiments.
2.2. Bacteriophage Isolation and Amplification
Almost three hundred and forty environmental samples (such as water samples from hospital and municipal wastewater, rivers, lakes, seas, fountains, households, and yards) from various geographical locations, from the collection of HIIET, were used to isolate new bacteriophages against Enterobacter spp. Water samples were stored as raw, incubated (i.e., mixed with peptone water and incubated at 37 °C before filtering), or 20 times concentrated (by tangential filtration with the use of a Vivaflow 200 ultrafiltration kit with a Hydrosart membrane and a MWCO 30 kDa operating at a pressure of 3 bar). A phage search based on the method described by Ślopek et al. (1983) [33] was used. For this purpose, bacterial suspensions were made in sugar broth (in the composition: meat extract 0.4 g, enzymatic hydrolysate of casein 5.4 g, yeast hydrolysate 1.7 g, Bacto Peptone 4.0 g, NaCl 3.5 g, glucose 10 g—per 1000 mL of H2O) and incubated for 2 h at 37 °C to obtain an optical density at wavelength λ = 600 nm (OD600) 0.4–0.6 (bacteria in logarithmic (midlog) phase growth), measured spectrophotometrically (BioSpectrometer basic, Eppendorf, Hamburg, Germany). The bacterial suspensions were subsequently poured onto plates with solidified agar intended for the cultivation of Gram-negative bacteria (pH = 7.4; composition in % w/v: distilled water, 1.2% agar (bioMerieux, Durham, NC, USA), 1.0% tryptone—enzymatic casein hydrolysate (bioMerieux, Durham, NC, USA), 0.5% NaCl (POCH, Gliwice, Poland), 0.3% meat extract (bioMerieux, Durham, NC, USA), 0.3% Na2HPO4 (POCH, Gliwice, Poland)), dried for a few minutes at room temperature (RT) and then for 1 h at 37 °C. Then, each environmental water sample was spotted on the plates, which were incubated for 3.5 h at 37 °C. Plates were left at 4 °C overnight and the results were read the next day. Complete/semicomplete lysis of the bacteria or single plaques at the site where the sample was spotted were considered a positive result. To isolate the phage, 200 µL of a two-hour culture of the host bacteria was suspended in 10 mL of peptone water (in the composition: meat extract 0.4 g, enzymatic hydrolyzate of casein 5.4 g, yeast hydrolyzate 1.7 g, Bacto Peptone 4.0 g, NaCl 3.5 g—per 1000 mL of H2O), and then 200 µL of the specific water sample was added, vortexed, and placed in an incubator (37 °C) overnight. The samples were filtered using syringe bacteriological filters with a pore diameter of 0.22 µm (Millipore, Burlington, MA, USA). Their serial dilutions (100–10−8) were completed and 50 µL of each were spotted on the previously prepared plates with bacterial lawn (routine test dilution—RTD method). After incubation (3.5 h at 37 °C and then overnight at 4 °C), the number of plaques was counted. Phage dilutions, in which the 10–40 plaques were present, were then used in the double-layer agar method [34,35,36,37]. Briefly, 100 µL of the host bacteria and 200 µL of the given phage lysate’s dilution were added to 2 mL of melted 0.7% agar, mixed, and poured onto plates with already solidified agar. Plates were incubated for 4 h at 37 °C, then at 4 °C, and the phage titers were calculated the next day.
2.3. Assessment of the Lytic Spectrum of Phages
For the evaluation of the lytic spectrum of each phage, the bacterial suspensions of each of 20 Enterobacter spp. strains were used, with the subsequent procedure being very similar to that used in the search for new bacteriophages. Each amplified phage lysate (40 µL), at a titer of approximately 107–108 PFU/mL, was spotted on the plates with a bacterial lawn, incubated under the same conditions, and assessed the next day according to the signs: CL—confluent lysis, SCL—semiconfluent lysis, +++—60 and more confluent plaques, ++—20–60 separate plaques, +—10–20 separate plaques, +/−—single plaques, OL—opaque lysis. The percentage of bacterial strains that were sensitive to specific phages made it possible to determine their lytic spectrum.
In order to investigate the specificity of the lytic activity of bacteriophages, phage typing was also performed on strains of other species from the ESKAPE group: E. faecalis (n = 2), S. aureus (n = 2), K. pneumoniae (n = 1), A. baumannii (n = 3), P. aeruginosa (n = 2), and Escherichia coli (n = 2).
2.4. Assessment of Optimal Multiplicity of Infection (MOI) for Phages
During the experiments, the best MOI was assessed to optimize bacteriophage amplification. In the first stage, plots of dependence of bacterial concentration (colony-forming unit per milliliter—CFU/mL) on the OD600 that were determined spectrophotometrically were made for bacterial hosts. Briefly, different dilutions (0, 2, 3, 5, 8, and 10-fold) of the bacterial suspension were prepared and the OD600 of each sample was measured. Then, serial dilutions of each sample (from 100 to 10−8) were made and 100 µL of the selected samples was spotted on an agar plate and spread over the entire surface. Plates were incubated at 37 °C for 11 h, then bacterial colonies were counted and bacterial titers that depend on optical density (OD600) were determined. In the next step, two-hour bacterial suspensions were made, and the bacterial concentrations were tested using the formulas generated from the plots (dependence of the bacterial titer in CFU/mL on OD600). Dilutions were then made to obtain a titer of about 3.0 × 106 CFU/mL. Different titers of phage lysates were also prepared. To 10 mL of peptone water, we added 200 µL of a suspension of the appropriate bacterial strain and 200 µL of a specific phage dilution (103, 104, 105, 106, 107, 108, or 109 PFU/mL) and mixed them. After overnight incubation at 37 °C, all samples were filtered and RTD in triplicate were performed for each of them to determine phage titers.
2.5. Planktonic Bacterial Lysis Assay
The experiment was performed similarly to those described elsewhere [38]. The lytic efficiency of the two new bacteriophages was determined using suspensions with different phage and bacteria ratios (at the most effective MOI determined in the previous experiment: 0.001, 0.01, 0.1, and 1.0). Briefly, the two-hour bacterial suspension was diluted to a titer of 106 CFU/mL and samples with different MOIs were prepared. In these samples, as well as the samples with bacterial suspension (positive control) and only phage lysates (negative control), the OD600 were measured spectrophotometrically at 0, 1, 2, 3, 4, 6, 8, and 10 h of incubation at 37 °C. Two independent experiments were conducted, and in each of them, sample measurements were performed in triplicate.
2.6. Determination of the Phage Adsorption Time
To determine the kinetics of phage adsorption to their hosts and to estimate the adsorption rate constant, we used the procedures described elsewhere [39,40], with some modifications. Briefly, 10 mL of phage lysate (107 PFU/mL) was added to 10 mL of bacterial suspension (108 CFU/mL) at MOI = 0.1, and then incubated at 37 °C. At time 0 and after specified time intervals, 1 mL of the sample was taken and filtered through a syringe filter. Its serial dilutions were made and the titer of free, unadsorbed phages in the solution was determined. The number of phage particles at time 0 (immediately after mixing bacteria and viruses) represented 100% of the unadsorbed phages. Two independent experiments were conducted, and in each of them, sample measurements were performed in triplicate. The adsorption rate constants were calculated according to the following mathematical formula:
where k is the adsorption rate constant (in mL/min); B is the concentration of bacterial cells; and t is the time interval in which the titer falls from P0 (original phage titer) to P (final phage titer) [41].
2.7. Long-Term Study of Phage Stability under Various Temperature Conditions
Phage lysates in the determined titers (1.62 × 1010 PFU/mL for Entb_43; 3.67 × 107 PFU/mL for Entb_45—the difference is that a higher titer could not be obtained at the beginning of the experiment) were placed in different temperature conditions: 37 °C, 22–24 °C (room temperature—RT), 4 °C, and frozen at −70 °C with or without addition of 20% glycerol (Difco). Experiments were performed over 6 months, and samples were taken after 1, 2, 4, 10, 14, 18, and 24 weeks, respectively. Additionally, to assess if defrosting affected the titer of bacteriophages stored at −70 °C, the same sample was defrosted several times at different time intervals. Phage titers were determined using RTD and the double-layer agar method in triplicate.
2.8. Investigation of Phage Stability in Solutions of Different pH and Disinfectants
To evaluate the stability of phages in solutions with a different pH, we tested the liquid culture medium—peptone water at a different pH, at a range from 3 to 12 (established with 5 M HCl or 10 M NaOH). Phage suspensions in pH = 7.2 (standard culture medium) were used as controls. For this purpose, 200 µL of phage lysate (109 PFU/mL) was added to 1800 µL of a specific solution (at 1:9 ratio) and incubated for 1 h at 37 °C in a water bath. After this period, serial dilutions of phages were prepared, and their titers were determined using RTD in triplicate. In addition, the stability of bacteriophages in various disinfectants or surfactants (including 70% ethanol, 10% dish soap, 10% liquid soap, 10% hand wash gel, and two antibacterial and antiviral liquid skin disinfectants diluted 2 times) was assessed. Using a similar method to that presented above, 200 µL of phage lysates were added to 1800 µL of specific solution and incubated at room temperature. Phage titers were determined after 5, 10, 20, and 30 min of incubation. All experiments were performed in triplicate. Results are presented as a percentage of active phages [40]. A similar method was used to test the phage stability with commercially available colloidal silver and copper solutions (Nano-Tech, Polska) at a concentration of 50 mg/kg (ppm), also used as antibacterial and antifungal agents. In this case, an additional 24 h of incubation was performed.
2.9. Stability of Phage Lysates in Urine Sample In Vitro
The fresh urine sample collected from the midstream was obtained from a healthy donor. The urine sample presented pH = 6, and a density of 1.015. The glucose, ketones, proteins, and nitrites were absent from the urine sample, and flat epithelial cells were sparse. The sample was filtered through 0.22 µm pore size filters. As in previous experiments, phages were incubated with the sample with a ratio of 1:9 at 37 °C for 30 min, 1 h, and 24 h. Phage titers after defined incubation times were determined by RTD. The experiment was performed in triplicate. Phage titer at the start of the experiment (time zero) was used as a comparison for the statistical analysis.
2.10. Transmission Electron Microscopy (TEM)
Phage lysates were prepared in a LB (Luria–Bertani) medium in high titers (Entb_43: 2.3 × 1010 PFU/mL; Entb_45: 1.22 × 109 PFU/mL) and filtered two times. Samples were concentrated using ultracentrifugation (1 h, 7 °C, 25,000 × g) and, after removing the supernatant, 0.1 M ammonium acetate was added and centrifuged in the same conditions. In the next step, a drop of phage lysate was placed on a 400 mesh copper grid (Athene). After being contrasted with 2% uranyl acetate, preparations were dried and analyzed using the Zeiss EM900 TEM (at an acceleration voltage 80 kV). Images of phages were obtained using Kodak/Carestream Electron Microscope Film 4489. Phages’ morphology and dimensions were defined.
2.11. Phage DNA Isolation Procedure and DNA Sequencing
Phage lysates at a volume of 20 mL in high titers (Entb_43: 2.3 × 1010 PFU/mL; Entb_45: 1.22 × 109 PFU/mL) were concentrated with the use of Amicon Ultra Centrifugal Filters (Merck, NJ, USA). To isolate the phage nucleic acid (dsDNA), QIAamp MinElute Virus kits (Qiagen, Germantown, MD, USA) were used. Analysis of dsDNA concentrations was prepared using QuantiFluor dsDNA System (Quantus, Promega, Madison, WI, USA), which uses a fluorescent DNA-binding dye. The Illumina DNA Prep system was used to construct the sequencing library, following the assessment of their quality using a high sensitivity D1000 ScreenTape Assay for TapeStation System (Agilent, Santa Clara, CA, USA). Sequencing of phage genomic DNA was performed with MiSEQ Reagent Kit Nano V2 or MiSEQ Reagent Kit V3 (New Generation Sequencing, Illumina system).
Read sequences (fastq) (from Illumina technology sequencing) for both phages were subjected to the same general pipeline. Quality assessment was completed with the Trimmomatic software [42]. Reads passing the quality check were used by SPAdes software (3.12) to assemble reads into contigs and scaffolds [43,44,45,46,47]. For the Entb_43 phage, k-mer size was detected automatically, but for Entb_45 it was set to 21, 33, and 55 with additional coverage cutoff set to 50. Assembly artifacts were checked and removed if recognized as bacterial contamination. This allowed one dominant scaffold to be assembled which relates to other phages, and this was used as a partial genome for further analysis. BLASTn (megablast) was used to evaluate similarity to prokaryotic and phage genomes in currently existing databases.
Genome annotations were performed with the Prokka software [48,49] and PGAP (Prokaryotic Genome Assembly Pipeline) [50,51,52] using prokaryotic databases. This forced adherence to International Nucleotide Sequence Database Collaboration (NCBI, ENA and DDBJ) standards and up-to-date databases. The PGAP annotation process is described in detail on the NCBI webpage (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/process/, accessed on 12 May 2022) and both processes result in functional annotation of genomes. In short, this approach mixes alignment-based methods, functional assessments of elements directly from the sequence (with the help of hidden Markov Models), representative reference proteins/genomes, and software (such as GeneMarkS2+). Alignments of closely related genomes were calculated by Geneious Prime 2022.1.1 (http://www.geneious.com, accessed on 12 May 2022) [53] and progressiveMauve [54]. SD motifs (ribosome binding sites) were found by the XSTREME [55] tool connecting the MEME, STREME, and SEA software to perform comprehensive motif discovery in accordance with guidelines for bacteriophage annotation [56].
2.12. Statistical Analysis
The statistical analysis was performed using the GraphPad Prism 9 software. A one-way ANOVA was used to determine if any changing factors (stability in nanoparticles, in solutions with different pH and in urine) influenced the phage titer and a Dunnett’s multiple comparisons test determined the statistical significance of differences between groups. Statistical significance was considered at p < 0.05. All experiments were performed in at least three repetitions.
3. Results
3.1. Characterization of the Bacterial Strains
The twenty bacterial strains used in the experiments were mainly identified as E. cloacae (15/20), but also as E. hormaechei (4/20) and E. kobei (1/20) (Table 1). All tested strains were resistant to amoxicillin/clavulanic acid. The host strains turned out to be multidrug resistant (resistance to several antibiotics simultaneously)—e.g., E. cloacae 30345, which has been shown to be resistant to amoxicillin/clavulanic acid, ciprofloxacin, levofloxacin, and tobramycin, and E. cloacae 29796, which, in addition to the antibiotics mentioned above, is resistant to cefotaxime, cefepime, gentamicin, and trimethoprim/sulfamethoxazole. Single bacterial strains also showed resistance to fosfomycin (E. cloacae 3344 and E. hormaechei 30550), as well as to a wider range of antibiotics from different pharmacological groups—e.g., ciprofloxacin, cefepime, ertapenem, gentamicin, levofloxacin, piperacillin/tazobactam, and tobramycin (E. cloacae 30342).

Table 1.
Bacterial strains used in the experiments.
3.2. Isolated Bacteriophages
After screening over 300 environmental samples, we isolated three new, potentially lytic bacteriophages, specific for Enterobacter spp.: Entb_43 (vB_EclM-43), Entb_44 (vB_EclM-44), and Entb_45 (vB_EclM-45). As a result of genetic analysis, we proved that the Entb_44 phage has an almost identical genome to Entb_43, so we decided to further characterize only the Entb_43 and Entb_45 phages. Both phages show species-specificity for Enterobacter spp. only—no lysis of other Gram-negative and Gram-positive strains from the ESKAPE group was observed (Table 2). The broadest lytic spectrum was determined for the Entb_45 phage, which also had the highest species specificity—causing lysis on various species belonging to Enterobacter (E. cloacae, E. hormaechei, E. kobei) (Table 3).

Table 2.
Results of phage typing. CL—confluent lysis; SCL—semiconfluent lysis; +++—60 and more plaques; ++—20–60 plaques; OL—opaque lysis.

Table 3.
Characterization of the newly isolated Enterobacter-specific bacteriophages.
For two strains, which were hosts for phages: E. cloacae 30345 (host for Entb_43) and E. cloacae 29796 (host for Entb_45), plots of dependence of the bacterial titer (CFU/mL) on the OD600 were prepared.
3.3. Morphology of Entb_43 and Entb_45 Phages and Their Plaques
Bacteriophage Entb_43 formed small, clear plaques on the solidified agar plates (less than 1 mm in diameter) with no apparent “halo” effect. The Entb_45 phage created slightly larger clear plaques on the agar plates (approximately 1–2 mm in diameter), and after about 3 days a slight “halo” effect was noticeable.
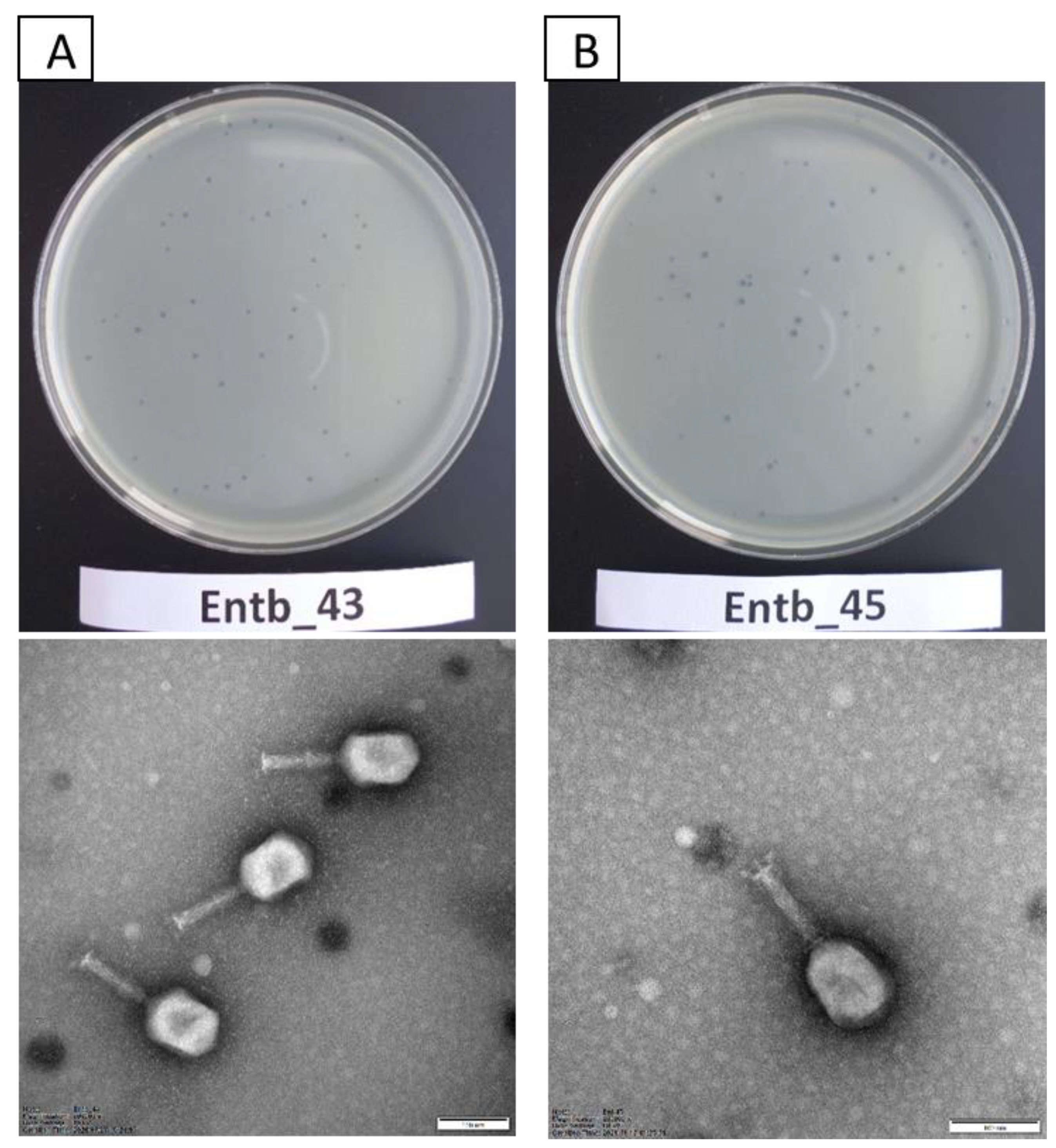
Studies with the use of TEM revealed that both newly isolated bacteriophages had the A2 morphotype. The morphology of the phages corresponds to that of myoviruses (Figure 1), which are characterized by contractile tails and icosahedral heads [57,58,59]. Entb_43, but not Entb_45, had a visible belt structure at the level of one-third of the tail. Dimensions of the capsids, tails, and base plates of each phage are included in Table 4.
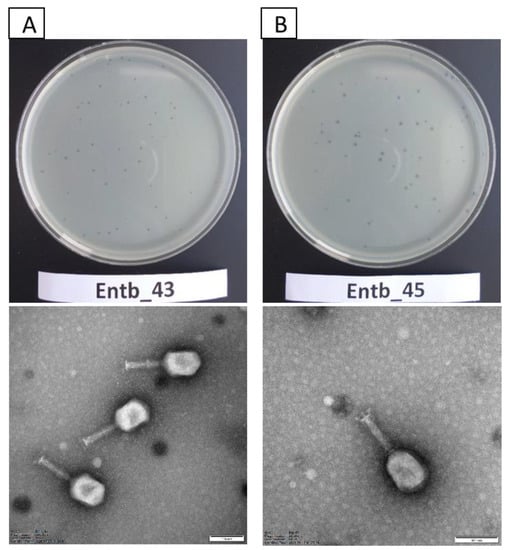
Figure 1.
Plaque morphology on agar plates (upper panel) and electron micrographs of phages (lower panel): (A) phage Entb_43 and (B) phage Entb_45. The photographs of the phages’ plaques were taken using a Samsung ST150F camera. Electronograms of the phages were taken using Kodak/Carestream Electron Microscope, at a magnification of (A) 200,000× or (B) 250,000×; bar in each micrograph indicates 100 nm.

Table 4.
Dimensions of bacteriophages. Average measurements of ten virions are shown.
3.4. Influence of Different MOI on the Effectiveness of Phage Amplification and Antimicrobial Effect
As shown in Table 5, the phage titers, after overnight cultivation at 37 °C, varied depending on the phage-to-bacteria ratio used (different MOI). In general, the lower MOI used (lower phage titer than bacterial titer), the higher final phage titer obtained. For both phages, the best phage-to-bacteria ratio was either 0.01 or 0.001.

Table 5.
Influence of different MOIs on the phage titers after amplification. The final phage titer results reflect the mean of the titers in at least triplicate.
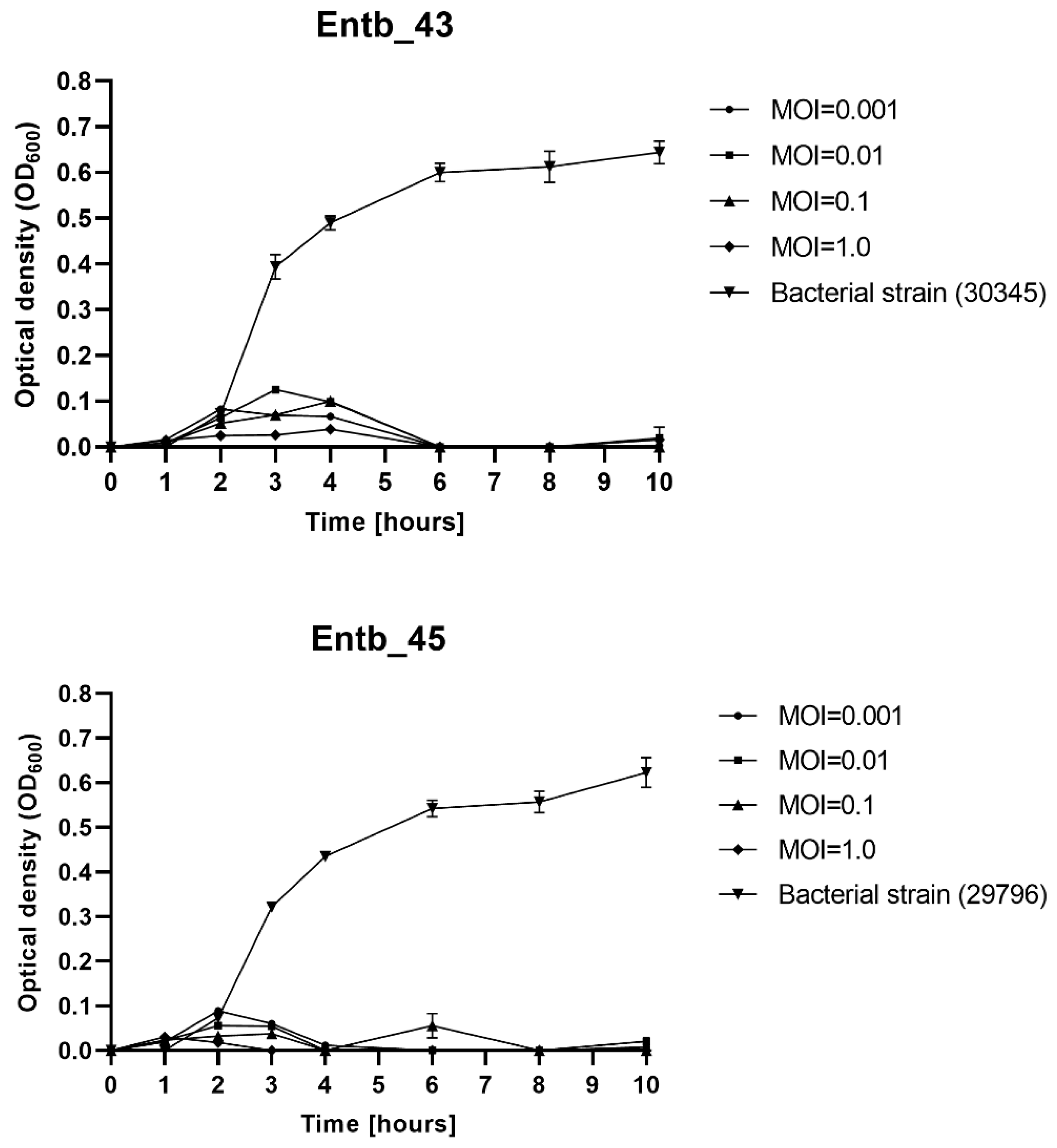
The MOI-dependent ability of phages to lyse planktonic bacteria was determined. As shown on Figure 2, each of the bacteriophages significantly inhibited the growth of the host bacteria. Bacterial lysis varied slightly on the used MOI; however, even a low MOI is sufficient to successfully eliminate bacterial cells, whereas the OD600 of the phage-untreated bacterial strains (positive controls) clearly increased over time. The optical density of the only phage lysates (negative controls) oscillated around 0.00 and did not change during the measurements (not shown in the graphs).
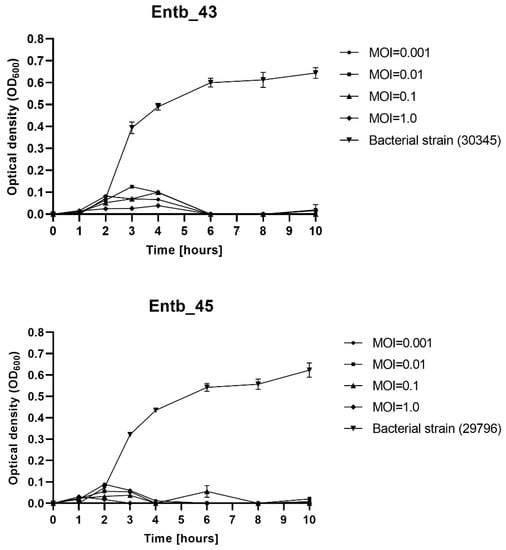
Figure 2.
Planktonic cell lysis assay for phage Entb_43 and phage Entb_45. Sample measurements were performed in triplicate in two independent experiments. Error bars represent the standard deviation (±SD) of the mean phage titers.
3.5. Phage Adsorption to Bacterial Cells
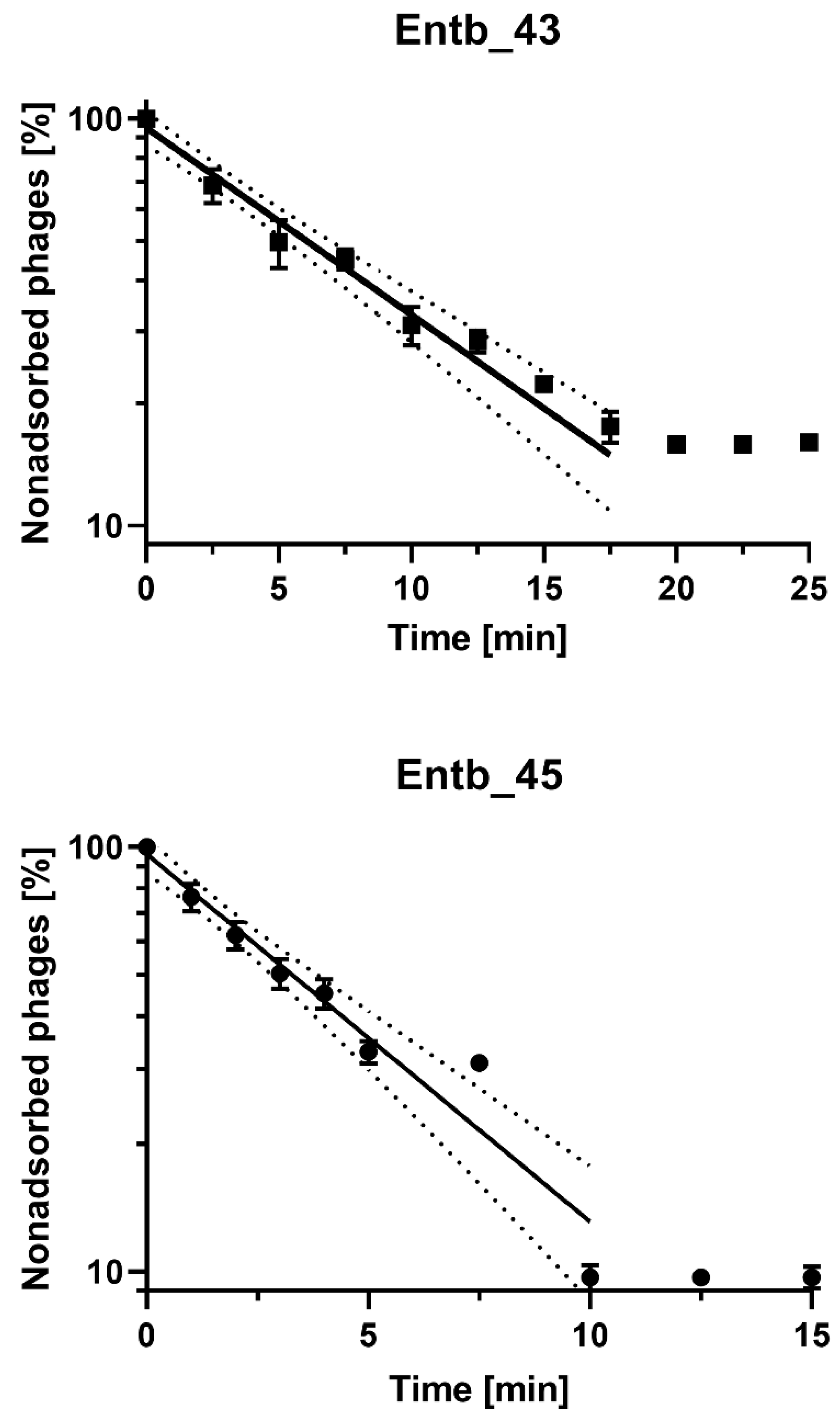
After the infection of E. cloacae 30345 with the Entb_43 phage (at MOI = 0.1), we obtained about 84% adsorption of the phage particles to the host cells at 37 °C in 20 min. However, the adsorption time of Entb_45 to E. cloacae 29796 was much faster, and after 10 min, almost 90% adsorption was achieved (Figure 3 and Supplementary Figure S1). The adsorption rate constants, which are included in Table 6, were also calculated.
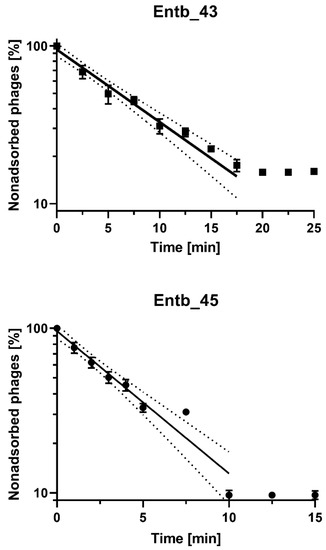
Figure 3.
Kinetics of Entb_43 phage adsorption on E. cloacae 30345 and Entb_45 phage on E. cloacae 29796 at MOI = 0.1. Error bars represent the standard deviation (± SD) of the mean phage titers. Lines represent model used for calculation of adsorption constant; dotted line is 95% confidence interval (CI).

Table 6.
Adsorption rate constant for phage Entb_43 and phage Entb_45.
3.6. Genome Sequencing
We present the genomes of two new phages: Enterobacter phage Entb_43 (174 kbp, acc. No. ON585039) and Enterobacter phage Entb_45 (173 kbp, acc. No. ON630910). Both phages were sequenced with Illumina technology and assembled into a single, gapless contig from reads passing through a quality check (see Materials and Methods for details). Phage genomes show a high degree of diversity between each other, with only 12% of the genome showing high similarity between two of them. These similarities were, however, limited (up to 75% of identity) and they were found mainly in regions coding for structural phage proteins. The presented genomes show poor genomic similarity to already known phages. In databases accessible through the International Nucleotide Sequence Database Collaboration (https://www.insdc.org/, accessed on 10 June 2022), only 8 and 7 phage genomes show a similarity above 70% to Entb_43 and Entb_45, respectively (Supplementary Tables S1 and S2). The first one is most closely related to the Klebsiella phage vB_KaeM KaAlpha (93% identical at the nucleotide level, acc. no. MN013084) and the second one is most closely related to the Enterobacter phages ENC9 (93% identical at the nucleotide level, acc. no. OL355124.1) and vB_EclM_CIP9 (94% identical at the nucleotide level, acc. no. NC_048849.1). Basic features of the presented phages are described in Table 7.

Table 7.
Basic features of sequenced phages.
For the genomes of Entb_43 and Entb_45, revealed after annotation 298 and 289 coding sequences, respectively, 44% and 58% of the proteins are designated as “hypothetical proteins”. One hypothetical protein is present in a tail gene cluster of Entb_43, but not in the Entb_45 phage (protein ID: M8345145). The highest similarity (except for hypothetical proteins) is shown for a putative GIY-YIG homing endonuclease (Yersinia phage vB_Yem_TG1, acc. no.: NC_028820.1), but no significant similarity to well-described proteins was found. Typically for Tevenvirinae, our annotation in both phages revealed no apparent integrase gene, suggesting a lytic life cycle. Genomes were linearized to the presumed position at the beginning of the identified gene coding the rIIA/B gene, as is standard for Tevenvirinae. Analysis of the Entb_43 genome revealed 19 tRNA, but only 9 tRNA in Entb_45. No similarity to antibiotic resistance genes were found in the CARD database (no strict hits) [60].
Based on genomic similarity, we classified phage Entb_43 to genus Karamvirus and Entb_45 phage to genus Kanagawavirus [61,62], which classifies both phages to the Tevenvirinae subfamily (Straboviridae family) in accordance with current ICTV guidelines [63]. Each phage genome showed a similarity of over 70% to representatives of only one genus. Similarity to the most closely similar phage is shown in Figure 4 and Figure 5. Similarity of the marker genes recommended by ICTV for Tevenvirinae shows that Entb_43 and Entb_45 are closely related to other representative phages from the Karamvirus and Kanagawavirus genera, respectively (Supplementary Tables S3 and S4) [64]. We identified Shine–Dalgarno sequences (ribosome binding site) in fragments 100 bp upstream of the START codon (Figure 6 and Figure 7) according to the recommendation by Turner et al. in the “Phage Annotation Guide: Guidelines for Assembly and High-Quality Annotation” [56].

Figure 4.
Graphical representation of phage Entb_43 (no. 1), aligned to the most similar known genome of Klebsiella phage vB_KaeM KaAlpha (no. 2, acc. no. MN013084). Green—100%, brown-green—between 30% and 100%, and red—0% local similarity. Lack of color shows a gap in alignment. Annotation is shown for reference. Coding sequences (CDS) are yellow, and tRNA coding sequences are pink.

Figure 5.
Graphical representation of phage Entb_45 (no. 1), aligned to the most similar known genome of Enterobacter phage ENC9 (no. 2, acc. no. OL355124.1). Green—100%, brown-green—between 30% and 100%, and red—0% local similarity. Lack of color shows a gap in alignment. Annotation is shown for reference. Coding sequences (CDS) are yellow, and tRNA coding sequences are pink.
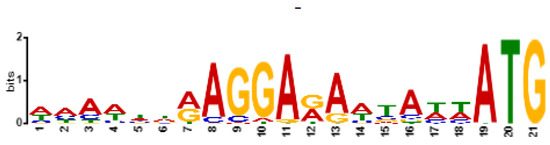
Figure 6.
Shine–Dalgarno motif (ribosome binding site) in sequences of 100 bp upstream of START codon (included) for phage Entb_43, identified by XSTREME software.

Figure 7.
Shine–Dalgarno motif (ribosome binding site) in sequences of 100 bp upstream of the START codon (included) for phage Entb_45, identified by XSTREME software.
3.7. Stability Studies of New Enterobacter-Specific Phages
In order to assess the effect of commonly used disinfectants and cleaning agents on the stability of bacteriophages, six different commercially available substances were used in the experiment (Table 8). In general, a significant reduction in phage titers was observed after incubation with the disinfectant that contained the active substances octenidinum dihydrochloridum and phenoxyethanol (1) (titer reduction of over 90% for phage Entb_43 and by 99.9% in the case of phage Entb_45, after 30 min of incubation), and was slightly less pronounced with 70% ethanol (reduction in the titer by nearly 70% for Entb_43 and by 97% for phage Entb_45, after 30 min of incubation). The remaining disinfectants and surfactants influenced the phage titers, but this was less significant.

Table 8.
Percentage of phages retaining activity after specified intervals of incubation with various disinfectants.
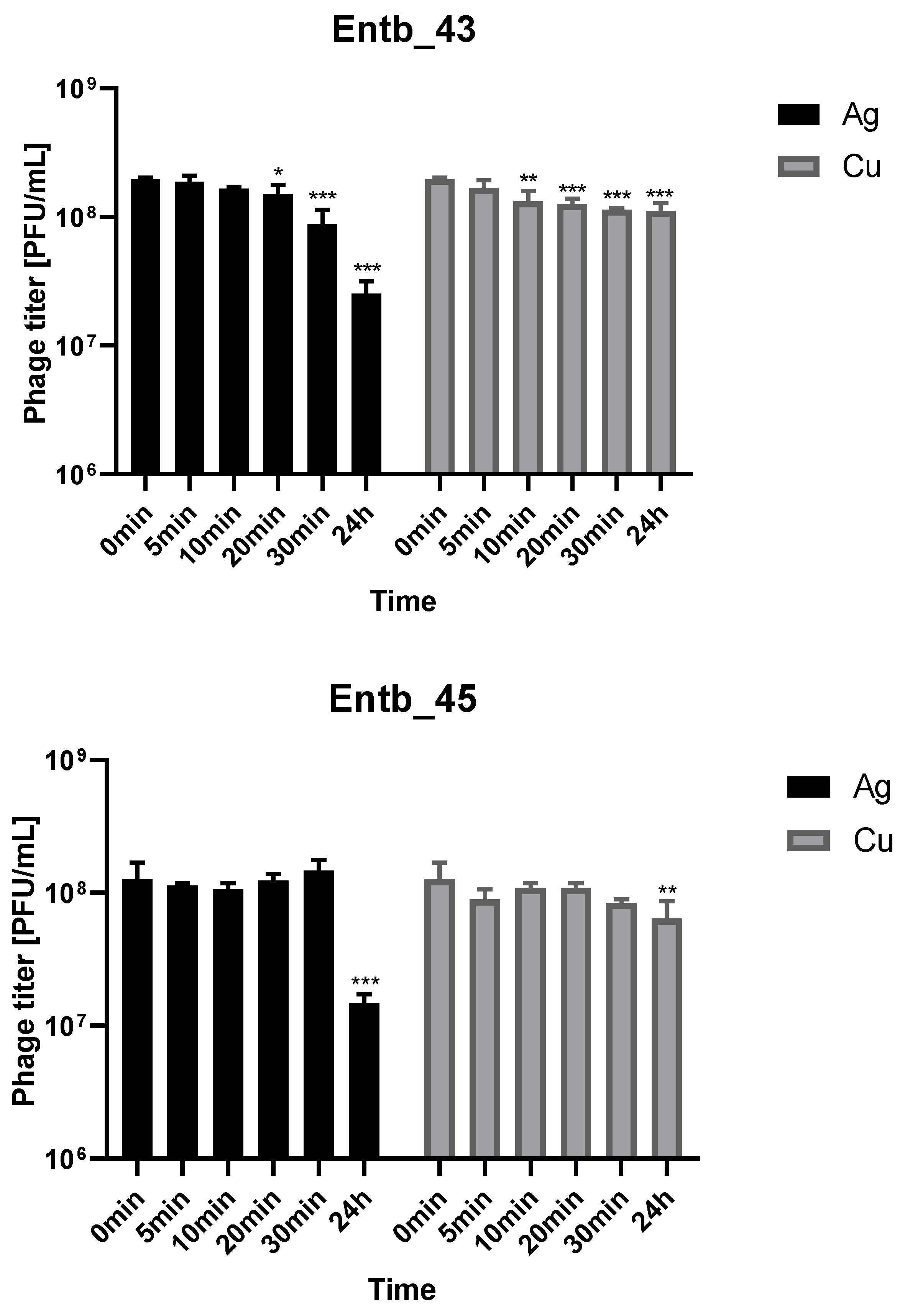
Metal nanoparticles such as copper and silver are used in a variety of ways as disinfectants. We decided to evaluate the stability of phage lysates in these solutions to assess if applying them together was possible. As shown in Figure 8, both bacteriophages retain a lot of active particles for 30 min of incubation with nanoparticle solutions at room temperature. On the other hand, 24 h incubation caused more significant reductions in phage titers in the case of silver colloid than copper.
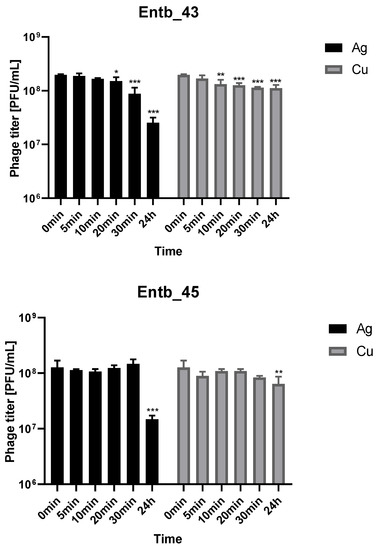
Figure 8.
Phage lysate stability in silver nanoparticles and copper nanoparticles. Samples at the start of the experiments (0 min) were treated as controls for statistical analysis. Error bars represent the standard deviation (±SD) of the mean phage titers. * p < 0.05; ** p < 0.01; *** p < 0.001.
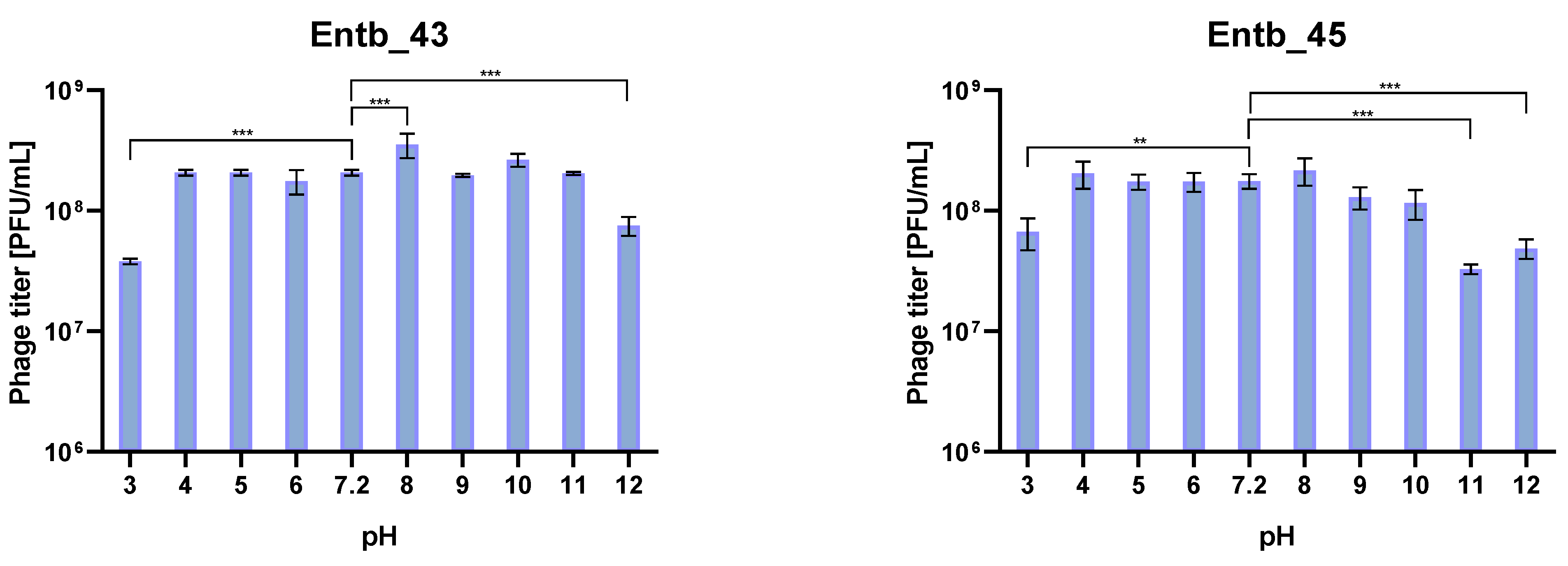
The isolated bacteriophages were also tested for stability in the culture medium (peptone water) with different pH values. As a control for comparisons, samples with peptone water with pH = 7.2 were used, which were treated the same as the other samples. As a result of the statistical analysis, it was observed that the most significant differences in phage titers after incubation occurred in the case of the most extreme pH values (pH = 12 for phages Entb_43 and Entb_45, pH = 11 for phage Entb_45, and pH = 3 for both phages). Interestingly, higher phage titers were obtained as a result of incubation with a medium for which the pH was 8, which may suggest more effective results of phage amplification in slightly more alkaline media than before (Figure 9).
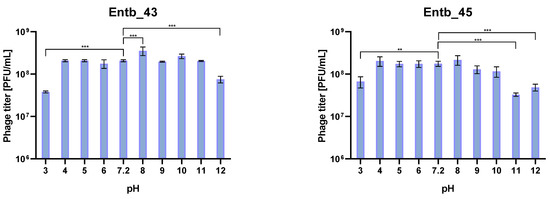
Figure 9.
Phage titers after one-hour incubation in different pH solutions. Samples incubated in a medium with pH = 7.2 were treated as controls. Error bars represent the standard deviation (±SD) of the mean phage titers. ** p < 0.01; *** p < 0.001.
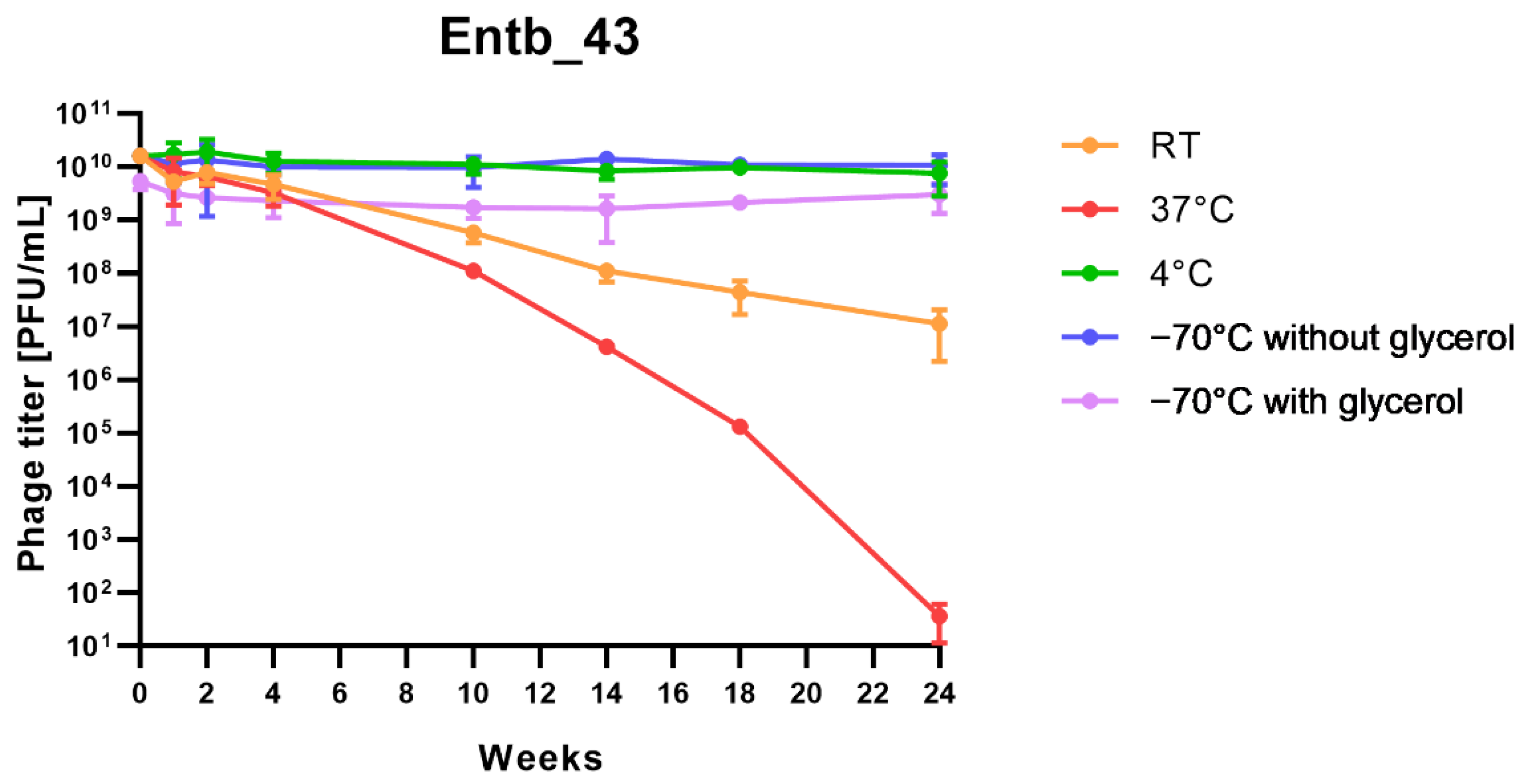
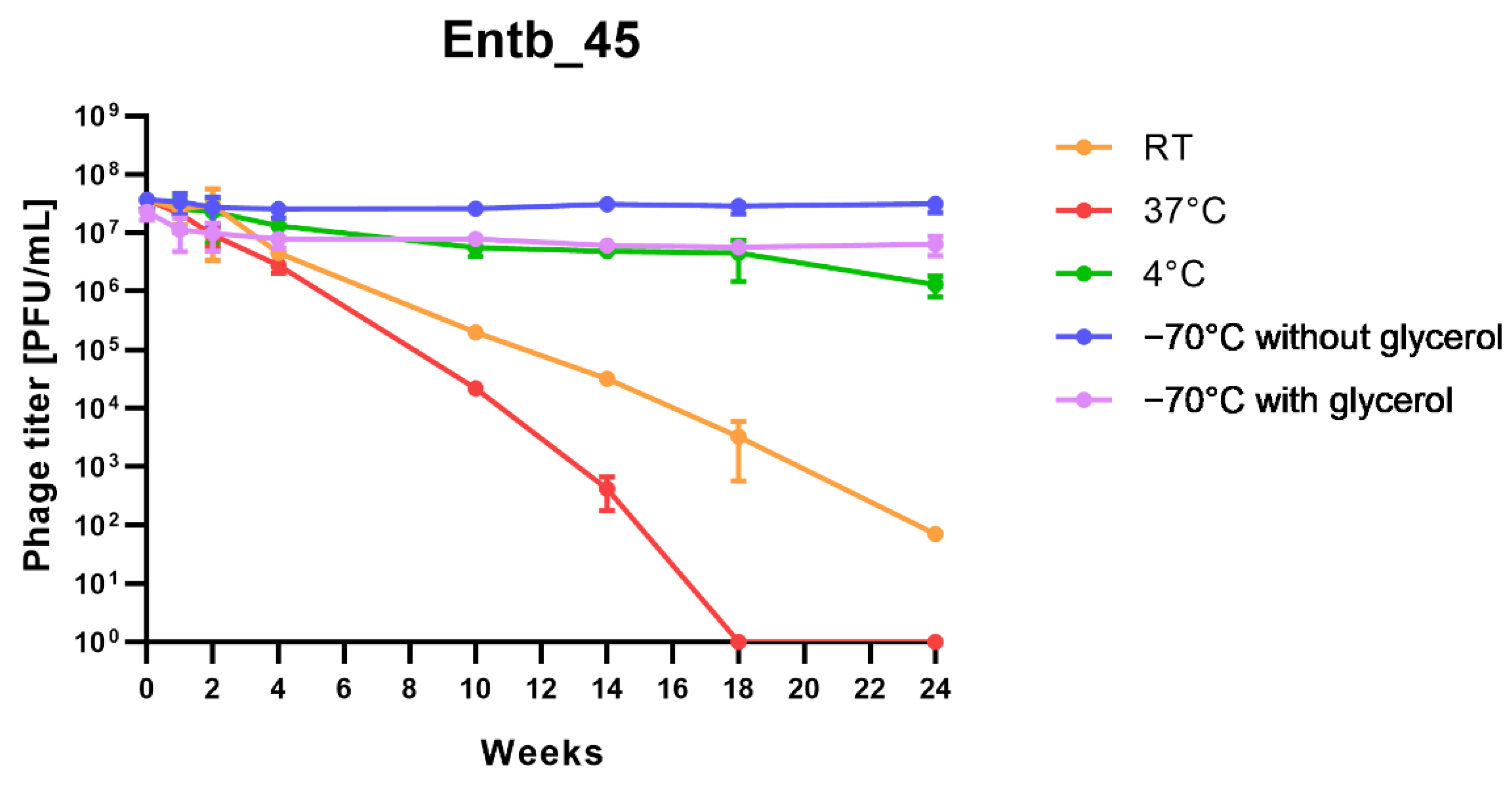
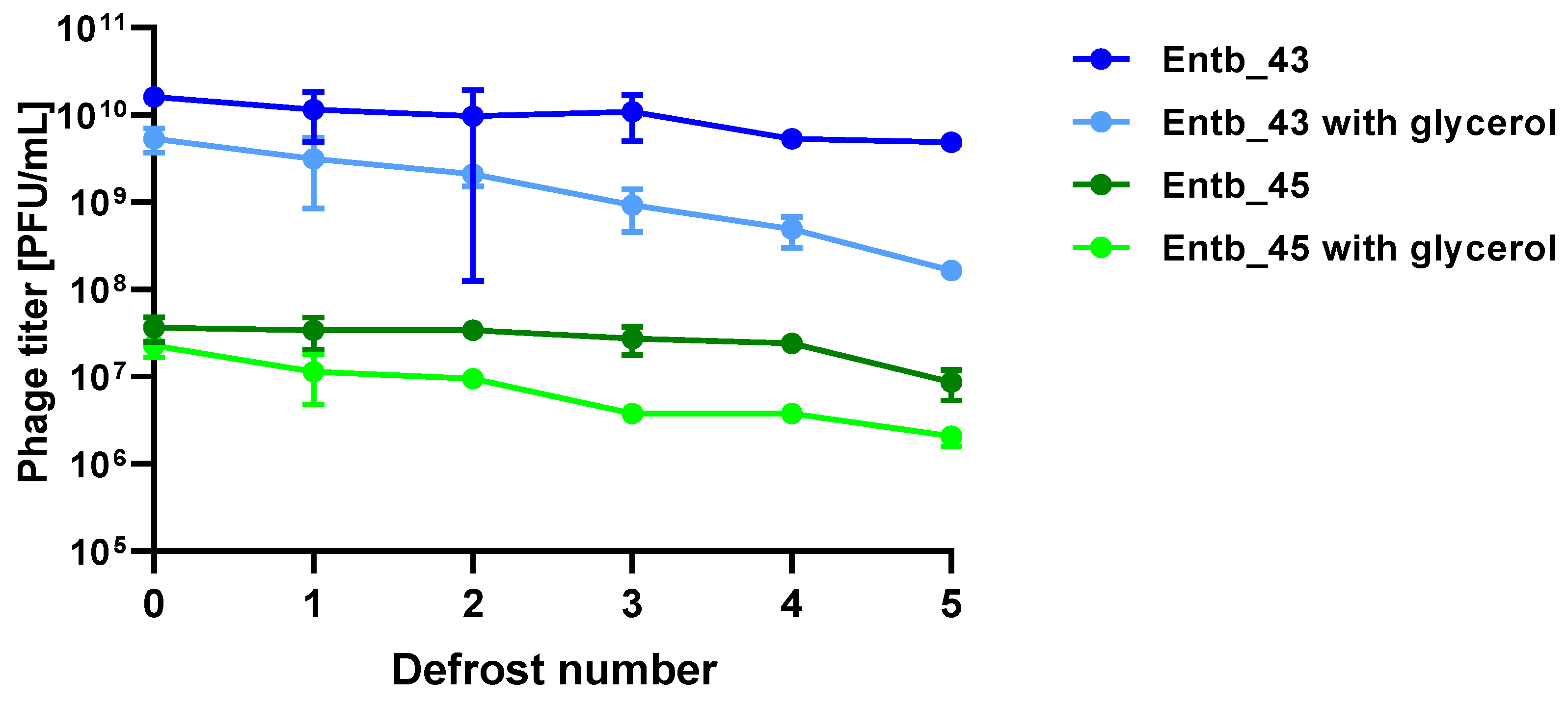
Stability of phages over six months (24 weeks) was clearly dependent on the temperature at which they were stored. As was shown in Figure 10, storage of bacteriophages at 4 °C, as well as in frozen form at −70 °C (with or without the addition of cryoprotective glycerol), did not significantly affect their titers (only the titer of Entb_45 stored at 4 °C, which initially had a lower titer, decreased by one order of magnitude on a logarithmic scale at the end of the experiment). Phage titers decreased successively at both room temperature and 37 °C. No active phage particles were found in the Entb_45 phage lysate after 18 weeks of storage at 37 °C, whereas the titer of the Entb_43 phage decreased from 1010 to 101 PFU/mL after 24 weeks. Moreover, the thaw-resistance of frozen phage lysates was demonstrated, because after defrosting the same samples five times and refreezing, the phage titers only decreased by about one order of magnitude (Figure 11).
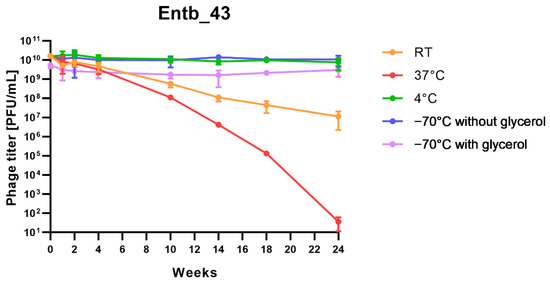
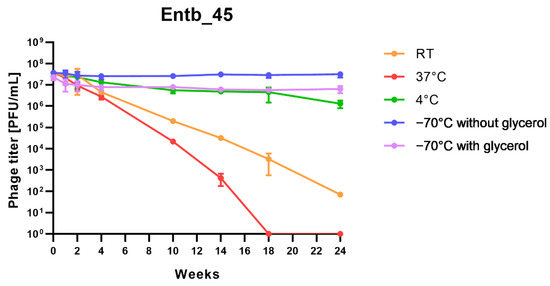
Figure 10.
Long-term stability of Entb_43 and Entb_45 phage lysates under various temperature conditions. Error bars represent the standard deviation (±SD) of the mean phage titers.
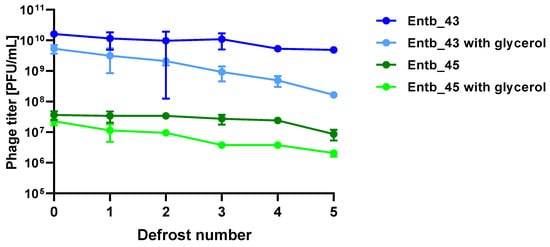
Figure 11.
Influence of repeated defrosting on titers of phage lysates with or without 20% glycerol. Error bars represent the standard deviation (±SD) of the mean phage titers.
3.8. Phage Stability in a Urine Sample
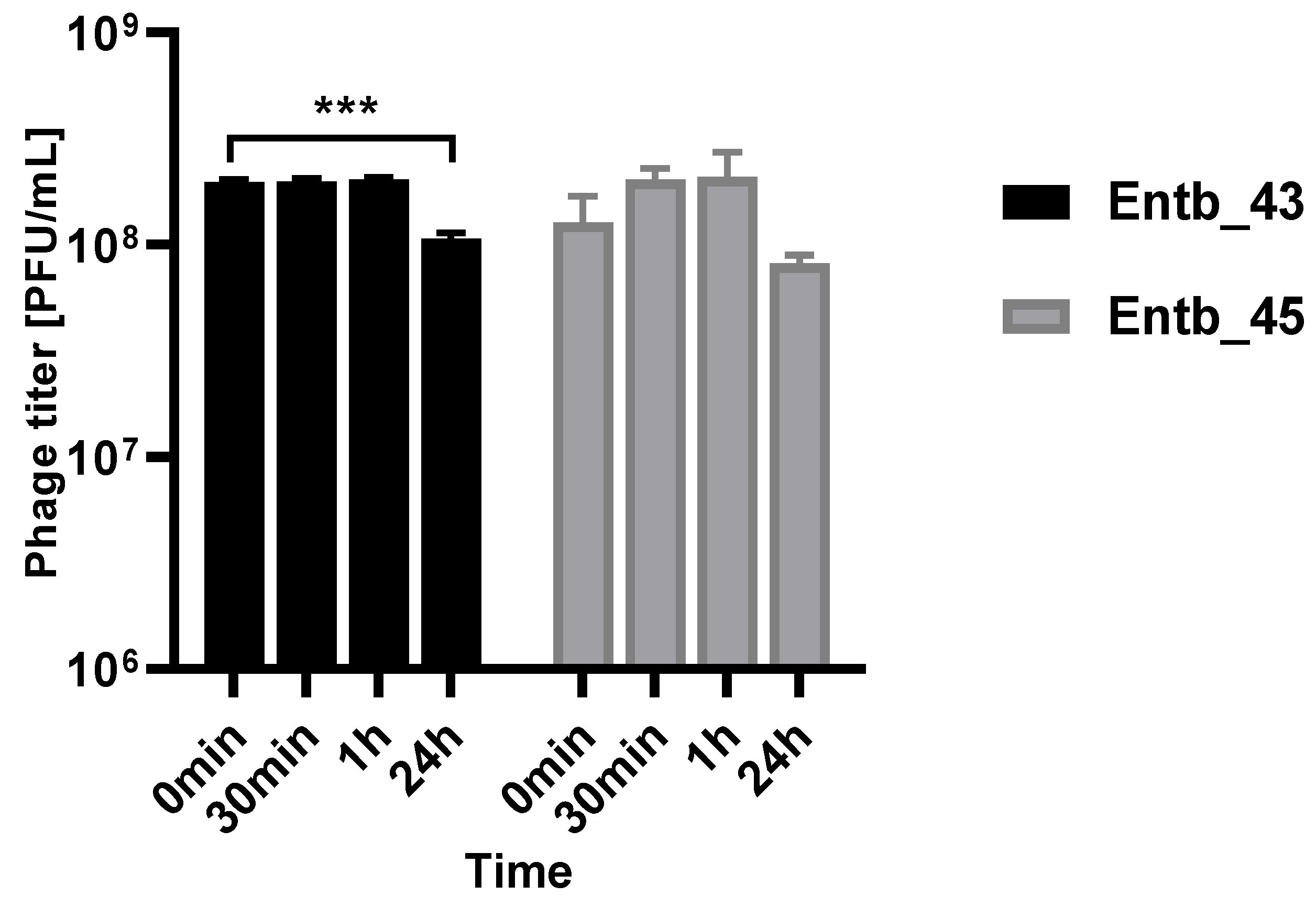
As mentioned before, Enterobacter spp. can cause severe urinary tract infections. A key feature of phage therapy in the intravesical administration of phages is their stability at the site of action, so we examined their ability to remain active in a urine sample (pH = 6) obtained from a healthy donor (Figure 12). It was noted that, after both half an hour and an hour of incubation in a fresh sample of urine, the phage titers remained at the same level as at the beginning of the experiment, which may suggest their potential efficacy without loss of activity at the infection site after intravesical administration.
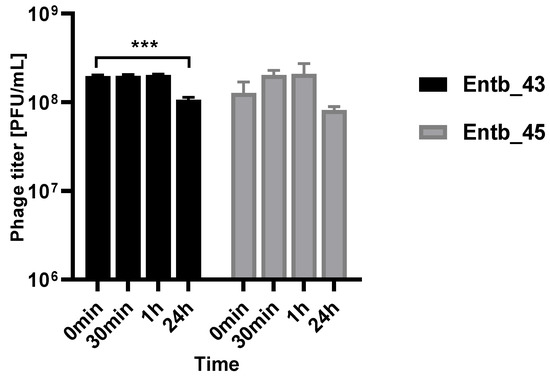
Figure 12.
Phage titers after incubation in a urine sample (after 30 min, 1 h, and 24 h of incubation at 37 °C). Samples at the start of the experiments (0 min) were treated as controls. Error bars represent the standard deviation (± SD) of the mean phage titers. *** p < 0.001.
4. Discussion
Because Enterobacter strains are one of the etiological factors of various life-threatening conditions, we decided to isolate and characterize new phages specific to these bacteria. Our characteristics concerned not only the standard life cycle parameters, the lytic spectrum, and the stability under various conditions, but also the detailed genomic characteristics (as suggested by Suh et al., 2021 [65]).
As a result of typing 339 samples from various sources, we were able to find three new bacteriophages (Entb_43, Entb_44, and Entb_45) showing an ability to lyse clinical Enterobacter strains. Despite the fact that the bacteriophages Entb_43 and Entb_44 were isolated from different environmental samples, a genetic analysis showed that they were too similar to be considered separate phages. Therefore, we decided to further characterize only Entb_43 and Entb_45.
Currently, 85 genomes of Enterobacter phages have been deposited in the gene bank (https://www.ncbi.nlm.nih.gov/nuccore, accessed on 26 May 2022), which may indicate the potential and the need to conduct research on these phages and their comprehensive characteristics. Bioinformatic analysis of the genomes of two of our new bacteriophages allowed us to describe them as double-stranded DNA and to determine their sizes, respectively, as 174,681 bp for Entb_43 and 172,771 bp for Entb_45. Similar results have already been reported for the Enterobacter-specific phage vB_EclM_CIP9 (defined as myovirus) with a 174,924 bp genome [66], as well as the EBP bacteriophage (also myovirus), which has a 179.1 kb genome size [67]. Interestingly, a phage active against E. cloacae was recently reported with a genome size of 51,894 bp and was assigned to a new genus, Eclunavirus [68]. Moreover, no rRNA or tRNA were found in its genome, whereas our phages had 19 and 9 tRNAs in their genomes, respectively. The similarity of Entb_43 to the Klebsiella-specific phage may be related to the similarity of these two types of bacteria. Until recently, Klebsiella aerogenes was considered Enterobacter aerogenes [9,69]. Moreover, genomic analyses provide data on the genetic relationships of these bacteria, as described, for example, for strains isolated from patients with a bloodstream infection (69% shared genes) [70].
The host range of phages estimated on the basis of the spot test was 40% and 60%, respectively. Both phages were also tested simultaneously: immediately after mixing, 24 h after, 48 h after, and one week after mixing, but no significant changes in the lytic spectrum were observed. The phage cocktail retained its lytic properties on strains against which at least one phage was active (data not presented herein). Interestingly, the myPSH1140 phage, described by Manohar et al. (2019), showed a much wider host range, reaching 100% of E. cloacae and 75% of E. hormaechei strains [71]. There are also Enterobacter-specific phages with an extremely narrow range of activity, lysing only one bacterial strain [72]. Instead, phages are also described as polyvalent, as presented by Finney et al. (2022), with phages lysing both Enterobacter spp. and Klebsiella aerogenes [73], or by Addablah et al. (2021), with phages infecting E. cloacae and E. coli simultaneously [74]. Jamal et al. (2018) reported the phage MJ2, which was able to inhibit the growth of E. cloacae, Achromobacter xylosoxidans, K. pneumoniae, and P. aeruginosa [75]. However, determination of the host range is not always clear and repeatable due to the different determination methods and the ability of phages to form plaques on the specific bacterial lawn [76].
Similar morphological characterization, as well as host range, was described for a lytic EBP bacteriophage by Asif et al. (2021) [67] and for three phages reported by Gibson et al. (2019) [77]. Nair et al. (2021) also described a myovirus, which was able to lyse both the planktonic form of bacteria and the biofilm produced by E. cloacae [78].
In the adsorption experiment, we used MOI = 0.1 for both phages (107 PFU/mL and 108 CFU/mL) and observed that phages achieved complete adsorption to host cells in 20 min (Entb_43) and 10 min (Entb_45). Conversely, phages that show more rapid adsorption were also described, such as the E. coli-specific phages vB-EcoS-95 (50% adsorbed phage particles in 2 min) [79] and vB_Eco4M-7 (completed adsorption of 90% after 1 min) [40], or the Klebsiella-specific phages (4 min and 6 min) [39]. Statistical analysis allowed us to define adsorption as a first-order kinetic process [41] (Figure 3), even though basic graphs on a nonlogarithmic scale might indicate biphasic adsorption (Supplementary Figure S1).
The best phage-to-bacteria ratio, both for estimating the optimal conditions for bacteriophage amplification and for lytic activity against planktonic bacteria, turned out to be 1:1000 or 1:100 (MOI = 0.001 or MOI = 0.01). Interestingly, studies using the new phage against Enterococcus faecalis producing biofilm in Foley catheters initially reported more efficacy of phage lytic activity at a higher MOI (MOI = 10). On the other hand, after 6 h and 24 h of exposing the bacterial biofilm to the phage, the preparations with a lower phage-to-bacteria ratio (MOI = 0.01 and MOI = 0.0001) were more effective [80]. However, we describe the MOI as the ratio of the number of phages to the number of bacteria at the start of the experiment, and it is suggested to also consider, inter alia, the changing densities of bacteria during the course of the experiment [81].
In the case of long-term storage of phage preparations, a temperature of 4 °C is usually recommended [82]. Our phage lysates (in peptone water) remained stable at this temperature throughout the six-month period of the experiment. The maintenance of stability by phages in frozen forms (−70 °C) indicates such a possibility for their storage. Interestingly, over the course of a month, only the titers of phages stored at the highest temperature (37 °C) decreased by only one order of magnitude. It is worth noting that studies on adaptive evolution regarding various thermal conditions are being designed to increase phage stability [83].
The ability of phages to remain active under unfavorable conditions may be very important for people working with these viruses, as well as for the industrial or pharmaceutical applications of phages [84]. An example of the industrial use of E. cloacae-specific bacteriophages is the attempt to use them as factors to reduce the bloating defect caused by these bacteria in cucumber fermentation [85]. The different values of acidity/alkalinity of the environment are of particular importance in the context of the human organism and other animals in the processes of health and disease. From the point of view of phage therapy, factors such as changes in the pH of skin and wounds [86], unfavorable pH in the stomach [87], or changes in the pH of the environment caused by growth of bacteria [88] can determine the effectiveness of treatments. Relatively good stability was observed in our phages after incubation in peptone water with a pH ranging from 4 to 11. Moreover, the observed decrease in titers as a result of a large deviation from physiological pH has been confirmed for most phages, including Enterobacter-specific [71,89,90,91,92] and other species (e.g., Escherichia coli-specific) [93]. Very similar tolerance to a wide pH range was also shown by the phages KKP 3262 and KKP 3263 described by Wójcicki et al. (2021) [94]. Interestingly, two of five E. hormaechei-specific phages isolated by Chen and coworkers (2022) showed an almost complete resistance to a more alkaline environment (such as pH = 12) [95].
The inactivation of phages as a result of contact with various disinfectants/surfactants may be of great importance in the quality of work involving them, and may also have an impact on the field of therapeutics when the use of surface disinfectants can support phage action (e.g., skin surface disinfection or coating the surface of catheters with metal nanoparticles). Compounds based on silver were applied in antimicrobial therapy until the 1940s when antibiotics were implemented [96]. The observed antimicrobial activity of silver may be the result of a direct toxic effect on bacterial cell walls [97]. When silver ions are released, the bacterial cell membrane and a variety of intracellular functions are disrupted. The small size of their particles, together with the large surface area of the dressing’s integrated silver nanoparticles, increase their solubility and cause a sustained release of silver ions in relatively nontoxic concentrations [96]. Coating the catheter’s surface with silver nanoparticles resulted in significant in vitro antimicrobial activity and also prevented biofilm formation by E. coli, Enterococcus, S. aureus, coagulase-negative staphylococci, P. aeruginosa, and Candida albicans [98]. Studies by Thomas et al. (2015) indicate a high potential in applying silver nanoparticles to urinary catheters for preventing catheter-associated infections, especially the attachment and colonization of bacteria [99,100]. The scientific literature also indicates that copper nanoparticles have antibacterial properties [101,102]. Therefore, it is important to evaluate phage–nanoparticle interactions due to data about the possibility of irreversible inactivation of phages (e.g., E. coli-specific) by nanoparticles [103,104]. The activity of our new phages, despite a statistically significant difference compared to the beginning of incubation, was well maintained for about 30 min of incubation at RT with both types of nanoparticles. This may indicate the possibility of a synergistic antibacterial effect and the need for further research into this phenomenon. Similarly, Abdelsattar et al. (2021) showed no significant differences in the titer of a Salmonella-specific phage after 4 h incubation at 37 °C with synthesized silver nanoparticles [105]. The 24 h incubation, especially in the case of silver, led to a more significant inactivation of our phages.
We also decided to test the phage stability in 10% dish soap and 10% liquid hand soap. In similar experiments, Necel et al. (2020) [40] found no decrease in two E. coli phage titers after 2 min and 5 min of incubation at RT, respectively. Our results indicated an initial reduction in titers for both phages by approximately 20–25% (after 5 min), while for half an hour, despite fluctuations in the titers, no reduction was observed. The commonly used skin disinfectant based on octenidinum dihydrochloridum and phenoxyethanol, as well as 70% ethanol, had the strongest effect in reducing the number of active phage particles, which may be important in skin or wound disinfection before phage administration. For ECML-117 and vB_Eco4M-7 phages, active phage particles were described as 0% and 0.01%, respectively, after one hour of incubation in 63% ethanol [40]. Likewise, such conditions caused a significant decrease in vB-EcoS-95 phage survival (0.8% active particles) [79]. Interestingly, Tomat et al. (2018) [106] reported E. coli phages that remained active after 24 h of incubation in 100% ethanol (decrease of titer by 2.5 PFU/mL). Recently, the possible negative effects of the uncontrolled spread of phages in food production processes have also been emphasized [107]. Information about the factors that deactivate them may therefore be useful in industrial fields.
Beta-lactam antibiotics, as well as fluoroquinolones and aminoglycosides, are the most common antimicrobials prescribed to treat infections caused by Enterobacteriaceae [108]. The bacteria we used demonstrated ESBL+ resistance, a mechanism that is characteristic of uropathogenic Enterobacter strains [109,110] and other Gram-negative bacteria [111]. More relevant data are emerging on the prospects of managing UTIs through the use of lytic bacteriophages, phage cocktails, enzymes derived from them, and genetically modified phages, as well as by using the synergistic effect of phages and antibiotics [112,113,114,115,116]. Promising clinical trials in this area are slowly emerging [117,118], but many issues still need to be refined [119]. There are reports of clinical successes in the treatment of UTIs as a result of intravenous phage cocktail administration, which, although it did not completely eradicate the pathogen, its succession resulted in a significant improvement in the patient’s condition [120]. As a preliminary study determining the possibility of using our phages in the treatment of UTI, we decided to evaluate their stability in a urine sample. The results turned out to be promising as bacteriophages were not inactivated by the components of the previously filtered urine. Pereira et al. (2016) described not only the good stability of Enterobacter-specific phages in a urine sample, but also an increase in their titer in the presence of bacteria [113].
The results described in this paper strongly suggest the therapeutic potential of the presented Enterobacter-specific bacteriophages.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v14071518/s1, Figure S1. Kinetics of Entb_43 phage adsorption on E. cloacae 30345 and Entb_45 phage on E. cloacae 29796 at MOI = 0.1. Table S1. List of all genomes with over 70% similarity to Enterobacter phage Entb_43 genome. Table S2. List of all genomes with over 70% similarity to Enterobacter phage Entb_45 genome. Table S3. Similarity of each protein product of marker genes from Enterobacter phage Entb_43 to the closest relative classified by ICTV. Table S4. Similarity of each protein product of marker genes from Enterobacter phage Entb_45 to the closest relative classified by ICTV.
Author Contributions
Conceptualization, M.C. and E.J.-M.; methodology, M.C., M.H., F.O. and E.J.-M.; software, M.H. and K.D.; validation, M.C., K.D. and E.J.-M.; investigation, M.C. and F.O.; data curation, M.C. and M.H.; writing—original draft preparation, M.C.; writing—review and editing, M.C., M.H., K.D., A.G. and E.J.-M.; visualization, M.C. and M.H.; supervision, K.D., A.G. and E.J.-M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by intramural funds from the Ludwik Hirszfeld Institute of Immunology and Experimental Therapy at the Polish Academy of Sciences.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The presented data are available on request from the corresponding author.
Acknowledgments
We would like to thank to Norbert Łodej and Karolina Hyczko for preparing and developing the electron micrographs of the studied phages. The transmission electron microscopy imaging and genome sequencing were performed within “Scientific Information Base Supporting Innovative Therapies—BINWIT” (POPC.02.03.01-00-0053/18.), cofunded by the European Regional Development Fund under the Operational Program Digital Poland “Sub-measure 2.3.1. Digital access to public sector information from administrative sources and learning resources”.
Conflicts of Interest
A. Górski is the co-inventor of patents owned by the L. Hirszfeld Institute and covered the phage preparations. Other authors declared that the research was conducted without any commercial or financial relationships that could be construed as potential conflicts of interest.
References
- Hormaeche, E.; Edwards, P.R. A proposed genus Enterobacter. Int. Bull. Bacteriol. Nomencl. Taxon. 1960, 10, 71–74. [Google Scholar] [CrossRef] [Green Version]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. Biomed Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.X.; Wang, C.Y.; Li, Y.Y.; Li, J.; Wan, Q.Q.; Chen, J.H.; Tay, F.R.; Niu, L.N. Considerations and Caveats in Combating ESKAPE Pathogens against Nosocomial Infections. Adv. Sci. 2019, 7, 1901872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef]
- Mulani, M.S.; Kamble, E.E.; Kumkar, S.N.; Tawre, M.S.; Pardesi, K.R. Emerging Strategies to Combat ESKAPE Pathogens in the Era of Antimicrobial Resistance: A Review. Front. Microbiol. 2019, 10, 539. [Google Scholar] [CrossRef]
- WHO Priority Pathogens List. Available online: https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 12 August 2021).
- Ramos-Vivas, J.; Chapartegui-González, I.; Fernández-Martínez, M.; González-Rico, C.; Fortún, J.; Escudero, R.; Marco, F.; Linares, L.; Montejo, M.; Aranzamendi, M.; et al. Biofilm formation by multidrug resistant Enterobacteriaceae strains isolated from solid organ transplant recipients. Sci. Rep. 2019, 9, 8928. [Google Scholar] [CrossRef] [Green Version]
- Cunha, B.A.; Theodoris, A.C.; Yannelli, B. Enterobacter cloacae graft infection/bacteremia in a hemodialysis patient. Am. J. Infect. Control. 2000, 28, 181–183. [Google Scholar] [CrossRef]
- Álvarez-Marín, R.; Lepe, J.A.; Gasch-Blasi, O.; Rodríguez-Martínez, J.M.; Calvo-Montes, J.; Lara-Contreras, R.; Martín-Gandul, C.; Tubau-Quintano, F.; Cano-García, M.E.; Rodríguez-López, F.; et al. Clinical characteristics and outcome of bacteraemia caused by Enterobacter cloacae and Klebsiella aerogenes: More similarities than differences. J. Glob. Antimicrob. Resist. 2021, 25, 351–358. [Google Scholar] [CrossRef]
- Boguniewicz, J.; Revell, P.A.; Scheurer, M.E.; Hulten, K.G.; Palazzi, D.L. Risk factors for microbiologic failure in children with Enterobacter species bacteremia. PLoS ONE 2021, 16, e0258114. [Google Scholar] [CrossRef]
- Khawcharoenporn, T.; Vasoo, S.; Singh, K. Urinary Tract Infections due to Multidrug-Resistant Enterobacteriaceae: Prevalence and Risk Factors in a Chicago Emergency Department. Emerg. Med. Int. 2013, 2013, 258517. [Google Scholar] [CrossRef] [Green Version]
- Akbari, M.; Bakhshi, B.; Najar Peerayeh, S. Particular Distribution of Enterobacter cloacae Strains Isolated from Urinary Tract Infection within Clonal Complexes. Iran. Biomed. J. 2016, 20, 49–55. [Google Scholar] [PubMed]
- Breathnach, A.S.; Riley, P.A.; Shad, S.; Jownally, S.M.; Law, R.; Chin, P.C.; Kaufmann, M.E.; Smith, E.J. An outbreak of wound infection in cardiac surgery patients caused by Enterobacter cloacae arising from cardioplegia ice. J. Hosp. Infect. 2006, 64, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, R.; Prudêncio, C. Post-surgical wound infections involving Enterobacteriaceae with reduced susceptibility to β-lactams in two Portuguese hospitals. Int. Wound J. 2010, 7, 508–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wang, L.; Yang, Y.; Lu, F.; Ma, X.; Xia, X.; Jiang, J. Enterobacter cloacae infection after anterior cervical decompression and fusion: Case study and literature review. Int. J. Clin. Exp. Med. 2015, 8, 3438–3446. [Google Scholar]
- Al Yazidi, L.S.; Hameed, H.; Isaacs, D.; Axt, M.; Kesson, A. Enterobacter cloacae osteoarticular infection without risk factors: Case report and review of the literature. J. Paediatr. Child Health 2018, 54, 915–917. [Google Scholar] [CrossRef]
- Cisse, H.; Vernet-Garnier, V.; Hentzien, M.; Bajolet, O.; Lebrun, D.; Bonnet, M.; Ohl, X.; Diallo, S.; Bani-Sadr, F. Treatment of bone and joint infections caused by Enterobacter cloacae with a fluoroquinolone-cotrimoxazole combination. Int. J. Antimicrob. Agents 2019, 54, 245–248. [Google Scholar] [CrossRef]
- Huang, J.; Xu, Q.; Liu, F.; Xiong, H.; Yang, J. Enterobacter cloacae infection of the shoulder in a 52-year-old woman without apparent predisposing risk factor: A case report and literature review. BMC Infect. Dis. 2021, 21, 13. [Google Scholar] [CrossRef]
- Novak, A.; Goic-Barisic, I.; Andrasevic, A.T.; Butic, I.; Radic, M.; Jelic, M.; Rubic, Z.; Tonkic, M. Monoclonal outbreak of VIM-1-carbapenemase-producing Enterobacter cloacae in intensive care unit, University Hospital Centre Split, Croatia. Microb. Drug Resist. 2014, 20, 399–403. [Google Scholar] [CrossRef]
- Van Maerken, T.; de Brabandere, E.; Noël, A.; Coorevits, L.; de Waegemaeker, P.; Ablorh, R.; Bouchez, S.; Herck, I.; Peperstraete, H.; Bogaerts, P.; et al. A recurrent and transesophageal echocardiography-associated outbreak of extended-spectrum β-lactamase-producing Enterobacter cloacae complex in cardiac surgery patients. Antimicrob. Resist. Infect. Control. 2019, 8, 152. [Google Scholar] [CrossRef]
- Noël, A.; Vastrade, C.; Dupont, S.; de Barsy, M.; Huang, T.D.; van Maerken, T.; Leroux-Roels, I.; Delaere, B.; Melly, L.; Rondelet, B.; et al. Nosocomial outbreak of extended-spectrum β-lactamase-producing Enterobacter cloacae among cardiothoracic surgical patients: Causes and consequences. J. Hosp. Infect. 2019, 102, 54–60. [Google Scholar] [CrossRef]
- Ferry, A.; Plaisant, F.; Ginevra, C.; Dumont, Y.; Grando, J.; Claris, O.; Vandenesch, F.; Butin, M. Enterobacter cloacae colonisation and infection in a neonatal intensive care unit: Retrospective investigation of preventive measures implemented after a multiclonal outbreak. BMC Infect. Dis. 2020, 20, 682. [Google Scholar] [CrossRef] [PubMed]
- Yurdakul, D.; Yazgan-Karataş, A.; Şahin, F. Enterobacter Strains Might Promote Colon Cancer. Curr. Microbiol. 2015, 71, 403–411. [Google Scholar] [CrossRef] [PubMed]
- García-Fulgueiras, V.; Bado, I.; Mota, M.I.; Robino, L.; Cordeiro, N.F.; Varela, A.; Algorta, G.; Gutkind, G.; Ayala, J.A.; Vignoli, R. Extended-spectrum β-lactamases and plasmid-mediated quinolone resistance in enterobacterial clinical isolates in the paediatric hospital of Uruguay. J. Antimicrob. Chemother. 2011, 66, 1725–1729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Huang, N.; Zhou, C.; Lin, Y.; Zhang, Y.; Wang, L.; Zheng, X.; Zhou, T.; Wang, Z. Molecular Mechanisms and Epidemiology of Carbapenem-Resistant Enterobacter cloacae Complex Isolated from Chinese Patients during 2004–2018. Infect. Drug Resist. 2021, 14, 3647–3658. [Google Scholar] [CrossRef] [PubMed]
- Annavajhala, M.K.; Gomez-Simmonds, A.; Uhlemann, A.C. Multidrug-Resistant Enterobacter cloacae Complex Emerging as a Global, Diversifying Threat. Front. Microbiol. 2019, 10, 44. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Fang, R.; Zhang, Y.; Chen, L.; Huang, N.; Yu, K.; Zhou, C.; Cao, J.; Zhou, T. Characterization of resistance mechanisms of Enterobacter cloacae Complex co-resistant to carbapenem and colistin. BMC Microbiol. 2021, 21, 208. [Google Scholar] [CrossRef]
- Ebomah, K.E.; Okoh, A.I. Enterobacter cloacae harbouring blaNDM-1, blaKPC, and blaOXA-48-like carbapenem-resistant genes isolated from different environmental sources in South Africa. Int. J. Environ. Sci. 2020, 78, 151–164. [Google Scholar] [CrossRef]
- Miftode, I.-L.; Pasare, M.-A.; Miftode, R.-S.; Nastase, E.; Plesca, C.E.; Lunca, C.; Miftode, E.-G.; Timpau, A.-S.; Iancu, L.S.; Dorneanu, O.S. What Doesn’t Kill Them Makes Them Stronger: The Impact of the Resistance Patterns of Urinary Enterobacterales Isolates in Patients from a Tertiary Hospital in Eastern Europe. Antibiotics 2022, 11, 548. [Google Scholar] [CrossRef]
- Brives, C.; Pourraz, J. Phage therapy as a potential solution in the fight against AMR: Obstacles and possible futures. Palgrave Commun. 2020, 6, 100. [Google Scholar] [CrossRef]
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef] [Green Version]
- Nale, J.Y.; Clokie, M.R. Preclinical data and safety assessment of phage therapy in humans. Curr. Opin. Biotechnol. 2021, 68, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Slopek, S.; Durlakowa, I.; Weber-Dabrowska, B.; Dabrowski, M.; Kucharewicz-Krukowska, A. Results of bacteriophage treatment of suppurative bacterial infections. III. Detailed evaluation of the results obtained in further 150 cases. Arch. Immunol. Ther. Exp. 1984, 32, 317–335. [Google Scholar]
- Gratia, A. Des relations numeriques entre bacteries lysogenes et particles de bacteriophage. Ann. Inst. Pasteur 1936, 57, 652–676. [Google Scholar]
- Adams, M.H. Enumeration of bacteriophage particles. In The Bacteriophages; Inter Science Publ.: New York, NY, USA, 1959. [Google Scholar]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 2009, 501, 69–76. [Google Scholar]
- Ács, N.; Gambino, M.; Brøndsted, L. Bacteriophage Enumeration and Detection Methods. Front. Microbiol. 2020, 11, 594868. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.; Kim, S.W.; Kim, S.G.; Kang, J.W.; Jung, W.J.; Lee, S.B.; Lee, Y.M.; Giri, S.S.; Chi, C.; Park, S.C. The Characterization of a Novel Phage, pPa_SNUABM_DT01, Infecting Pseudomonas aeruginosa. Microorganisms 2021, 9, 2040. [Google Scholar] [CrossRef]
- Laforêt, F.; Antoine, C.; Blasdel Reuter, B.; Detilleux, J.; Pirnay, J.-P.; Brisse, S.; Fall, A.; Duprez, J.-N.; Delcenserie, V.; Thiry, D. In Vitro and In Vivo Assessments of Two Newly Isolated Bacteriophages against an ST13 Urinary Tract Infection Klebsiella pneumoniae. Viruses 2022, 14, 1079. [Google Scholar] [CrossRef]
- Necel, A.; Bloch, S.; Nejman-Faleńczyk, B.; Grabski, M.; Topka, G.; Dydecka, A.; Kosznik-Kwaśnicka, K.; Grabowski, Ł.; Jurczak-Kurek, A.; Wołkowicz, T.; et al. Characterization of a bacteriophage, vB_Eco4M-7, that effectively infects many Escherichia coli O157 strains. Sci. Rep. 2020, 10, 3743. [Google Scholar] [CrossRef]
- Kropinski, A.M. Measurement of the rate of attachment of bacteriophage to cells. In Bacteriophages. Methods in Molecular Biology; Clokie, M.R., Kropinski, A.M., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 501, pp. 151–155. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. hybridSPAdes: An algorithm for hybrid assembly of short and long reads. Bioinformatics 2016, 32, 1009–1015. [Google Scholar] [CrossRef] [Green Version]
- Prjibelski, A.D.; Vasilinetc, I.; Bankevich, A.; Gurevich, A.; Krivosheeva, T.; Nurk, S.; Pham, S.; Korobeynikov, A.; Lapidus, A.; Pevzner, P.A. ExSPAnder: A universal repeat resolver for DNA fragment assembly. Bioinformatics 2014, 30, i293–i301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasilinetc, I.; Prjibelski, A.D.; Gurevich, A.; Korobeynikov, A.; Pevzner, P.A. Assembling short reads from jumping libraries with large insert sizes. Bioinformatics 2015, 31, 3262–3268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.; Korobeynikov, A.; Lapidus, A.; Prjibelsky, A.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Genomes and Mini-metagenomes from Highly Chimeric Reads. In Research in Computational Molecular Biology; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2013; Volume 7821, pp. 158–170. [Google Scholar]
- Cuccuru, G.; Orsini, M.; Pinna, A.; Sbardellati, A.; Soranzo, N.; Travaglione, A.; Uva, P.; Zanetti, G.; Fotia, G. Orione, a web-based framework for NGS analysis in microbiology. Bioinformatics 2014, 30, 1928–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; O’Neill, K.R.; Haft, D.H.; DiCuccio, M.; Chetvernin, V.; Badretdin, A.; Coulouris, G.; Chitsaz, F.; Derbyshire, M.K.; Durkin, A.S.; et al. RefSeq: Expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res. 2021, 49, D1020–D1028. [Google Scholar] [CrossRef]
- Haft, D.H.; DiCuccio, M.; Badretdin, A.; Brover, V.; Chetvernin, V.; O’Neill, K.; Li, W.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; et al. RefSeq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018, 46, D851–D860. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [Green Version]
- Grant, C.E.; Bailey, T.L. XSTREME: Comprehensive motif analysis of biological sequence datasets. bioRxiv 2021. [Google Scholar] [CrossRef]
- Turner, D.; Adriaenssens, E.M.; Tolstoy, I.; Kropinski, A.M. Phage Annotation Guide: Guidelines for Assembly and High-Quality Annotation. Phage 2021, 2, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, H.W. Bacteriophage classification. In Bacteriophages: Biology and Applications; Kutter, E., Sulakvelidze, A., Eds.; CRC Press: Boca Raton, FL, USA, 2005; pp. 67–89. [Google Scholar]
- Ackermann, H.W. Bacteriophage electron microscopy. Adv. Virus Res. 2012, 82, 1–32. [Google Scholar] [PubMed]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef] [PubMed]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Schoch, C.L.; Ciufo, S.; Domrachev, M.; Hotton, C.L.; Kannan, S.; Khovanskaya, R.; Leipe, D.; Mcveigh, R.; O’Neill, K.; Robbertse, B.; et al. NCBI Taxonomy: A comprehensive update on curation, resources and tools. Database 2020, 2020, baaa062. [Google Scholar] [CrossRef]
- ICTV Taxonomy. Available online: https://ictv.global/taxonomy/ (accessed on 26 May 2022).
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef] [Green Version]
- ICTV Proposal. Available online: https://talk.ictvonline.org/ictv/proposals/2021.082B.A.v1.Tevens_new_families.zip (accessed on 26 May 2022).
- Suh, G.A.; Lodise, T.P.; Tamma, P.D.; Knisely, J.M.; Alexander, J.; Aslam, S.; Barton, K.D.; Bizzell, E.; Totten, K.M.C.; Campbell, J.; et al. Antibacterial Resistance Leadership Group. Considerations for the Use of Phage Therapy in Clinical Practice. Antimicrob. Agents Chemother. 2022, 66, e02071-21. [Google Scholar] [CrossRef]
- Finney, A.G.; Perry, J.M.; Evans, D.R.; Westbrook, K.J.; McElheny, C.L.; Iovleva, A.; Doi, Y.; Shields, R.K.; van Tyne, D. Isolation and Characterization of Lytic Bacteriophages Targeting Diverse Enterobacter spp. Clinical Isolates. Ther. Appl. Res. 2022, 3, 50–58. [Google Scholar] [CrossRef]
- Wang, K.; Tamayo, M.G.; Penner, T.V.; Cook, B.W.M.; Court, D.A.; Theriault, S.S. Characterization of the Enterobacter Phage vB_EclM_CIP9. Microbiol. Resour. Announc. 2020, 9, e01600-19. [Google Scholar] [CrossRef] [Green Version]
- Asif, M.; Naseem, H.; Alvi, I.A.; Basit, A.; Rehman, S.U. Characterization of a lytic EBP bacteriophage with large size genome against Enterobacter cloacae. APMIS 2021, 129, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Li, Z.; Li, X.; Wang, L.; Xu, Y. Genome sequence analysis of a novel Enterobacter cloacae phage, Ec_L1, belonging to the genus Eclunavirus. Arch. Virol. 2020, 165, 1929–1932. [Google Scholar] [CrossRef] [PubMed]
- Jeon, M.; Huh, K.; Ko, J.H.; Cho, S.Y.; Huh, H.J.; Lee, N.Y.; Kang, C.I.; Chung, D.R.; Peck, K.R. Difference in the Clinical Outcome of Bloodstream Infections Caused by Klebsiella aerogenes and Enterobacter cloacae Complex. Open Forum Infect. Dis. 2021, 8, ofab390. [Google Scholar] [CrossRef] [PubMed]
- Wesevich, A.; Sutton, G.; Ruffin, F.; Park, L.P.; Fouts, D.E.; Fowler, V.G., Jr.; Thaden, J.T. Newly Named Klebsiella aerogenes (formerly Enterobacter aerogenes) Is Associated with Poor Clinical Outcomes Relative to Other Enterobacter Species in Patients with Bloodstream Infection. J. Clin. Microbiol. 2020, 58, e00582-20. [Google Scholar] [CrossRef] [PubMed]
- Manohar, P.; Tamhankar, A.J.; Lundborg, C.S.; Nachimuthu, R. Therapeutic Characterization and Efficacy of Bacteriophage Cocktails Infecting Escherichia coli, Klebsiella pneumoniae, and Enterobacter Species. Front. Microbiol. 2019, 10, 574. [Google Scholar] [CrossRef] [Green Version]
- Morozova, V.; Jdeed, G.; Kozlova, Y.; Babkin, I.; Tikunov, A.; Tikunova, N. A New Enterobacter cloacae Bacteriophage EC151 Encodes the Deazaguanine DNA Modification Pathway and Represents a New Genus within the Siphoviridae Family. Viruses 2021, 13, 1372. [Google Scholar] [CrossRef]
- Addablah, A.Y.A.; Kakou-Ngazoa, S.; Akpa, E.E.; M’Bourou Ndombi, F.; Adioumani, E.; Koudou, A.; N’Golo, D.C.; Sina, M.K.; Kouassi, S.K.; Aoussi, S.; et al. Investigation of Phages Infecting Escherichia coli Strains B and C, and Enterobacter cloacae in Sewage and Ebrié Lagoon, Côte d’Ivoire. PHAGE 2021, 2, 104–111. [Google Scholar]
- Jamal, M.; Andleeb, S.; Jalil, F.; Imran, M.; Nawaz, M.A.; Hussain, T.; Ali, M.; Ur Rahman, S.; Das, C.R. Isolation, characterization and efficacy of phage MJ2 against biofilm forming multi-drug resistant Enterobacter cloacae. Folia Microbiol. 2019, 64, 101–111. [Google Scholar] [CrossRef]
- Ross, A.; Ward, S.; Hyman, P. More Is Better: Selecting for Broad Host Range Bacteriophages. Front. Microbiol. 2016, 7, 1352. [Google Scholar] [CrossRef] [Green Version]
- Gibson, S.B.; Green, S.I.; Liu, C.G.; Salazar, K.C.; Clark, J.R.; Terwilliger, A.L.; Kaplan, H.B.; Maresso, A.W.; Trautner, B.W.; Ramig, R.F. Constructing and Characterizing Bacteriophage Libraries for Phage Therapy of Human Infections. Front. Microbiol. 2019, 10, 2537. [Google Scholar] [CrossRef] [Green Version]
- Nair, A.; Vyawahare, R.; Khairnar, K. Characterization of a novel, biofilm dispersing, lytic bacteriophage against drug-resistant Enterobacter cloacae. J. Appl. Microbiol. 2021, 132, 2721–2732. [Google Scholar] [CrossRef] [PubMed]
- Topka, G.; Bloch, S.; Nejman-Faleńczyk, B.; Gąsior, T.; Jurczak-Kurek, A.; Necel, A.; Dydecka, A.; Richert, M.; Węgrzyn, G.; Węgrzyn, A. Characterization of Bacteriophage vB-EcoS-95, Isolated From Urban Sewage and Revealing Extremely Rapid Lytic Development. Front. Microbiol. 2019, 9, 3326. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.W.; Yuan, L.; Zhou, W.Y.; Mgomi, F.C.; Zhang, Y.S.; Wang, Y.; Zheng, X.F.; Hu, Q.; Gao, L.; Rao, S.Q.; et al. Isolation and genomic characterization of P.A-5, a novel virulent bacteriophage against Enterobacter hormaechei. Microb. Pathog. 2021, 152, 104767. [Google Scholar] [CrossRef] [PubMed]
- Topka-Bielecka, G.; Nejman-Faleńczyk, B.; Bloch, S.; Dydecka, A.; Necel, A.; Węgrzyn, A.; Węgrzyn, G. Phage-Bacteria Interactions in Potential Applications of Bacteriophage vB_EfaS-271 against Enterococcus faecalis. Viruses 2021, 13, 318. [Google Scholar] [CrossRef]
- Abedon, S.T. Phage therapy dosing: The problem(s) with multiplicity of infection (MOI). Bacteriophage 2016, 6, e1220348. [Google Scholar] [CrossRef] [Green Version]
- Duyvejonck, H.; Merabishvili, M.; Vaneechoutte, M.; de Soir, S.; Wright, R.; Friman, V.P.; Verbeken, G.; de Vos, D.; Pirnay, J.P.; van Mechelen, E.; et al. Evaluation of the Stability of Bacteriophages in Different Solutions Suitable for the Production of Magistral Preparations in Belgium. Viruses 2021, 13, 865. [Google Scholar] [CrossRef]
- Kering, K.K.; Zhang, X.; Nyaruaba, R.; Yu, J.; Wei, H. Application of Adaptive Evolution to Improve the Stability of Bacteriophages during Storage. Viruses 2020, 12, 423. [Google Scholar] [CrossRef] [Green Version]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The influence of external factors on bacteriophages. Folia Microbiol. 2011, 56, 191–200. [Google Scholar] [CrossRef] [Green Version]
- Thompson, S. Isolation and Characterization of Bacteriophages Infecting Enterobacter cloacae to Reduce Bloater Defect in Cucumber Fermentations. Master’s Thesis, Kennesaw State University, Kennesaw, GA, USA, 2020. [Google Scholar]
- Kuo, S.H.; Shen, C.J.; Shen, C.F.; Cheng, C.M. Role of pH Value in Clinically Relevant Diagnosis. Diagnostics 2020, 10, 107. [Google Scholar] [CrossRef] [Green Version]
- Beasley, D.E.; Koltz, A.M.; Lambert, J.E.; Fierer, N.; Dunn, R.R. The Evolution of Stomach Acidity and Its Relevance to the Human Microbiome. PLoS ONE 2015, 10, e0134116. [Google Scholar]
- Sánchez-Clemente, R.; Igeño, M.I.; Población, A.G.; Guijo, M.I.; Merchán, F.; Blasco, R. Study of pH Changes in Media during Bacterial Growth of Several Environmental Strains. Proceedings 2018, 2, 1297. [Google Scholar]
- Zhao, J.; Zhang, Z.; Tian, C.; Chen, X.; Hu, L.; Wei, X.; Li, H.; Lin, W.; Jiang, A.; Feng, R.; et al. Characterizing the Biology of Lytic Bacteriophage vB_EaeM_φEap-3 Infecting Multidrug-Resistant Enterobacter aerogenes. Front. Microbiol. 2019, 10, 420. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; Wei, X.; Ma, Y.; Yin, Z.; Li, H.; Lin, W.; Wang, X.; Li, C.; Shen, Z.; Zhao, R.; et al. Isolation and characterization of a bacteriophage phiEap-2 infecting multidrug resistant Enterobacter aerogenes. Sci. Rep. 2016, 6, 28338. [Google Scholar] [CrossRef] [PubMed]
- Khawaja, K.A.; Abbas, Z.; ur Rehman, S. Isolation and characterization of lytic phages TSE1-3 against Enterobacter cloacae. Open Life Sci. 2016, 11, 287–292. [Google Scholar] [CrossRef] [Green Version]
- Montso, P.K.; Mlambo, V.; Ateba, C.N. Characterization of Lytic Bacteriophages Infecting Multidrug-Resistant Shiga Toxigenic Atypical Escherichia coli O177 Strains Isolated from Cattle Feces. Front. Public Health 2019, 7, 355. [Google Scholar] [CrossRef] [PubMed]
- Wójcicki, M.; Średnicka, P.; Błażejak, S.; Gientka, I.; Kowalczyk, M.; Emanowicz, P.; Świder, O.; Sokołowska, B.; Juszczuk-Kubiak, E. Characterization and Genome Study of Novel Lytic Bacteriophages against Prevailing Saprophytic Bacterial Microflora of Minimally Processed Plant-Based Food Products. Int. J. Mol. Sci. 2021, 22, 12460. [Google Scholar] [CrossRef]
- Chen, C.W.; Yuan, L.; Zhang, Y.S.; Mgomi, F.C.; Wang, Y.; Yang, Z.Q.; Jiao, X.A. Comparison of biological and genomic characteristics of five virulent bacteriophages against Enterobacter hormaechei. Microb. Pathog. 2022, 162, 105375. [Google Scholar] [CrossRef]
- Tobin, E.; Brenner, S. Nanotechnology Fundamentals Applied to Clinical Infectious Diseases and Public Health. Open Forum Infect. Dis. 2021, 8, ofab583. [Google Scholar] [CrossRef]
- Durán, N.; Durán, M.; de Jesus, M.B.; Seabra, A.B.; Fávaro, W.J.; Nakazato, G. Silver nanoparticles: A new view on mechanistic aspects on antimicrobial activity. Nanomedicine 2016, 12, 789–799. [Google Scholar] [CrossRef]
- Roe, D.; Karandikar, B.; Bonn-Savage, N.; Gibbins, B.; Roullet, J.B. Antimicrobial surface functionalization of plastic catheters by silver nanoparticles. J. Antimicrob. Chemother. 2008, 61, 869–876. [Google Scholar] [CrossRef]
- Thomas, R.; Soumya, K.R.; Mathew, J.; Radhakrishnan, E.K. Inhibitory effect of silver nanoparticle fabricated urinary catheter on colonization efficiency of Coagulase Negative Staphylococci. J. Photochem. Photobiol. B 2015, 149, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Prateeksha, P.; Bajpai, R.; Rao, C.V.; Upreti, D.K.; Barik, S.K.; Singh, B.N. Chrysophanol-Functionalized Silver Nanoparticles for Anti-Adhesive and Anti-Biofouling Coatings to Prevent Urinary Catheter-Associated Infections. ACS Appl. Nano Mater. 2021, 4, 1512–1528. [Google Scholar] [CrossRef]
- Ermini, M.L.; Voliani, V. Antimicrobial Nano-Agents: The Copper Age. ACS Nano 2021, 15, 6008–6029. [Google Scholar] [CrossRef] [PubMed]
- Crisan, M.C.; Teodora, M.; Lucian, M. Copper Nanoparticles: Synthesis and Characterization, Physiology, Toxicity and Antimicrobial Applications. Appl. Sci. 2021, 12, 141. [Google Scholar] [CrossRef]
- Richter, Ł.; Paszkowska, K.; Cendrowska, U.; Olgiati, F.; Silva, P.J.; Gasbarri, M.; Guven, Z.P.; Paczesny, J.; Stellacci, F. Broad-spectrum nanoparticles against bacteriophage infections. Nanoscale 2021, 13, 18684–18694. [Google Scholar] [CrossRef]
- Sinclair, T.R.; van den Hengel, S.K.; Raza, B.G.; Rutjes, S.A.; de Roda Husman, A.M.; Peijnenburg, W.J.G.M.; Roesink, H.E.D.W.; de Vos, W.M. Surface chemistry-dependent antiviral activity of silver nanoparticles. Nanotechnology 2021, 32, 365101. [Google Scholar] [CrossRef]
- Abdelsattar, A.S.; Nofal, R.; Makky, S.; Safwat, A.; Taha, A.; El-Shibiny, A. The Synergistic Effect of Biosynthesized Silver Nanoparticles and Phage ZCSE2 as a Novel Approach to Combat Multidrug-Resistant Salmonella enterica. Antibiotics 2021, 10, 678. [Google Scholar] [CrossRef]
- Tomat, D.; Balagué, C.; Aquili, V.; Verdini, R.; Quiberoni, A. Resistance of phages lytic to pathogenic Escherichia coli to sanitisers used by the food industry and in home settings. Int. J. Food Sci. Technol. 2018, 53, 533–540. [Google Scholar] [CrossRef]
- Sommer, J.; Trautner, C.; Witte, A.K.; Fister, S.; Schoder, D.; Rossmanith, P.; Mester, P.J. Don’t Shut the Stable Door after the Phage Has Bolted-The Importance of Bacteriophage Inactivation in Food Environments. Viruses 2019, 11, 468. [Google Scholar] [CrossRef] [Green Version]
- Teklu, D.S.; Negeri, A.A.; Legese, M.H.; Bedada, T.L.; Woldemariam, H.K.; Tullu, K.D. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa, Ethiopia. Antimicrob. Resist. Infect. Control. 2019, 8, 39. [Google Scholar] [CrossRef]
- Osthoff, M.; Mcguinness, S.L.; Wagen, A.Z.; Eisen, D.P. Urinary tract infections due to extended-spectrum beta-lactamase-producing Gram-negative bacteria: Identification of risk factors and outcome predictors in an Australian tertiary referral hospital. Int. J. Infect. Dis. 2015, 34, 79–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alevizakos, M.; Nasioudis, D.; Mylonakis, E. Urinary tract infections caused by ESBL-producing Enterobacteriaceae in renal transplant recipients: A systematic review and meta-analysis. Transpl. Infect. Dis. 2017, 19, e12759. [Google Scholar] [CrossRef] [PubMed]
- Castanheira, M.; Simner, P.J.; Bradford, P.A. Extended-spectrum β-lactamases: An update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist. 2021, 3, dlab092. [Google Scholar] [CrossRef]
- Sybesma, W.; Zbinden, R.; Chanishvili, N.; Kutateladze, M.; Chkhotua, A.; Ujmajuridze, A.; Mehnert, U.; Kessler, T.M. Bacteriophages as Potential Treatment for Urinary Tract Infections. Front. Microbiol. 2016, 7, 465. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Pereira, C.; Santos, L.; Klumpp, J.; Almeida, A. Potential of phage cocktails in the inactivation of Enterobacter cloacae--An in vitro study in a buffer solution and in urine samples. Virus Res. 2016, 211, 199–208. [Google Scholar] [CrossRef]
- Chegini, Z.; Khoshbayan, A.; Vesal, S.; Moradabadi, A.; Hashemi, A.; Shariati, A. Bacteriophage therapy for inhibition of multi drug-resistant uropathogenic bacteria: A narrative review. Ann. Clin. Microbiol. Antimicrob. 2021, 20, 30. [Google Scholar] [CrossRef]
- Malik, S.; Sidhu, P.K.; Rana, J.S.; Nehra, K. Managing urinary tract infections through phage therapy: A novel approach. Folia Microbiol. 2020, 65, 217–231. [Google Scholar] [CrossRef]
- De Miguel, T.; Rama, J.L.R.; Sieiro, C.; Sánchez, S.; Villa, T.G. Bacteriophages and Lysins as Possible Alternatives to Treat Antibiotic-Resistant Urinary Tract Infections. Antibiotics 2020, 9, 466. [Google Scholar] [CrossRef]
- Leitner, L.; Sybesma, W.; Chanishvili, N.; Goderdzishvili, M.; Chkhotua, A.; Ujmajuridze, A.; Schneider, M.P.; Sartori, A.; Mehnert, U.; Bachmann, L.M.; et al. Bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: A randomized, placebo-controlled, double-blind clinical trial. BMC Urol. 2017, 17, 90. [Google Scholar] [CrossRef] [Green Version]
- Leitner, L.; Ujmajuridze, A.; Chanishvili, N.; Goderdzishvili, M.; Chkonia, I.; Rigvava, S.; Chkhotua, A.; Changashvili, G.; McCallin, S.; Schneider, M.P.; et al. Intravesical bacteriophages for treating urinary tract infections in patients undergoing transurethral resection of the prostate: A randomised, placebo-controlled, double-blind clinical trial. Lancet Infect. Dis. 2021, 21, 427–436. [Google Scholar] [CrossRef]
- Górski, A.; Borysowski, J.; Międzybrodzki, R. Phage Therapy: Towards a Successful Clinical Trial. Antibiotics 2020, 9, 827. [Google Scholar] [CrossRef] [PubMed]
- Terwilliger, A.; Clark, J.; Karris, M.; Hernandez-Santos, H.; Green, S.; Aslam, S.; Maresso, A. Phage Therapy Related Microbial Succession Associated with Successful Clinical Outcome for a Recurrent Urinary Tract Infection. Viruses 2021, 13, 2049. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).