African Swine Fever Virus Manipulates the Cell Cycle of G0-Infected Cells to Access Cellular Nucleotides
Abstract
:1. Introduction
2. Material and Methods
2.1. Animals
2.2. Virus
2.3. Alveolar Macrophage Culture
2.4. PAM Infection
2.5. Nucleotides in Cultural Medium
2.6. Dexamethasone in Cultural Medium
2.7. Histopathological Studies
2.8. Safranin and Indigo-Picro-Carmine Staining Technique
2.9. Image Cytometry of the PAM/Cell Cycle Phase
2.10. DNA Quantification
2.11. ELISA Analysis
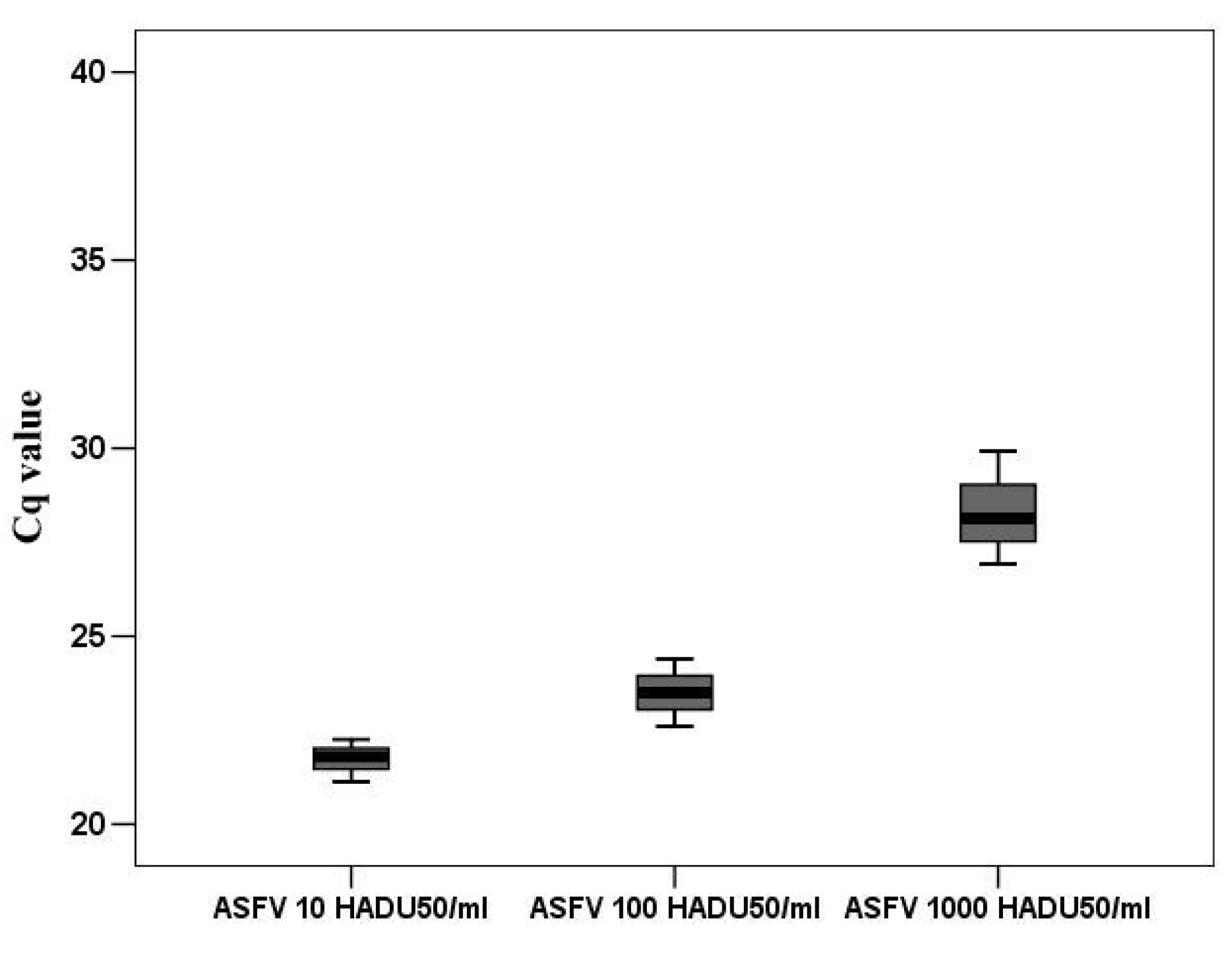
2.12. Gene Expression Analysis by Quantitative Real-Time PCR
2.13. Statistical Analysis
3. Results
3.1. Cell Cycle Changes in Tissues of ASFV Infected Pig
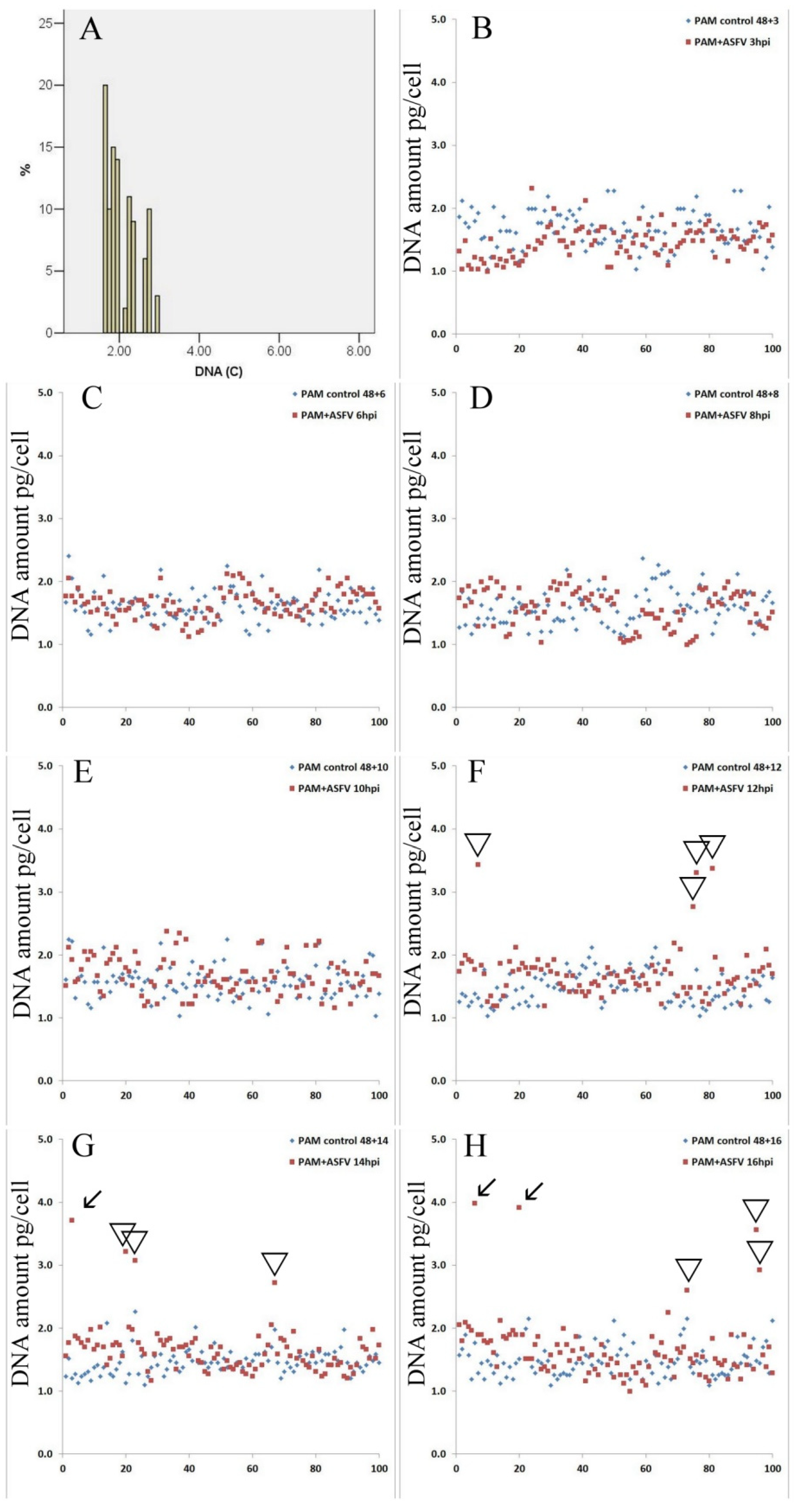
3.2. Nuclear DNA Synthesis in Infected PAM
3.3. ASFV (Arm07) Increases Expression of Cyclin A and Cyclin E in PAM Cells
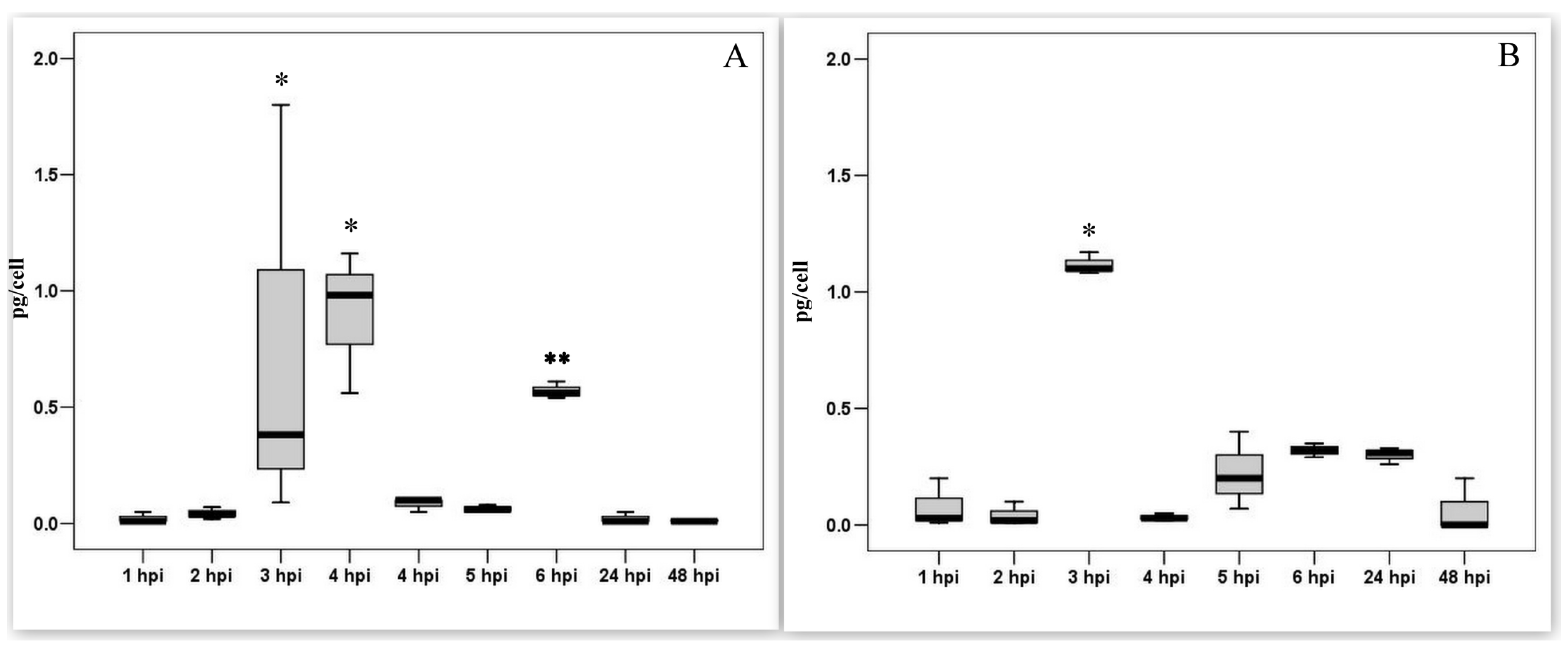
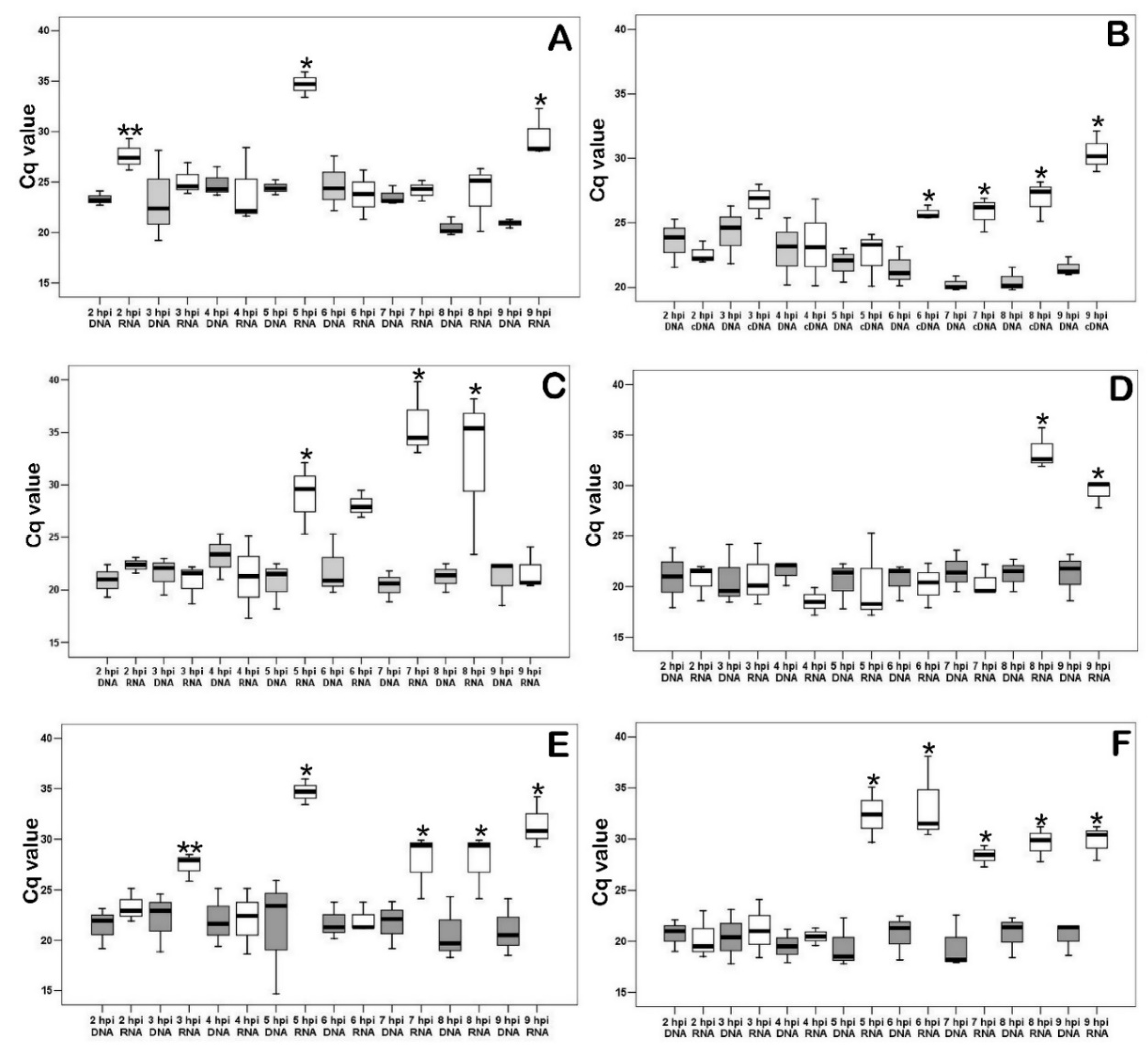
3.4. Measurement of the Transcriptional Activity of Viral Genes Involved in DNA Synthesis during the Pre-Synthesis of Cellular DNA
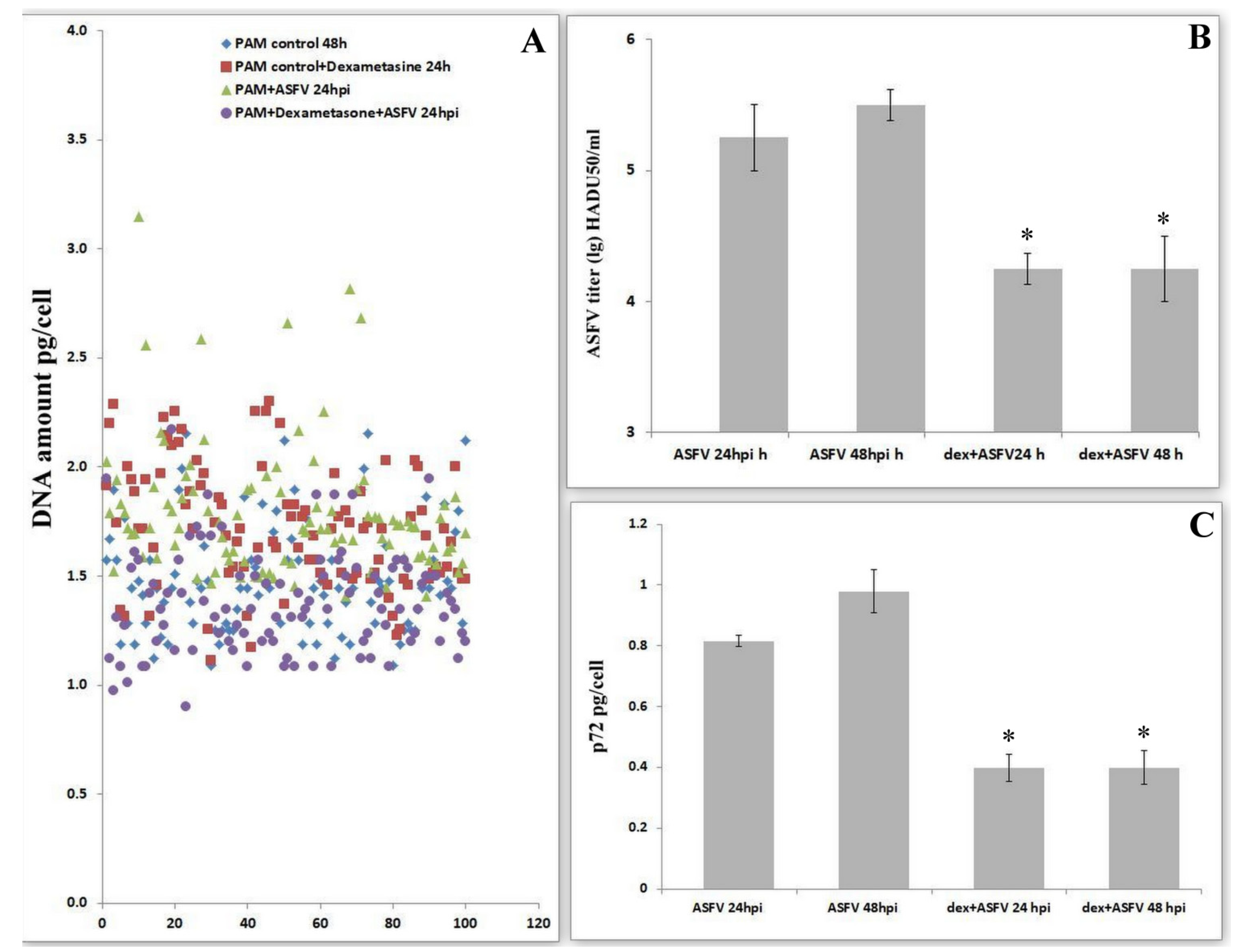
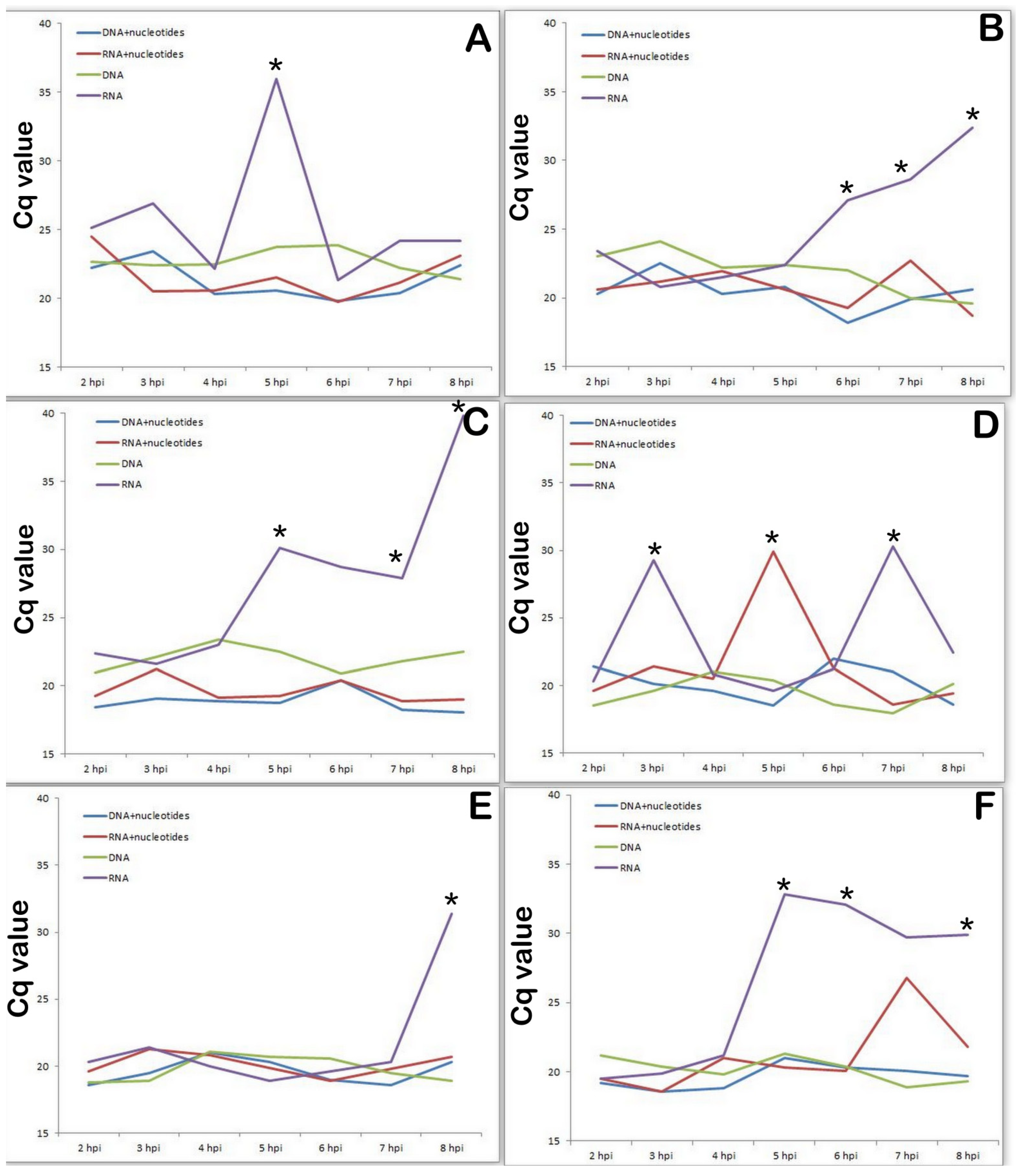
3.5. Changes in Transcriptional Activity of the Viral Genes Involved in DNA Synthesis Depend on the Presence of Nucleotides in Medium
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chapman, D.A.G.; Tcherepanov, V.; Upton, C.; Dixon, L.K. Comparison of the genome sequences of non-pathogenic and pathogenic African swine fever virus isolates. J. Gen. Virol. 2008, 89, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Bastos, A.D.; Arnot, L.F.; Jacquier, M.D.; Maree, S. A host species-informative internal control for molecular assessment of African swine fever virus infection rates in the African sylvatic cycleOrnithodorosvector. Med. Vet. Entomol. 2009, 23, 399–409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thelander, L.; Reichard, P. Reduction of Ribonucleotides. Annu. Rev. Biochem. 1979, 48, 133–158. [Google Scholar] [CrossRef] [PubMed]
- Bagga, S.; Bouchard, M.J. Cell Cycle Regulation During Viral Infection. Methods Mol. Biol. 2014, 1170, 165–227. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.M.; Salas, M.L.; Santarén, J.F. African swine fever virus-induced polypeptides in porcine alveolar macrophages and in Vero cells: Two-dimensional gel analysis. Proteomics 2001, 1, 1447–1456. [Google Scholar] [CrossRef]
- Dixon, L.K.; Chapman, D.A.; Netherton, C.L.; Upton, C. African swine fever virus replication and genomics. Virus Res. 2013, 173, 3–14. [Google Scholar] [CrossRef]
- Moore, D.M.; Zsak, L.; Neilan, J.G.; Lu, Z.; Rock, D.L. The African Swine Fever Virus Thymidine Kinase Gene Is Required for Efficient Replication in Swine Macrophages and for Virulence in Swine. J. Virol. 1998, 72, 10310–10315. [Google Scholar] [CrossRef] [Green Version]
- Sanford, B.; Holinka, L.; O’Donnell, V.; Krug, P.; Carlson, J.; Alfano, M.; Carrillo, C.; Wu, P.; Lowe, A.; Risatti, G.; et al. Deletion of the thymidine kinase gene induces complete attenuation of the Georgia isolate of African swine fever virus. Virus Res. 2016, 213, 165–171. [Google Scholar] [CrossRef]
- A Baylis, S.; Banham, A.H.; Vydelingum, S.; Dixon, L.K.; Smith, G.L. African swine fever virus encodes a serine protein kinase which is packaged into virions. J. Virol. 1993, 67, 4549–4556. [Google Scholar] [CrossRef] [Green Version]
- Yáñez, R.J.; Rodríguez, J.M.; Rodriguez, J.F.; Salas, M.L.; Viñuela, E. African swine fever virus thymidylate kinase gene: Sequence and transcriptional mapping. J. Gen. Virol. 1993, 74, 1633–1638. [Google Scholar] [CrossRef]
- Oliveros, M.; Garcia-Escudero, R.; Alejo, A.; Viñuela, E.; Salas, M.L.; Salas, J. African Swine Fever Virus dUTPase Is a Highly Specific Enzyme Required for Efficient Replication in Swine Macrophages. J. Virol. 1999, 73, 8934–8943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enjuanes, L.; Cubero, I.; Vinuela, E. Sensitivity of Macrophages from Different Species to African Swine Fever (ASF) Virus. J. Gen. Virol. 1977, 34, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Karalova, E.M.; Sargsyan, K.V.; Hampikian, G.K.; Voskanyan, H.E.; Abroyan, L.O.; Avetisyan, A.S.; Hakobyan, L.A.; Arzumanyan, H.H.; Zakaryan, H.S.; Karalyan, Z.A. Phenotypic and cytologic studies of lymphoid cells and monocytes in primary culture of porcine bone marrow during infection of African swine fever virus. Vitr. Cell Dev. Biol. Anim. 2010, 47, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Karalyan, Z.; Zakaryan, H.; Arzumanyan, H.; Sargsyan, K.; Voskanyan, H.; Hakobyan, L.; Abroyan, L.; Avetisyan, A.; Karalova, E. Pathology of porcine peripheral white blood cells during infection with African swine fever virus. BMC Vet. Res. 2012, 8, 18. [Google Scholar] [CrossRef] [Green Version]
- Forman, A.; Wardley, R.; Norley, S. Interactions of porcine alveolar macrophages and bone marrow cells with African swine fever virus and virus-infected cells. Vet. Microbiol. 1983, 8, 163–177. [Google Scholar] [CrossRef]
- Carvalhal, A.V.; Santos, S.S.; Carrondo, M.J.T. Extracellular purine and pyrimidine catabolism in cell culture. Biotechnol. Prog. 2011, 27, 1373–1382. [Google Scholar] [CrossRef]
- Chen, Y.H.; Desai, P.; Shiao, R.T.; Lavelle, D.; Haleem, A.; Chen, J. Inhibition of myeloma cell growth by dexamethasone and all-trans retinoic acid: Synergy through modulation of interleukin-6 autocrine loop at multiple sites. Blood 1996, 87, 314–323. [Google Scholar] [CrossRef] [Green Version]
- Alvarez, Y.; Valladares, Y. Differential Staining of the Cell Cycle. Nat. New Biol. 1972, 238, 279–280. [Google Scholar] [CrossRef]
- Swain, D.; De, D.N. Differential Staining of the Cell Cycle of Plant Cells Using Safranin and Indigo-Picrocarmine. Stain. Technol. 1990, 65, 197–204. [Google Scholar] [CrossRef]
- Gaub, J.; Auer, G.; Zetterberg, A. Quantitative cytochemical aspects of a combined Feulgen-Naphthol Yellow S staining procedure for the simultaneous determination of nuclear and cytoplasmic proteins and DNA in mammalian cells. Exp. Cell Res. 1975, 92, 323–332. [Google Scholar] [CrossRef]
- Karalyan, Z.A.; Izmailyan, R.A.; Abroyan, L.O.; Avetisyan, A.S.; Hakobyan, L.A.; Zakaryan, H.S.; Karalova, E.M. Evaluation of Viral Genome Copies Within Viral Factories on Different DNA Viruses. J. Histochem. Cytochem. 2018, 66, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Lassalle, B.; Gautron, J.; Martelly, I.; Le Moigne, A. Image analysis of rat satellite cell proliferation in vitro. Cytotechnology 1989, 2, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Ginzinger, D.G. Gene quantification using real-time quantitative PCR: An emerging technology hits the mainstream. Exp. Hematol. 2002, 30, 503–512. [Google Scholar] [CrossRef]
- Yin, J.L.; Shackel, N.A.; Zekry, A.; McGuinness, P.H.; Richards, C.; Van Der Putten, K.; Mccaughan, G.; Eris, J.M.; Bishop, G.A. Real-time reverse transcriptase–polymerase chain reaction (RT–PCR) for measurement of cytokine and growth factor mRNA expression with fluorogenic probes or SYBR Green I. Immunol. Cell Biol. 2001, 79, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loyer, P.; Corlu, A.; Desdouets, C. Regulation of the Hepatocyte Cell Cycle: Signaling Pathways and Protein Kinases. Int. J. Hepatol. 2012, 2012, 592354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goya, L.; Maiyar, A.C.; Ge, Y.; Firestone, G.L. Glucocorticoids induce a G1/G0 cell cycle arrest of Con8 rat mammary tumor cells that is synchronously reversed by steroid withdrawal or addition of transforming growth factor-alpha. Mol. Endocrinol. 1993, 7, 1121–1132. [Google Scholar] [CrossRef] [Green Version]
- Mattern, J.; Büchler, M.W.; Herr, I. Cell Cycle Arrest by Glucocorticoids May Protect Normal Tissue and Solid Tumors from Cancer Therapy. Cancer Biol. Ther. 2007, 6, 1341–1350. [Google Scholar] [CrossRef] [Green Version]
- Tatoyan, M.; Ter-Pogossyan, Z.; Semerjyan, A.; Gevorgyan, V.; Karalyan, N.Y.; Sahakyan, C.; Mkrtchyan, G.; Gazaryan, H.; Avagyan, H.; Karalyan, Z. Serum Concentrations of Vascular Endothelial Growth Factor, Stromal Cell-Derived Factor, Nitric Oxide and Endothelial DNA Proliferation in Development of Microvascular Pathology in Acute African Swine Fever. J. Comp. Pathol. 2019, 167, 50–59. [Google Scholar] [CrossRef]
- Rodríguez, J.M.; Salas, M.L. African swine fever virus transcription. Virus Res. 2013, 173, 15–28. [Google Scholar] [CrossRef]
- Boursnell, M.; Shaw, K.; Yáñez, R.J.; Viñuela, E.; Dixon, L. The sequences of the ribonucleotide reductase genes from African swine fever virus show considerable homology with those of the orthopoxvirus, vaccinia virus. Virology 1991, 184, 411–416. [Google Scholar] [CrossRef]
- Lane, A.N.; Fan, T.W.-M. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015, 43, 2466–2485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandez, A.; Pérez, J.; Mulas, J.M.D.L.; Carrasco, L.; Dominguez, J.; Sierra, M. Localization of African swine fever viral antigen, swine IgM, IgG and C1q in lung and liver tissues of experimentally infected pigs. J. Comp. Pathol. 1992, 107, 81–90. [Google Scholar] [CrossRef]
- Gómez-Villamandos, J.C.; Hervás, J.; Méndez, A.; Carrasco, L.; Mulas, J.M.D.L.; Villeda, C.J.; Wilkinson, P.J.; Sierra, M.A. Experimental African swine fever: Apoptosis of lymphocytes and virus replication in other cells. J. Gen. Virol. 1995, 76, 2399–2405. [Google Scholar] [CrossRef] [PubMed]
- Karger, A.; Pérez-Núñez, D.; Urquiza, J.; Hinojar, P.; Alonso, C.; Freitas, F.B.; Revilla, Y.; Le Potier, M.-F.; Montoya, M. An Update on African Swine Fever Virology. Viruses 2019, 11, 864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Freitas, F.B.; Frouco, G.; Martins, C.; Ferreira, F. African swine fever virus encodes for an E2-ubiquitin conjugating enzyme that is mono- and di-ubiquitinated and required for viral replication cycle. Sci. Rep. 2018, 8, 3471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapinas, K.; Grandy, R.; Ghule, P.; Medina, R.; Becker, K.; Pardee, A.; Zaidi, S.K.; Lian, J.; Stein, J.; van Wijnen, A.; et al. The abbreviated pluripotent cell cycle. J. Cell. Physiol. 2012, 228, 9–20. [Google Scholar] [CrossRef] [Green Version]
- Matson, J.; Cook, J.G. Cell cycle proliferation decisions: The impact of single cell analyses. FEBS J. 2016, 284, 362–375. [Google Scholar] [CrossRef] [Green Version]
- Xia, N.; Wang, H.; Liu, X.; Shao, Q.; Ao, D.; Xu, Y.; Jiang, S.; Luo, J.; Zhang, J.; Chen, N.; et al. African Swine Fever Virus Structural Protein p17 Inhibits Cell Proliferation through ER Stress—ROS Mediated Cell Cycle Arrest. Viruses 2020, 13, 21. [Google Scholar] [CrossRef]
- Castelló, A.; Quintas, A.; Sánchez, E.G.; Sabina, P.; Nogal, M.; Carrasco, L.; Revilla, Y. Regulation of Host Translational Machinery by African Swine Fever Virus. PLoS Pathog. 2009, 5, e1000562. [Google Scholar] [CrossRef] [Green Version]
- Shen, Z.; Chen, C.; Yang, Y.; Xie, Z.; Ao, Q.; Lv, L.; Zhang, S.; Chen, H.; Hu, R.; Chen, H.; et al. Novel Function of African Swine Fever Virus pE66L in Inhibition of Host Translation by the PKR/eIF2α Pathway. J. Virol. 2021, 95, e01872-20. [Google Scholar] [CrossRef]
- Martin-Hernandez, A.M.; Camacho, A.; Prieto, J.; del Campo, A.; Tabarés, E. Isolation and characterization of TK-deficient mutants of African swine fever virus. Virus Res. 1995, 36, 67–75. [Google Scholar] [CrossRef]
- Rodríguez, J.M.; Almazan, F.; Viñuela, E.; Rodriguez, J.F. Genetic manipulation of African swine fever virus: Construction of recombinant viruses expressing the β-galactosidase gene. Virology 1992, 188, 67–76. [Google Scholar] [CrossRef]
- Cackett, G.; Matelska, D.; Sýkora, M.; Portugal, R.; Malecki, M.; Bähler, J.; Dixon, L.; Werner, F. The African Swine Fever Virus Transcriptome. J. Virol. 2020, 94, e00119-20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prichard, M.N. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev. Med. Virol. 2009, 19, 215–229. [Google Scholar] [CrossRef]
- Kanatani, Y.; Kasukabe, T.; Okabe-Kado, J.; Hayashi, S.; Yamamoto-Yamaguchi, Y.; Motoyoshi, K.; Nagata, N.; Honma, Y. Trans-forming growth factor beta and dexamethasone cooperatively enhance c-jun gene expression and inhibit the growth of human monocytoid leukemia cells. Cell Growth Differ. 1996, 7, 187–196. [Google Scholar]
- Kanatani, Y.; Kasukabe, T.; Okabe-Kado, J.; Yamamoto-Yamaguchi, Y.; Nagata, N.; Motoyoshi, K.; Honma, Y. Role of CD14 expression in the differentiation-apoptosis switch in human monocytic leukemia cells treated with 1alpha,25-dihydroxyvitamin D3 or dexamethasone in the presence of transforming growth factor beta1. Cell Growth Differ. Mol. Biol. J. Am. Assoc. Cancer Res. 1999, 10, 705–712. [Google Scholar]
- Bryan, B.A.; Dyson, O.F.; Akula, S.M. Identifying cellular genes crucial for the reactivation of Kaposi’s sarcoma-associated herpesvirus latency. J. Gen. Virol. 2006, 87, 519–529. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Avagyan, H.R.; Hakobyan, S.A.; Poghosyan, A.A.; Bayramyan, N.V.; Arzumanyan, H.H.; Abroyan, L.O.; Avetisyan, A.S.; Hakobyan, L.A.; Karalova, E.M.; Karalyan, Z.A. African Swine Fever Virus Manipulates the Cell Cycle of G0-Infected Cells to Access Cellular Nucleotides. Viruses 2022, 14, 1593. https://doi.org/10.3390/v14081593
Avagyan HR, Hakobyan SA, Poghosyan AA, Bayramyan NV, Arzumanyan HH, Abroyan LO, Avetisyan AS, Hakobyan LA, Karalova EM, Karalyan ZA. African Swine Fever Virus Manipulates the Cell Cycle of G0-Infected Cells to Access Cellular Nucleotides. Viruses. 2022; 14(8):1593. https://doi.org/10.3390/v14081593
Chicago/Turabian StyleAvagyan, Hranush R., Sona A. Hakobyan, Arpine A. Poghosyan, Nane V. Bayramyan, Hranush H. Arzumanyan, Liana O. Abroyan, Aida S. Avetisyan, Lina A. Hakobyan, Elena M. Karalova, and Zaven A. Karalyan. 2022. "African Swine Fever Virus Manipulates the Cell Cycle of G0-Infected Cells to Access Cellular Nucleotides" Viruses 14, no. 8: 1593. https://doi.org/10.3390/v14081593
APA StyleAvagyan, H. R., Hakobyan, S. A., Poghosyan, A. A., Bayramyan, N. V., Arzumanyan, H. H., Abroyan, L. O., Avetisyan, A. S., Hakobyan, L. A., Karalova, E. M., & Karalyan, Z. A. (2022). African Swine Fever Virus Manipulates the Cell Cycle of G0-Infected Cells to Access Cellular Nucleotides. Viruses, 14(8), 1593. https://doi.org/10.3390/v14081593





