Phosphomimetic S207D Lysyl–tRNA Synthetase Binds HIV-1 5′UTR in an Open Conformation and Increases RNA Dynamics
Abstract
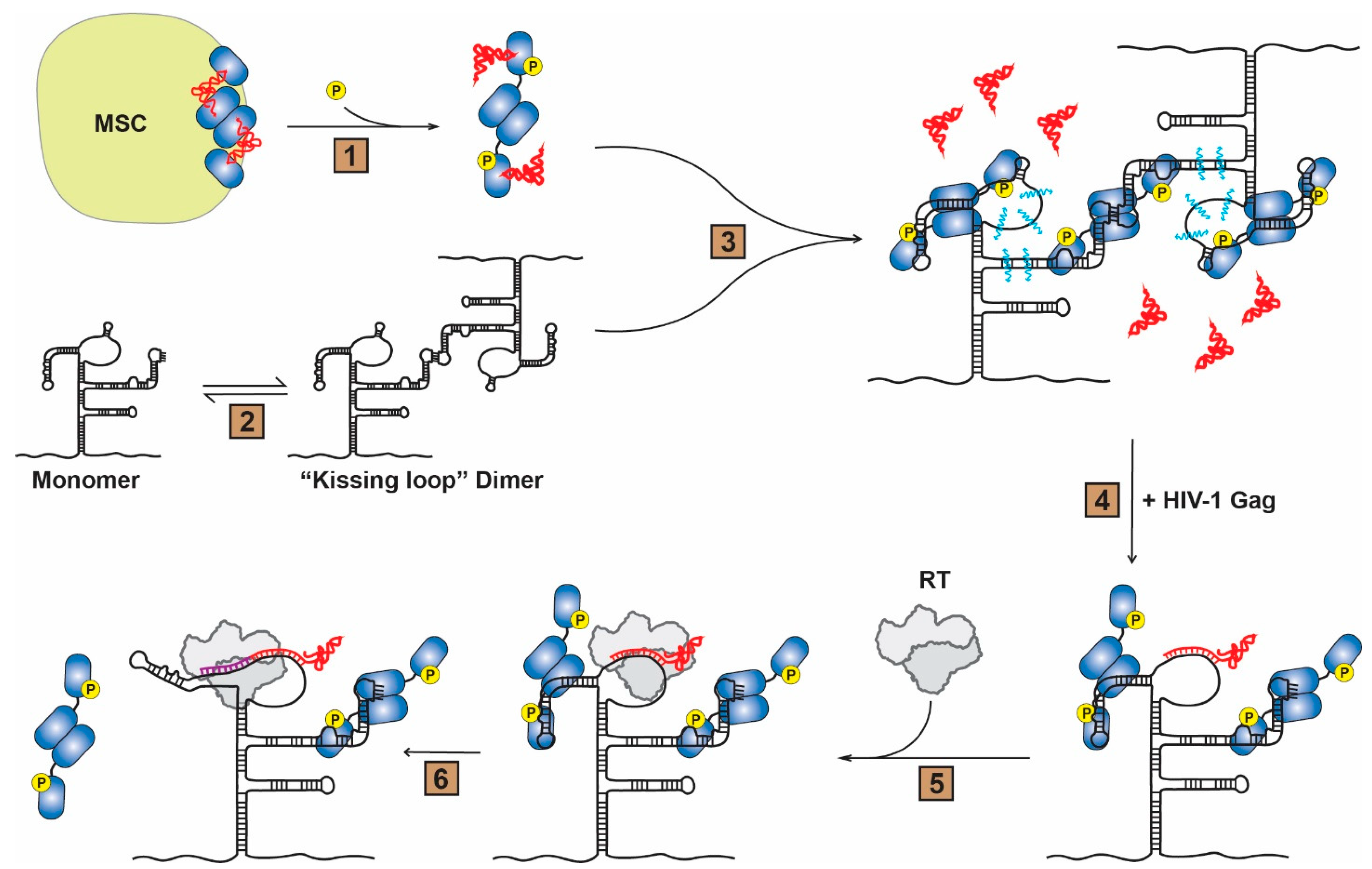
:1. Introduction
2. Materials and Methods
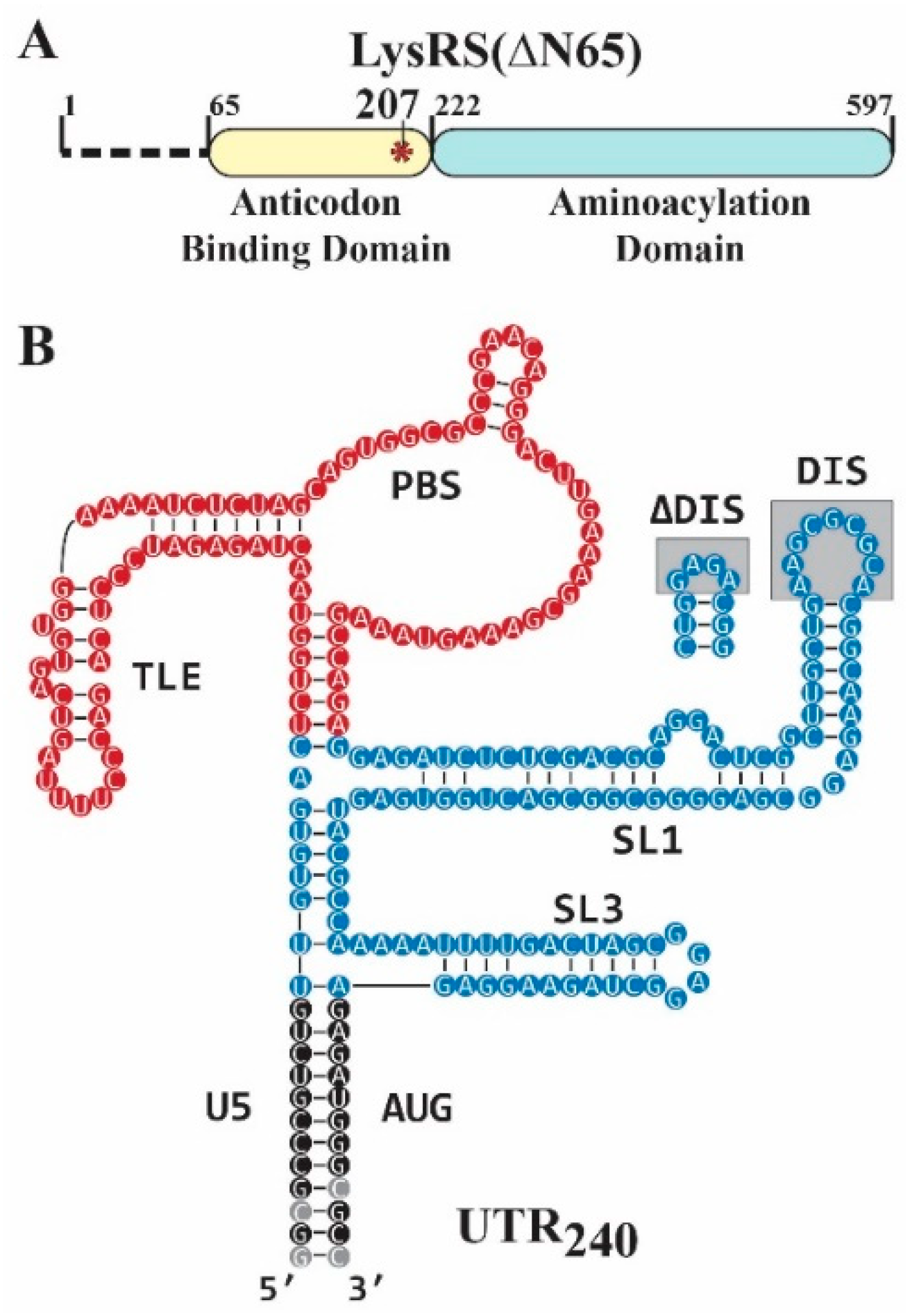
2.1. Preparation of Recombinant LysRS Variants
2.2. Preparation and Purification of RNAs and DNA-Primer-Annealed RNA Complexes
2.3. Fluorescence Anisotropy (FA) Binding Assays
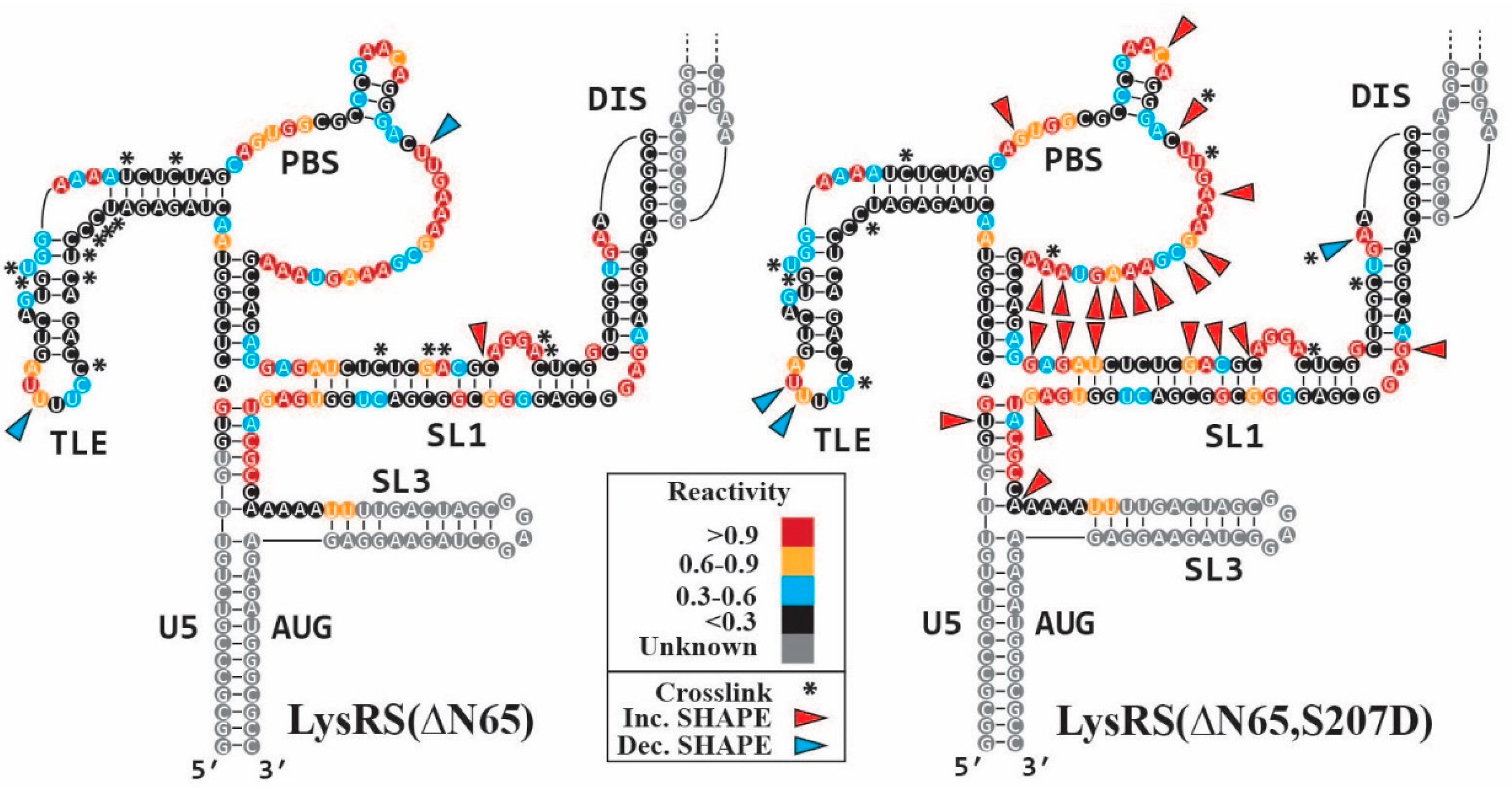
2.4. UV Crosslinking with SHAPE (XL-SHAPE) Experiments
2.5. Small-Angle X-ray Scattering (SAXS) Analysis
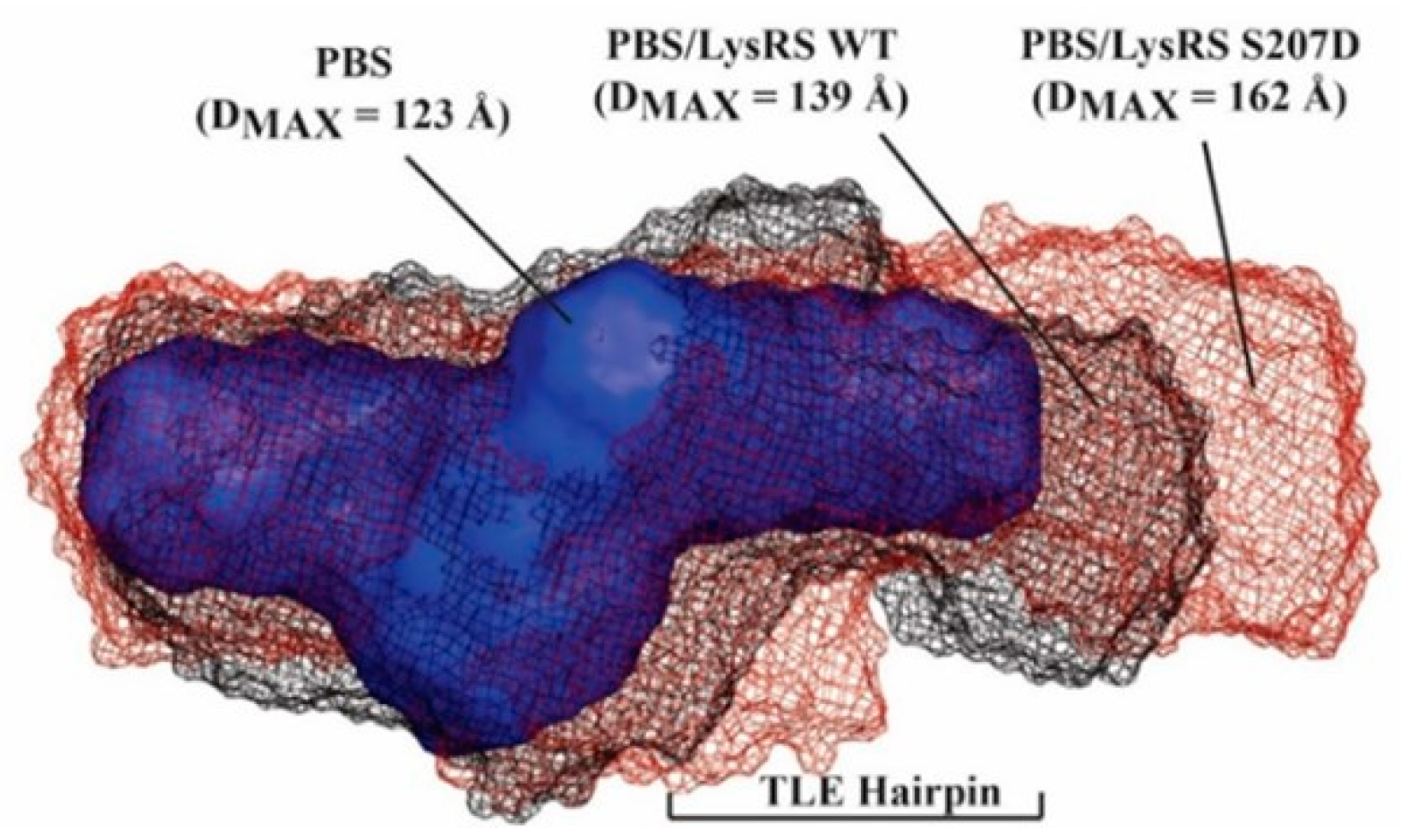
2.6. SEC–SAXS Analysis of Primer-Annealed and LysRS Bound PBS/TLE Complexes
3. Results
3.1. The DIS Contributes to gRNA Binding of Both WT and S207D LysRS(ΔN65)
3.2. S207D LysRS(ΔN65) Binds the PBS/TLE in an Open Conformation
3.3. S207D LysRS(ΔN65) Binding Induces gRNA Conformational Flexibility
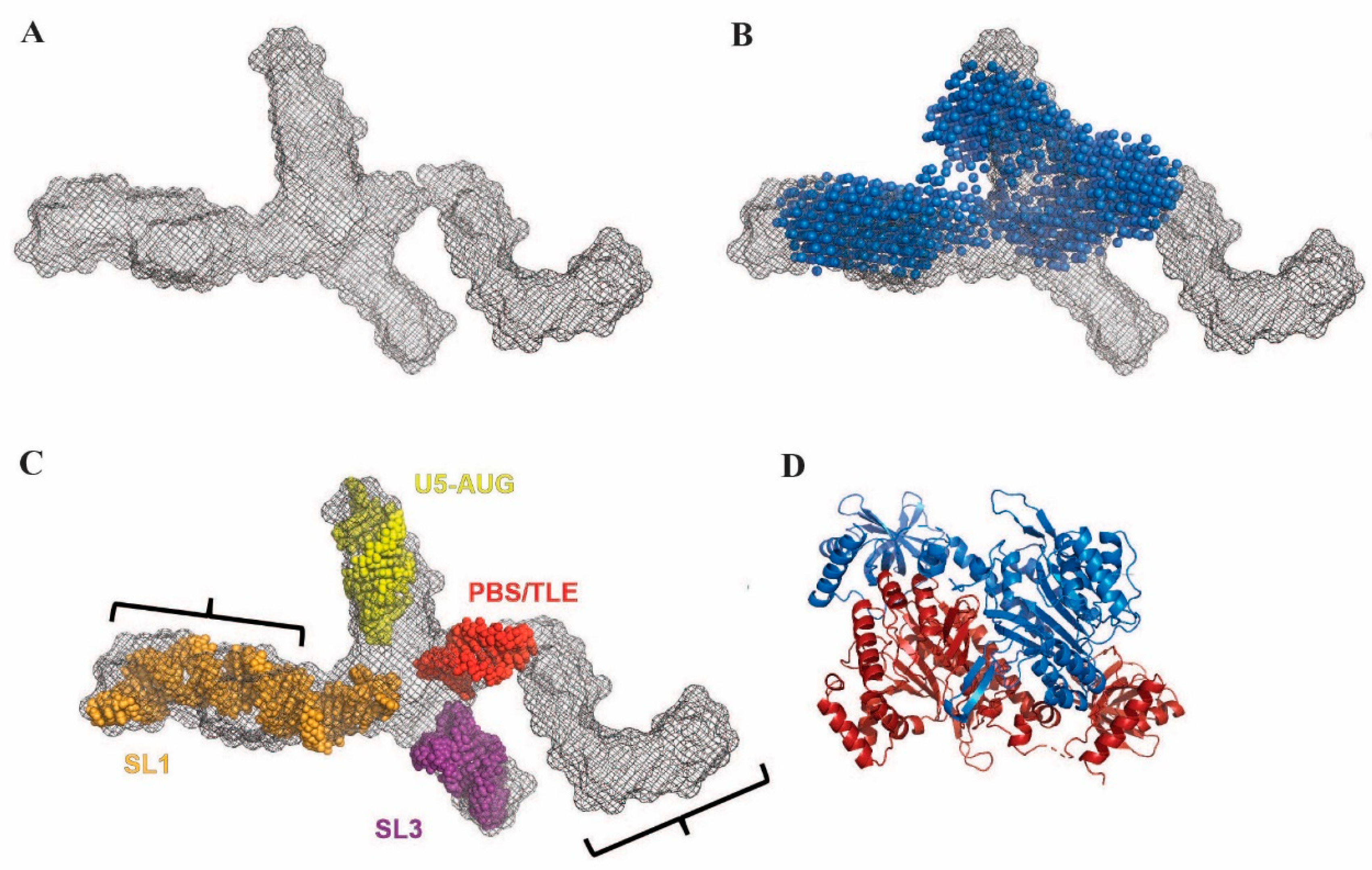
3.4. The Monomeric UTR240(ΔDIS) Adopts an Extended Cruciform-like Conformation
3.5. Disruption of TLE Structure by Primer Extension Releases Bound LysRS
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jiang, M.; Mak, J.; Ladha, A.; Cohen, E.; Klein, M.; Rovinski, B.; Kleiman, L. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 1993, 67, 3246–3253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pavon-Eternod, M.; Wei, M.; Pan, T.; Kleiman, L. Profiling non-lysyl tRNAs in HIV-1. RNA 2010, 16, 267–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stapulionis, R.; Deutscher, M.P. A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl. Acad. Sci. USA 1995, 92, 7158–7161. [Google Scholar] [CrossRef] [Green Version]
- Evans, M.E.; Clark, W.C.; Zheng, G.; Pan, T. Determination of tRNA aminoacylation levels by high-throughput sequencing. Nucleic Acids Res. 2017, 45, e133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cen, S.; Khorchid, A.; Javanbakht, H.; Gabor, J.; Stello, T.; Shiba, K.; Musier-Forsyth, K.; Kleiman, L. Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J. Virol. 2001, 75, 5043–5048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.; Cen, S.; Niu, M.; Javanbakht, H.; Kleiman, L. Specific inhibition of the synthesis of human lysyl-tRNA synthetase results in decreases in tRNA(Lys) incorporation, tRNA(3)(Lys) annealing to viral RNA, and viral infectivity in human immunodeficiency virus type 1. J. Virol. 2003, 77, 9817–9822. [Google Scholar] [CrossRef] [Green Version]
- Javanbakht, H.; Cen, S.; Musier-Forsyth, K.; Kleiman, L. Correlation between tRNALys3 aminoacylation and its incorporation into HIV-1. J. Biol. Chem. 2002, 277, 17389–17396. [Google Scholar] [CrossRef] [Green Version]
- Cen, S.; Javanbakht, H.; Niu, M.; Kleiman, L. Ability of wild-type and mutant lysyl-tRNA synthetase to facilitate tRNA(Lys) incorporation into human immunodeficiency virus type 1. J. Virol. 2004, 78, 1595–1601. [Google Scholar] [CrossRef] [Green Version]
- Tolkunova, E.; Park, H.; Xia, J.; King, M.P.; Davidson, E. The human lysyl-tRNA synthetase gene encodes both the cytoplasmic and mitochondrial enzymes by means of an unusual alternative splicing of the primary transcript. J. Biol. Chem. 2000, 275, 35063–35069. [Google Scholar] [CrossRef] [Green Version]
- Kaminska, M.; Shalak, V.; Francin, M.; Mirande, M. Viral hijacking of mitochondrial lysyl-tRNA synthetase. J. Virol. 2007, 81, 68–73. [Google Scholar] [CrossRef] [Green Version]
- Ofir-Birin, Y.; Fang, P.; Bennett, S.P.; Zhang, H.M.; Wang, J.; Rachmin, I.; Shapiro, R.; Song, J.; Dagan, A.; Pozo, J.; et al. Structural switch of lysyl-tRNA synthetase between translation and transcription. Mol. Cell 2013, 49, 30–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hei, Z.; Wu, S.; Liu, Z.; Wang, J.; Fang, P. Retractile lysyl-tRNA synthetase-AIMP2 assembly in the human multi-aminoacyl-tRNA synthetase complex. J. Biol. Chem. 2019, 294, 4775–4783. [Google Scholar] [CrossRef] [PubMed]
- Halwani, R.; Cen, S.; Javanbakht, H.; Saadatmand, J.; Kim, S.; Shiba, K.; Kleiman, L. Cellular distribution of Lysyl-tRNA synthetase and its interaction with Gag during human immunodeficiency virus type 1 assembly. J. Virol. 2004, 78, 7553–7564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, P.; Zhang, H.M.; Shapiro, R.; Marshall, A.G.; Schimmel, P.; Yang, X.L.; Guo, M. Structural context for mobilization of a human tRNA synthetase from its cytoplasmic complex. Proc. Natl. Acad. Sci. USA 2011, 108, 8239–8244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duchon, A.A.; St Gelais, C.; Titkemeier, N.; Hatterschide, J.; Wu, L.; Musier-Forsyth, K. HIV-1 Exploits a Dynamic Multi-aminoacyl-tRNA Synthetase Complex To Enhance Viral Replication. J. Virol. 2017, 91, e01240-17. [Google Scholar] [CrossRef] [Green Version]
- Watts, J.M.; Dang, K.K.; Gorelick, R.J.; Leonard, C.W.; Bess, J.W., Jr.; Swanstrom, R.; Burch, C.L.; Weeks, K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 2009, 460, 711–716. [Google Scholar] [CrossRef] [Green Version]
- Jones, C.P.; Saadatmand, J.; Kleiman, L.; Musier-Forsyth, K. Molecular mimicry of human tRNALys anti-codon domain by HIV-1 RNA genome facilitates tRNA primer annealing. RNA 2013, 19, 219–229. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Comandur, R.; Jones, C.P.; Tsang, P.; Musier-Forsyth, K. Anticodon-like binding of the HIV-1 tRNA-like element to human lysyl-tRNA synthetase. RNA 2016, 22, 1828–1835. [Google Scholar] [CrossRef]
- Jones, C.P.; Cantara, W.A.; Olson, E.D.; Musier-Forsyth, K. Small-angle X-ray scattering-derived structure of the HIV-1 5′ UTR reveals 3D tRNA mimicry. Proc. Natl. Acad. Sci. USA 2014, 111, 3395–3400. [Google Scholar] [CrossRef] [Green Version]
- Kleiman, L.; Cen, S. The tRNALys packaging complex in HIV-1. Int. J. Biochem. Cell Biol. 2004, 36, 1776–1786. [Google Scholar] [CrossRef]
- Jin, D.; Musier-Forsyth, K. Role of host tRNAs and aminoacyl-tRNA synthetases in retroviral replication. J. Biol. Chem. 2019, 294, 5352–5364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiba, K.; Stello, T.; Motegi, H.; Noda, T.; Musier-Forsyth, K.; Schimmel, P. Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescues Escherichia coli double-defective mutant. J. Biol. Chem. 1997, 272, 22809–22816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiu, J.; Tillett, D.; Dawes, I.W.; March, P.E. Site-directed, Ligase-Independent Mutagenesis (SLIM) for highly efficient mutagenesis of plasmids greater than 8kb. J. Microbiol. Methods 2008, 73, 195–198. [Google Scholar] [CrossRef] [PubMed]
- Pagano, J.M.; Clingman, C.C.; Ryder, S.P. Quantitative approaches to monitor protein-nucleic acid interactions using fluorescent probes. RNA 2011, 17, 14–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rye-McCurdy, T.; Rouzina, I.; Musier-Forsyth, K. Fluorescence anisotropy-based salt-titration approach to characterize protein-nucleic acid interactions. Methods Mol. Biol. 2015, 1259, 385–402. [Google Scholar]
- Stewart-Maynard, K.M.; Cruceanu, M.; Wang, F.; Vo, M.N.; Gorelick, R.J.; Williams, M.C.; Rouzina, I.; Musier-Forsyth, K. Retroviral nucleocapsid proteins display nonequivalent levels of nucleic acid chaperone activity. J. Virol. 2008, 82, 10129–10142. [Google Scholar] [CrossRef] [Green Version]
- Wilkinson, K.A.; Gorelick, R.J.; Vasa, S.M.; Guex, N.; Rein, A.; Mathews, D.H.; Giddings, M.C.; Weeks, K.M. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008, 6, e96. [Google Scholar] [CrossRef] [Green Version]
- Cantara, W.A.; Hatterschide, J.; Wu, W.; Musier-Forsyth, K. RiboCAT: A new capillary electrophoresis data analysis tool for nucleic acid probing. RNA 2017, 23, 240–249. [Google Scholar] [CrossRef] [Green Version]
- Steen, K.A.; Rice, G.M.; Weeks, K.M. Fingerprinting noncanonical and tertiary RNA structures by differential SHAPE reactivity. J. Am. Chem. Soc. 2012, 134, 13160–13163. [Google Scholar] [CrossRef] [Green Version]
- Hura, G.L.; Menon, A.L.; Hammel, M.; Rambo, R.P.; Poole, F.L., II; Tsutakawa, S.E.; Jenney, F.E., Jr.; Classen, S.; Frankel, K.A.; Hopkins, R.C.; et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat. Methods 2009, 6, 606–612. [Google Scholar] [CrossRef]
- Cantara, W.A.; Olson, E.D.; Musier-Forsyth, K. Analysis of RNA structure using small-angle X-ray scattering. Methods 2017, 113, 46–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Konarev, P.V.; Petoukhov, M.V.; Volkov, V.V.; Svergun, D.I. ATSAS2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2006, 39, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Acerbo, A.S.; Cook, M.J.; Gillilan, R.E. Upgrade of MacCHESS facility for X-ray scattering of biological macromolecules in solution. J. Synchrotron Radiat. 2015, 22, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, J.B.; Gillilan, R.E.; Skou, S. BioXTAS RAW: Improvements to a free open-source program for small-angle X-ray scattering data reduction and analysis. J. Appl. Crystallogr. 2017, 50, 1545–1553. [Google Scholar] [CrossRef] [Green Version]
- Skou, S.; Gillilan, R.E.; Ando, N. Synchrotron-based small-angle X-ray scattering of proteins in solution. Nat. Protoc. 2014, 9, 1727–1739. [Google Scholar] [CrossRef]
- Francin, M.; Mirande, M. Identity elements for specific aminoacylation of a tRNA by mammalian lysyl-tRNA synthetase bearing a nonspecific tRNA-interacting factor. Biochemistry 2006, 45, 10153–10160. [Google Scholar] [CrossRef]
- Comandur, R.; Olson, E.D.; Musier-Forsyth, K. Conservation of tRNA mimicry in the 5′-untranslated region of distinct HIV-1 subtypes. RNA 2017, 23, 1850–1859. [Google Scholar] [CrossRef] [Green Version]
- Keane, S.C.; Heng, X.; Lu, K.; Kharytonchyk, S.; Ramakrishnan, V.; Carter, G.; Barton, S.; Hosic, A.; Florwick, A.; Santos, J.; et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science 2015, 348, 917–921. [Google Scholar] [CrossRef] [Green Version]
- Merino, E.J.; Wilkinson, K.A.; Coughlan, J.L.; Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 2005, 127, 4223–4231. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Merino, E.J.; Weeks, K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006, 1, 1610–1616. [Google Scholar] [CrossRef]
- McGinnis, J.L.; Dunkle, J.A.; Cate, J.H.; Weeks, K.M. The mechanisms of RNA SHAPE chemistry. J. Am. Chem. Soc. 2012, 134, 6617–6624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenyon, J.C.; Prestwood, L.J.; Lever, A.M. A novel combined RNA-protein interaction analysis distinguishes HIV-1 Gag protein binding sites from structural change in the viral RNA leader. Sci. Rep. 2015, 5, 14369. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Harada, B.T.; Miller, J.T.; Le Grice, S.F.; Zhuang, X. Initiation complex dynamics direct the transitions between distinct phases of early HIV reverse transcription. Nat. Struct. Mol. Biol. 2010, 17, 1453–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathew, E.; Mirza, A.; Menhart, N. Liquid-chromatography-coupled SAXS for accurate sizing of aggregating proteins. J. Synchrotron Radiat. 2004, 11, 314–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Nikolaitchik, O.; Singh, J.; Wright, A.; Bencsics, C.E.; Coffin, J.M.; Ni, N.; Lockett, S.; Pathak, V.K.; Hu, W.S. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. USA 2009, 106, 13535–13540. [Google Scholar] [CrossRef] [Green Version]
- Goldschmidt, V.; Paillart, J.C.; Rigourd, M.; Ehresmann, B.; Aubertin, A.M.; Ehresmann, C.; Marquet, R. Structural variability of the initiation complex of HIV-1 reverse transcription. J. Biol. Chem. 2004, 279, 35923–35931. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.S.; Hughes, S.H. HIV-1 reverse transcription. Cold Spring Harb. Perspect. Med. 2012, 2, a006882. [Google Scholar] [CrossRef] [Green Version]
- Kleiman, L.; Jones, C.P.; Musier-Forsyth, K. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010, 584, 359–365. [Google Scholar] [CrossRef] [Green Version]
- Brigham, B.S.; Kitzrow, J.P.; Reyes, J.C.; Musier-Forsyth, K.; Munro, J.B. Intrinsic conformational dynamics of the HIV-1 genomic RNA 5′ UTR. Proc. Natl. Acad. Sci. USA 2019, 116, 10372–10381. [Google Scholar] [CrossRef] [Green Version]
- Huthoff, H.; Berkhout, B. Two alternating structures of the HIV-1 leader RNA. RNA 2001, 7, 143–157. [Google Scholar] [CrossRef] [Green Version]
- Abbink, T.E.; Ooms, M.; Haasnoot, P.C.; Berkhout, B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry 2005, 44, 9058–9066. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Heng, X.; Garyu, L.; Monti, S.; Garcia, E.L.; Kharytonchyk, S.; Dorjsuren, B.; Kulandaivel, G.; Jones, S.; Hiremath, A.; et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 2011, 334, 242–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolaitchik, O.A.; Liu, S.; Kitzrow, J.P.; Liu, Y.; Rawson, J.M.O.; Shakya, S.; Cheng, Z.; Pathak, V.K.; Hu, W.S.; Musier-Forsyth, K. Selective packaging of HIV-1 RNA genome is guided by the stability of 5′ untranslated region polyA stem. Proc. Natl. Acad. Sci. USA 2021, 118, e2114494118. [Google Scholar] [CrossRef]
- Ye, L.; Gribling-Burrer, A.S.; Bohn, P.; Kibe, A.; Bortlein, C.; Ambi, U.B.; Ahmad, S.; Olguin-Nava, M.; Smith, M.; Caliskan, N.; et al. Short- and long-range interactions in the HIV-1 5′ UTR regulate genome dimerization and packaging. Nat. Struct. Mol. Biol. 2022, 29, 306–319. [Google Scholar] [CrossRef] [PubMed]
- Barat, C.; Lullien, V.; Schatz, O.; Keith, G.; Nugeyre, M.T.; Gruninger-Leitch, F.; Barre-Sinoussi, F.; LeGrice, S.F.; Darlix, J.L. HIV-1 reverse transcriptase specifically interacts with the anticodon domain of its cognate primer tRNA. EMBO J. 1989, 8, 3279–3285. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.P.; Datta, S.A.; Rein, A.; Rouzina, I.; Musier-Forsyth, K. Matrix domain modulates HIV-1 Gag’s nucleic acid chaperone activity via inositol phosphate binding. J. Virol. 2011, 85, 1594–1603. [Google Scholar] [CrossRef] [Green Version]

| Kd (nM ± SD) | ||
|---|---|---|
| RNA | WT LysRS(ΔN65) | S207D LysRS(ΔN65) |
| tRNALys3 | 407 ± 33 a | 470 ± 190 b |
| PBS/TLE105 | 330 ± 115 | 315 ± 130 |
| UTR240 | 146 ± 47 | 139 ± 45 |
| UTR240(ΔDIS) | 315 ± 142 | 482 ± 103 |
| RNA-DNA Complex | Kd (nM ± SD) | |||
|---|---|---|---|---|
| RNA | DNA Primer | WT LysRS(ΔN65) | S207D LysRS(ΔN65) | TLE Rotation |
| PBS/TLE105 | antiPBS18 | 240 ± 95 | 417 ± 90 | 0° |
| PBS/TLE105 | antiPBS18+3 | 488 ± 171 | 305 ± 75 | ~65° |
| PBS/TLE105 | antiPBS18+6 | 865 ± 379 | 590 ± 182 | ~145° |
| PBS/TLE105 | antiPBS18+11 | 1008 ± 639 | 949 ± 148 | ~225° |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cantara, W.A.; Pathirage, C.; Hatterschide, J.; Olson, E.D.; Musier-Forsyth, K. Phosphomimetic S207D Lysyl–tRNA Synthetase Binds HIV-1 5′UTR in an Open Conformation and Increases RNA Dynamics. Viruses 2022, 14, 1556. https://doi.org/10.3390/v14071556
Cantara WA, Pathirage C, Hatterschide J, Olson ED, Musier-Forsyth K. Phosphomimetic S207D Lysyl–tRNA Synthetase Binds HIV-1 5′UTR in an Open Conformation and Increases RNA Dynamics. Viruses. 2022; 14(7):1556. https://doi.org/10.3390/v14071556
Chicago/Turabian StyleCantara, William A., Chathuri Pathirage, Joshua Hatterschide, Erik D. Olson, and Karin Musier-Forsyth. 2022. "Phosphomimetic S207D Lysyl–tRNA Synthetase Binds HIV-1 5′UTR in an Open Conformation and Increases RNA Dynamics" Viruses 14, no. 7: 1556. https://doi.org/10.3390/v14071556
APA StyleCantara, W. A., Pathirage, C., Hatterschide, J., Olson, E. D., & Musier-Forsyth, K. (2022). Phosphomimetic S207D Lysyl–tRNA Synthetase Binds HIV-1 5′UTR in an Open Conformation and Increases RNA Dynamics. Viruses, 14(7), 1556. https://doi.org/10.3390/v14071556






