Usutu Virus Infects Human Placental Explants and Induces Congenital Defects in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cell Lines
2.3. USUV Strains, Viral Stock Production and Cell Infection
2.4. Placental Histocultures
2.5. Mouse Experiments
2.6. Immunofluorescence
2.7. Intracellular Staining and Flow Cytometry Analysis
2.8. Analysis of Cell Growth and Viability
2.9. TUNEL Assay
2.10. RNA Extraction
2.11. RT-qPCR Analysis
2.12. RT2 Profiler PCR Arrays
2.13. Histology and Immunohistochemistry
2.14. Measurement of Viral Burden In Vivo
2.15. Statistical Analyses
3. Results
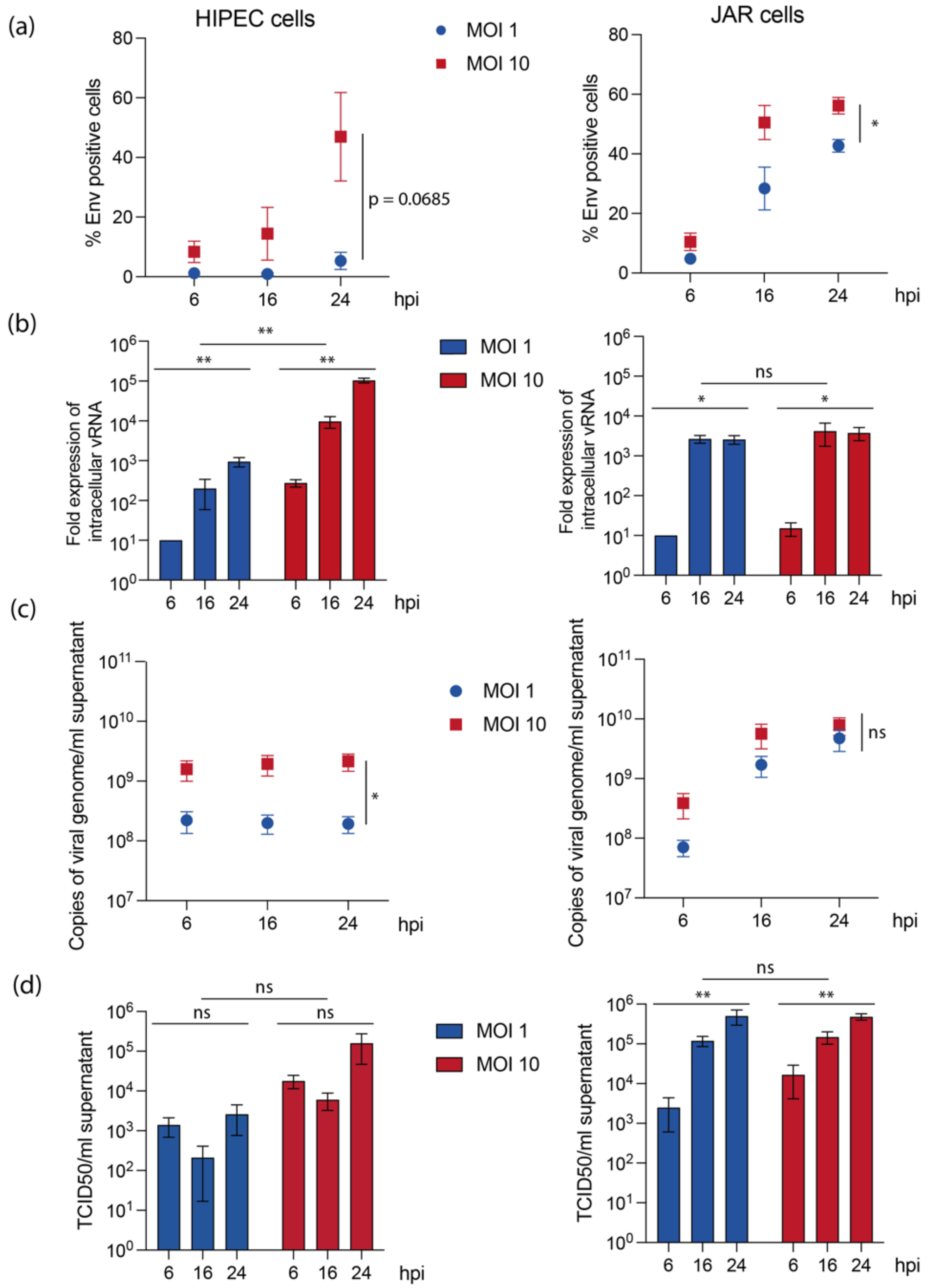
3.1. Human Placental Cell Lines Exhibit Variable Permissiveness to USUV Replication
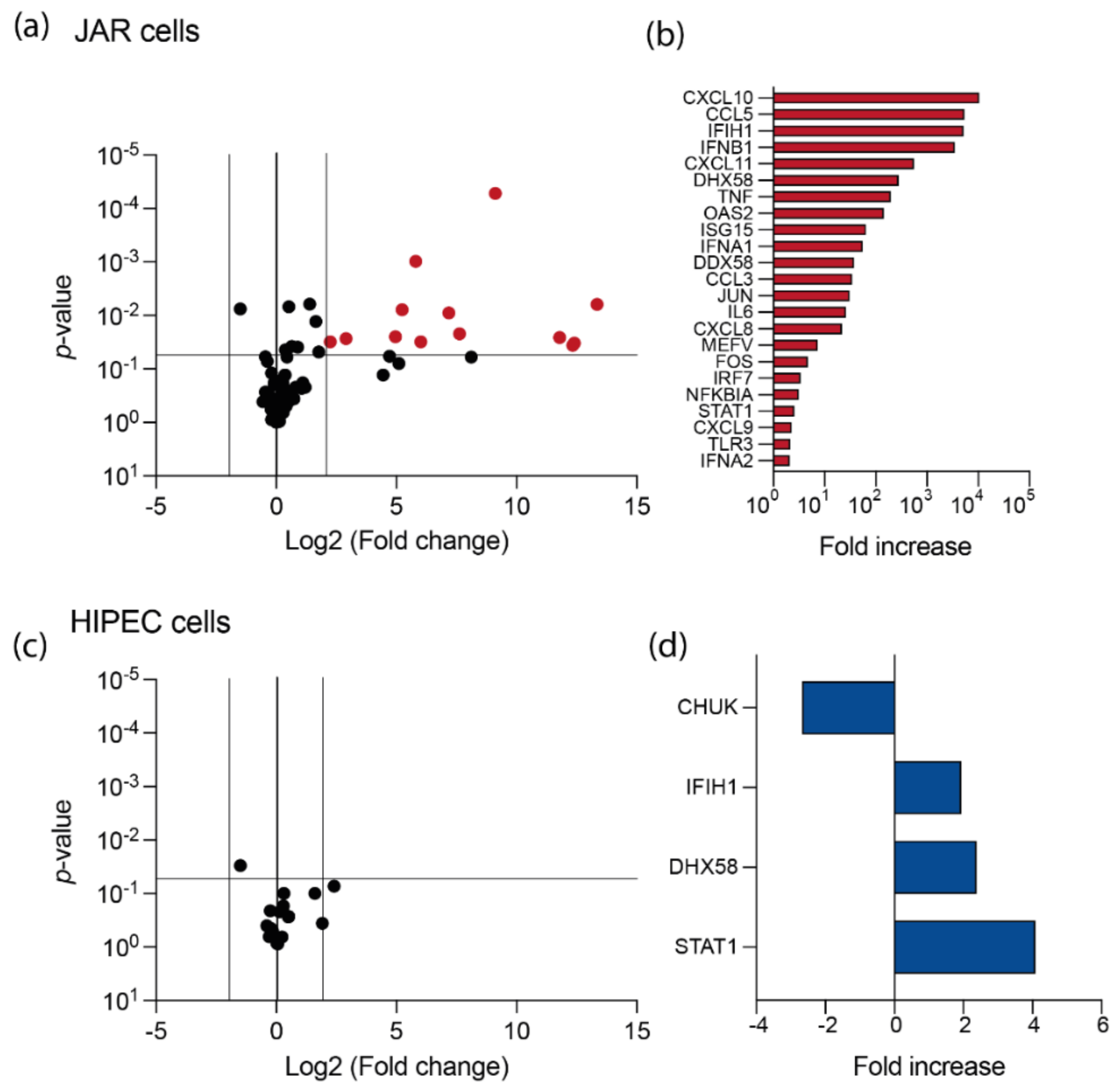
3.2. USUV Elicits a Strong Antiviral Response in JAR Cell Line
3.3. Human Placental Tissues Are Permissive to USUV Replication
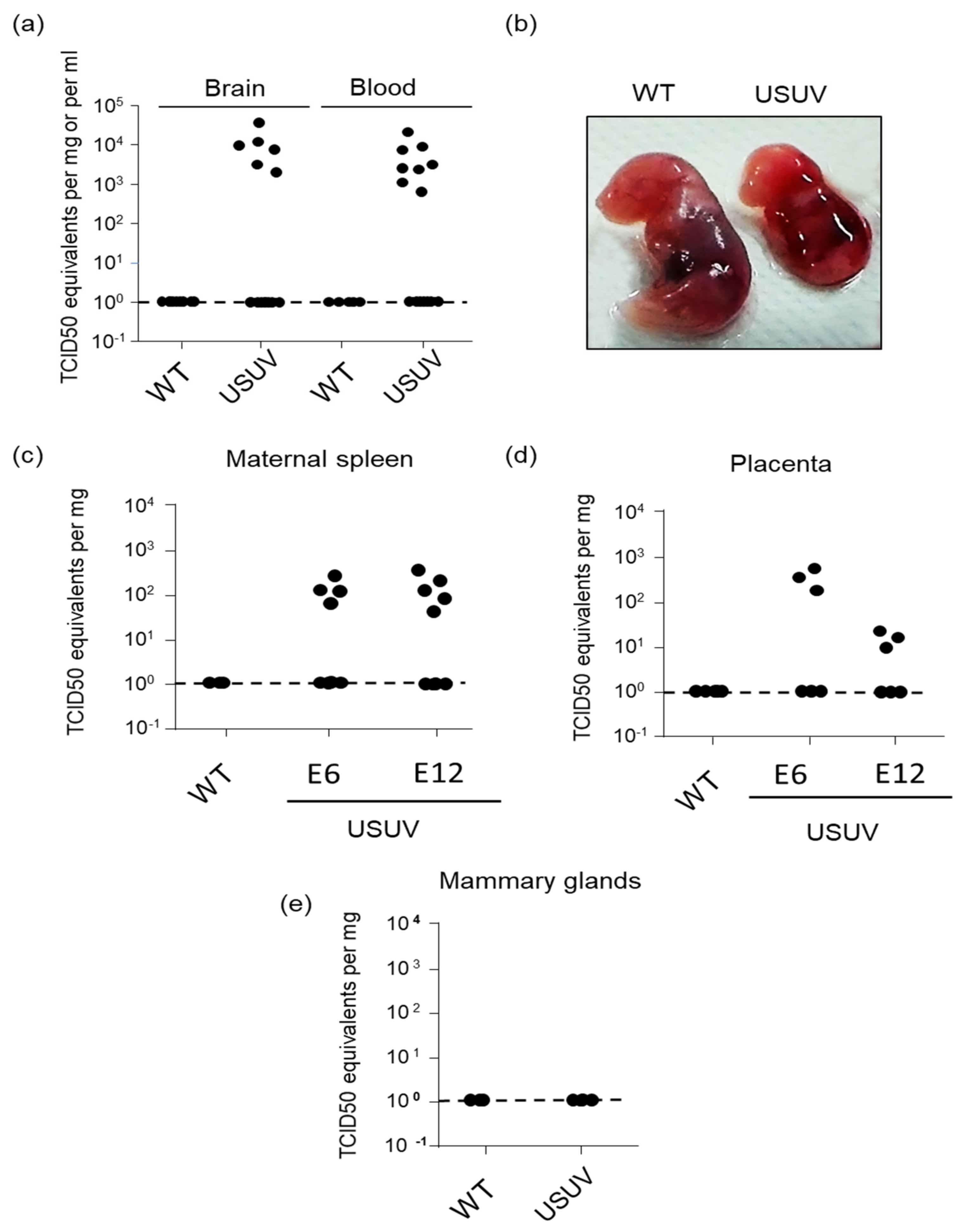
3.4. USUV Can Achieve Congenital Infection in Immunocompetent Mice and Causes Occasional Fetal Demise
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weissenböck, H.; Kolodziejek, J.; Url, A.; Lussy, H.; Rebel-Bauder, B.; Nowotny, N. Emergence of Usutu virus, an African mosquito-borne Flavivirus of the Japanese encephalitis virus group, central Europe. Emerg. Infect. Dis. 2002, 8, 652. [Google Scholar] [CrossRef] [PubMed]
- Bakonyi, T.; Gould, E.A.; Kolodziejek, J.; Weissenböck, H.; Nowotny, N. Complete genome analysis and molecular characterization of Usutu virus that emerged in Austria in 2001: Comparison with the South African Strain SAAR-1776 and other flaviviruses. Virology 2004, 328, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Cadar, D.; Lühken, R.; van der Jeugd, H.; Garigliany, M.; Ziegler, U.; Keller, M.; Lahoreau, J.; Lachmann, L.; Becker, N.; Kik, M.; et al. Widespread activity of multiple lineages of Usutu virus, Western Europe, 2016. Eurosurveillance 2017, 22, 30452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McIntosh BM Usutu (SAAr 1776); nouvel arbovirus du groupe B. Int Cat Arboviruses 1985, 3, 1059–1060.
- Woodall, J. The viruses isolated from arthropods at the East African Virus Research Institute in the 26 years ending December 1963. Proc. E Afr. Acad. 1964, 2, 141–146. [Google Scholar]
- Calisher, C.H.; Gould, E.A. Taxonomy of the virus family Flaviviridae. Adv. Virus Res. 2003, 59, 1–19. [Google Scholar] [CrossRef]
- Poidinger, M.; Hall, R.A.; Mackenzie, J.S. Molecular characterization of the Japanese encephalitis serocomplex of the flavivirus genus. Virology 1996, 218, 417–421. [Google Scholar] [CrossRef]
- Clé, M.; Beck, C.; Salinas, S.; Lecollinet, S.; Gutierrez, S.; Van de Perre, P.; Baldet, T.; Foulongne, V.; Simonin, Y. Usutu virus: A new threat? Epidemiol. Infect. 2019, 147, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Nikolay, B.; Diallo, M.; Boye, C.S.B.; Sall, A.A. Usutu Virus in Africa. Vector-Borne Zoonotic Dis. 2011, 11, 1417–1423. [Google Scholar] [CrossRef]
- Pecorari, M.; Longo, G.; Gennari, W.; Grottola, A.; Sabbatini, A.M.T.; Tagliazucchi, S. First human case of usutu virus neuro invasive infection, Italy, August-September 2009. Eurosurveillance 2009, 14, 19446. [Google Scholar] [CrossRef]
- Cavrini, F.; Gaibani, P.; Longo, G.; Pierro, A.M.; Rossini, G.; Bonilauri, P.; Gerundi, G.E.; Di Benedetto, F.; Pasetto, A.; Girardis, M.; et al. Usutu virus infection in a patient who underwent orthotropic liver transplantation, Italy, August-September 2009. Euro Surveill. 2009, 14, 19448. [Google Scholar] [CrossRef]
- Vilibic-Cavlek, T.; Savic, V.; Sabadi, D.; Peric, L.; Barbic, L.; Klobucar, A.; Miklausic, B.; Tabain, I.; Santini, M.; Vucelja, M.; et al. Prevalence and molecular epidemiology of West Nile and Usutu virus infections in Croatia in the “One health” context, 2018. Transbound. Emerg. Dis. 2019, 66, 1946–1957. [Google Scholar] [CrossRef]
- Simonin, Y.; Sillam, O.; Carles, M.J.; Gutierrez, S.; Gil, P.; Constant, O. Human Usutu virus infection with atypical neurologic presentation, Montpellier, France, 2016. Emerg. Infect. Dis. 2018, 24, 875. [Google Scholar] [CrossRef] [Green Version]
- Carletti, F.; Colavita, F.; Rovida, F.; Percivalle, E.; Baldanti, F.; Ricci, I.; De Liberato, C.; Rosone, F.; Messina, F.; Lalle, E.; et al. Expanding Usutu virus circulation in Italy: Detection in the Lazio region, central Italy, 2017 to 2018. Euro Surveill. 2019, 24, 1800649. [Google Scholar] [CrossRef]
- Gaibani, P.; Cavrini, F.; Gould, E.A.; Rossini, G.; Pierro, A.; Landini, M.P.; Sambri, V. Comparative Genomic and Phylogenetic Analysis of the First Usutu Virus Isolate from a Human Patient Presenting with Neurological Symptoms. PLoS ONE 2013, 8, e64761. [Google Scholar] [CrossRef] [Green Version]
- Cavrini, F.; Pepa, M.E.D.; Gaibani, P.; Pierro, A.M.; Rossini, G.; Landini, M.P.; Sambri, V. A rapid and specific real-time RT-PCR assay to identify Usutu virus in human plasma, serum, and cerebrospinal fluid. J. Clin. Virol. 2011, 50, 221–223. [Google Scholar] [CrossRef]
- Grottola, A.; Marcacci, M.; Tagliazucchi, S.; Gennari, W.; Di Gennaro, A.; Orsini, M.; Monaco, F.; Marchegiano, P.; Marini, V.; Meacci, M.; et al. Usutu virus infections in humans: A retrospective analysis in the municipality of Modena, Italy. Clin. Microbiol. Infect. 2017, 23, 33–37. [Google Scholar] [CrossRef] [Green Version]
- Caracciolo, I.; Mora-Cardenas, E.; Aloise, C.; Carletti, T.; Segat, L.; Burali, M.S.; Chiarvesio, A.; Totis, V.; Avšič-županc, T.; Mastrangelo, E.; et al. Comprehensive response to Usutu virus following first isolation in blood donors in the Friuli Venezia Giulia region of Italy: Development of recombinant NS1-based serology and sensitivity to antiviral drugs. PLoS Negl. Trop. Dis. 2020, 14, e0008156. [Google Scholar] [CrossRef]
- Percivalle, E.; Cassaniti, I.; Sarasini, A.; Rovida, F.; Adzasehoun, K.M.G.; Colombini, I.; Isernia, P.; Cuppari, I.; Baldanti, F. West Nile or Usutu Virus? A Three-Year Follow-Up of Humoral and Cellular Response in a Group of Asymptomatic Blood Donors. Viruses 2020, 12, 157. [Google Scholar] [CrossRef] [Green Version]
- Pacenti, M.; Sinigaglia, A.; Martello, T.; de Rui, M.E.; Franchin, E.; Pagni, S.; Peta, E.; Riccetti, S.; Milani, A.; Montarsi, F.; et al. Clinical and virological findings in patients with Usutu virus infection, northern Italy, 2018. Euro Surveill. 2019, 24, 1900180. [Google Scholar] [CrossRef]
- Nagy, A.; Mezei, E.; Nagy, O.; Bakonyi, T.; Csonka, N.; Kaposi, M.; Koroknai, A.; Szomor, K.; Rigó, Z.; Molnár, Z.; et al. Extraordinary increase in West Nile virus cases and first confirmed human Usutu virus infection in Hungary, 2018. Euro Surveill. 2019, 24, 1900038. [Google Scholar] [CrossRef] [Green Version]
- Santini, M.; Vilibic-Cavlek, T.; Barsic, B.; Barbic, L.; Savic, V.; Stevanovic, V.; Listes, E.; Di Gennaro, A.; Savini, G. First cases of human Usutu virus neuroinvasive infection in Croatia, August, September 2013: Clinical and laboratory features. J. Neurovirol. 2014, 21, 92–97. [Google Scholar] [CrossRef]
- Platt, D.J.; Smith, A.M.; Arora, N.; Diamond, M.S.; Coyne, C.B.; Miner, J.J. Zika virus-related neurotropic flaviviruses infect human placental explants and cause fetal demise in mice. Sci. Transl. Med. 2018, 10, eaao7090. [Google Scholar] [CrossRef] [Green Version]
- Blázquez, A.B.; Sáiz, J.C. West Nile virus (WNV) transmission routes in the murine model: Intrauterine, by breastfeeding and after cannibal ingestion. Virus Res. 2010, 151, 240–243. [Google Scholar] [CrossRef]
- Zanluca, C.; de Noronha, L.; Duarte dos Santos, C.N. Maternal-fetal transmission of the zika virus: An intriguing interplay. Tissue Barriers 2018, 6, e1402143. [Google Scholar] [CrossRef] [Green Version]
- del Campo, M.; Feitosa, I.M.L.; Ribeiro, E.M.; Horovitz, D.D.G.; Pessoa, A.L.S.; França, G.V.A.; García-Alix, A.; Doriqui, M.J.R.; Wanderley, H.Y.C.; Sanseverino, M.V.T.; et al. The phenotypic spectrum of congenital Zika syndrome. Am. J. Med. Genet. A 2017, 173, 841–857. [Google Scholar] [CrossRef] [Green Version]
- Microcephaly in Infants, Pernambuco State, Brazil, 2015. Emerg. Infect. Dis. 2016, 22, 1090–1093. [CrossRef]
- Cao, B.; Diamond, M.S.; Mysorekar, I.U. Maternal-Fetal Transmission of Zika Virus: Routes and Signals for Infection. J. Interferon Cytokine Res. 2017, 37, 287–294. [Google Scholar] [CrossRef] [Green Version]
- Quicke, K.M.; Bowen, J.R.; Johnson, E.L.; McDonald, C.E.; Ma, H.; O’Neal, J.T.; Rajakumar, A.; Wrammert, J.; Rimawi, B.H.; Pulendran, B.; et al. Zika Virus Infects Human Placental Macrophages. Cell Host Microbe 2016, 20, 83–90. [Google Scholar] [CrossRef] [Green Version]
- Weisblum, Y.; Oiknine-Djian, E.; Vorontsov, O.M.; Haimov-Kochman, R.; Zakay-Rones, Z.; Meir, K.; Shveiky, D.; Elgavish, S.; Nevo, Y.; Roseman, M.; et al. Zika Virus Infects Early- and Midgestation Human Maternal Decidual Tissues, Inducing Distinct Innate Tissue Responses in the Maternal-Fetal Interface. J. Virol. 2019, 93, e01451-19. [Google Scholar] [CrossRef] [Green Version]
- Tabata, T.; Petitt, M.; Puerta-Guardo, H.; Michlmayr, D.; Harris, E.; Pereira, L. Zika Virus Replicates in Proliferating Cells in Explants From First-Trimester Human Placentas, Potential Sites for Dissemination of Infection. J. Infect. Dis. 2018, 217, 1202–1213. [Google Scholar] [CrossRef] [Green Version]
- Tabata, T.; Petitt, M.; Puerta-Guardo, H.; Michlmayr, D.; Wang, C.; Fang-Hoover, J.; Harris, E.; Pereira, L. Zika Virus Targets Different Primary Human Placental Cells, Suggesting Two Routes for Vertical Transmission. Cell Host Microbe 2016, 20, 155–166. [Google Scholar] [CrossRef] [Green Version]
- Guzeloglu-Kayisli, O.; Guo, X.; Tang, Z.; Semerci, N.; Ozmen, A.; Larsen, K.; Mutluay, D.; Guller, S.; Schatz, F.; Kayisli, U.A.; et al. Zika Virus-Infected Decidual Cells Elicit a Gestational Age-Dependent Innate Immune Response and Exaggerate Trophoblast Zika Permissiveness: Implication for Vertical Transmission. J. Immunol. 2020, 205, 3083–3094. [Google Scholar] [CrossRef]
- Chen, J.; Liang, Y.; Yi, P.; Xu, L.; Hawkins, H.K.; Rossi, S.L.; Soong, L.; Cai, J.; Menon, R.; Sun, J. Outcomes of Congenital Zika Disease Depend on Timing of Infection and Maternal-Fetal Interferon Action. Cell Rep. 2017, 21, 1588–1599. [Google Scholar] [CrossRef] [Green Version]
- Gurung, S.; Reuter, N.; Preno, A.; Dubaut, J.; Nadeau, H.; Hyatt, K.; Singleton, K.; Martin, A.; Parks, W.T.; Papin, J.F.; et al. Zika virus infection at mid-gestation results in fetal cerebral cortical injury and fetal death in the olive baboon. PLoS Pathog. 2019, 15, e1007507. [Google Scholar] [CrossRef] [Green Version]
- Jagger, B.W.; Miner, J.J.; Cao, B.; Arora, N.; Smith, A.M.; Kovacs, A.; Mysorekar, I.U.; Coyne, C.B.; Diamond, M.S. Gestational Stage and IFN-λ Signaling Regulate ZIKV Infection In Utero. Cell Host Microbe 2017, 22, 366–376. [Google Scholar] [CrossRef] [Green Version]
- Pavan, L.; Tarrade, A.; Hermouet, A.; Delouis, C.; Titeux, M.; Vidaud, M.; Thérond, P.; Evain-Brion, D.; Fournier, T. Human invasive trophoblasts transformed with simian virus 40 provide a new tool to study the role of PPARgamma in cell invasion process. Carcinogenesis 2003, 24, 1325–1336. [Google Scholar] [CrossRef] [Green Version]
- Kärber, G. Beitrag zur kollektiven Behandlung pharmakologischer Reihenversuche. Naunyn. Schmiedebergs. Arch. Exp. Pathol. Pharmakol. 1931, 162, 480–483. [Google Scholar] [CrossRef]
- Bergamelli, M.; Martin, H.; Bénard, M.; Ausseil, J.; Mansuy, J.M.; Hurbain, I.; Mouysset, M.; Groussolles, M.; Cartron, G.; Tanguy le Gac, Y.; et al. Human Cytomegalovirus Infection Changes the Pattern of Surface Markers of Small Extracellular Vesicles Isolated From First Trimester Placental Long-Term Histocultures. Front. Cell Dev. Biol. 2021, 9, 2281. [Google Scholar] [CrossRef]
- Hamel, R.; Dejarnac, O.; Wichit, S.; Ekchariyawat, P.; Neyret, A.; Natthanej, L.; Perera-Lecoin, M.; Surasombatpattana, P.; Talignani, L.; Thomas, F.; et al. Biology of Zika Virus Infection in Human Skin Cells. J. Virol. 2015, 17, 8880–8896. [Google Scholar] [CrossRef] [Green Version]
- Nikolay, B.; Weidmann, M.; Dupressoir, A.; Faye, O.; Boye, C.S.; Diallo, M.; Sall, A.A. Development of a Usutu virus specific real-time reverse transcription PCR assay based on sequenced strains from Africa and Europe. J. Virol. Methods 2014, 197, 51–54. [Google Scholar] [CrossRef] [Green Version]
- Ruddon, R.W.; Hanson, C.A.; Bryan, A.H.; Putterman, G.J.; White, E.L.; Perini, F.; Meade, K.S.; Aldenderfer, P.H. Synthesis and secretion of human chorionic gonadotropin subunits by cultured human malignant cells. J. Biol. Chem. 1980, 255, 1000–1007. [Google Scholar] [CrossRef]
- Grümmer, R.; Hohn, H.P.; Mareel, M.M.; Denker, H.W. Adhesion and invasion of three human choriocarcinoma cell lines into human endometrium in a three-dimensional organ culture system. Placenta 1994, 15, 411–429. [Google Scholar] [CrossRef]
- Lopez, H.; Benard, M.; Saint-Aubert, E.; Baron, M.; Martin, H.; Al Saati, T.; Plantavid, M.; Duga-Neulat, I.; Berrebi, A.; Cristini, C.; et al. Novel model of placental tissue explants infected by cytomegalovirus reveals different permissiveness in early and term placentae and inhibition of indoleamine 2,3-dioxygenase activity. Placenta 2011, 32, 522–530. [Google Scholar] [CrossRef]
- Miner, J.J.; Cao, B.; Govero, J.; Smith, A.M.; Fernandez, E.; Cabrera, O.H.; Garber, C.; Noll, M.; Klein, R.S.; Noguchi, K.K.; et al. Zika Virus Infection during Pregnancy in Mice Causes Placental Damage and Fetal Demise. Cell 2016, 165, 1081–1091. [Google Scholar] [CrossRef] [Green Version]
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B.; Freitas, B.C.; Dias, J.L.M.; Guimarães, K.P.; Benazzato, C.; Almeida, N.; Pignatari, G.C.; Romero, S.; et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef] [Green Version]
- O’Leary, D.R.; Kuhn, S.; Kniss, K.L.; Hinckley, A.F.; Rasmussen, S.A.; Pape, W.J.; Kightlinger, L.K.; Beecham, B.D.; Miller, T.K.; Neitzel, D.F.; et al. Birth outcomes following West Nile Virus infection of pregnant women in the United States: 2003–2004. Pediatrics 2006, 117, e537–e545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Intrauterine West Nile Virus Infection—New York, 2002. JAMA 2003, 289, 295. [CrossRef] [Green Version]
- Chaturvedi, U.C.; Mathur, A.; Chandra, A.; Das, S.K.; Tandon, H.O.; Singh, U.K. Transplacental infection with Japanese encephalitis virus. J. Infect. Dis. 1980, 141, 712–715. [Google Scholar] [CrossRef]
- Li, C.; Xu, D.; Ye, Q.; Hong, S.; Jiang, Y.; Liu, X.; Zhang, N.; Shi, L.; Qin, C.F.; Xu, Z. Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell 2016, 19, 672. [Google Scholar] [CrossRef] [Green Version]
- Fujisaki, Y.; Miura, Y.; Sugimori, T.; Murakami, Y.; Miura, K. Experimental studies on vertical infection of mice with Japanese encephalitis virus. IV. Effect of virus strain on placental and fetal infection. Natl. Inst. Anim. Health Q. 1983, 23, 21–26. [Google Scholar]
- Mathur, A.; Arora, K.L.; Chaturvedi, U.C. Congenital infection of mice with Japanese encephalitis virus. Infect. Immun. 1981, 34, 26–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burns, K.F. Congenital Japanese B encephalitis infection of swine. Proc. Soc. Exp. Biol. Med. 1950, 75, 621–625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, A.A.; Hanson, R.P. Intrauterine infection of mice with St. Louis encephalitis virus: Immunological, physiological, neurological, and behavioral effects on progeny. Infect. Immun. 1975, 12, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Andersen, A.A.; Hanson, R.P. Experimental transplacental transmission of st. Louis encephalitis virus in mice. Infect. Immun. 1970, 2, 320–325. [Google Scholar] [CrossRef] [Green Version]
- Julander, J.G.; Winger, Q.A.; Olsen, A.L.; Day, C.W.; Sidwell, R.W.; Morrey, J.D. Treatment of West Nile virus-infected mice with reactive immunoglobulin reduces fetal titers and increases dam survival. Antivir. Res. 2005, 65, 79–85. [Google Scholar] [CrossRef]
- Julander, J.G.; Winger, Q.A.; Rickords, L.F.; Shi, P.Y.; Tilgner, M.; Binduga-Gajewska, I.; Sidwell, R.W.; Morrey, J.D. West Nile virus infection of the placenta. Virology 2006, 347, 175–182. [Google Scholar] [CrossRef] [Green Version]
- Rothbauer, M.; Patel, N.; Gondola, H.; Siwetz, M.; Huppertz, B.; Ertl, P. A comparative study of five physiological key parameters between four different human trophoblast-derived cell lines. Sci. Rep. 2017, 7, 5892. [Google Scholar] [CrossRef]
- Lee, C.Q.E.; Gardner, L.; Turco, M.; Zhao, N.; Murray, M.J.; Coleman, N.; Rossant, J.; Hemberger, M.; Moffett, A. What Is Trophoblast? A Combination of Criteria Define Human First-Trimester Trophoblast. Stem Cell Rep. 2016, 6, 257–272. [Google Scholar] [CrossRef] [Green Version]
- Salinas, S.; Constant, O.; Desmetz, C.; Barthelemy, J.; Lemaitre, J.-M.; Milhavet, O.; Nagot, N.; Foulongne, V.; Perrin, F.E.; Saiz, J.-C.; et al. Deleterious effect of Usutu virus on human neural cells. PLoS Negl. Trop. Dis. 2017, 11, e0005913. [Google Scholar] [CrossRef]
- El Costa, H.; Gouilly, J.; Mansuy, J.M.; Chen, Q.; Levy, C.; Cartron, G.; Veas, F.; Al-Daccak, R.; Izopet, J.; Jabrane-Ferrat, N. ZIKA virus reveals broad tissue and cell tropism during the first trimester of pregnancy. Sci. Rep. 2016, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pierson, T.C.; Diamond, M.S. The emergence of Zika virus and its new clinical syndromes. Nature 2018, 560, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Honein, M.A.; Dawson, A.L.; Petersen, E.E.; Jones, A.M.; Lee, E.H.; Yazdy, M.M.; Ahmad, N.; Macdonald, J.; Evert, N.; Bingham, A.; et al. Birth Defects Among Fetuses and Infants of US Women With Evidence of Possible Zika Virus Infection During Pregnancy. JAMA 2017, 317, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Arora, N.; Sadovsky, Y.; Dermody, T.S.; Coyne, C.B. Microbial Vertical Transmission during Human Pregnancy. Cell Host Microbe 2017, 21, 561–567. [Google Scholar] [CrossRef] [PubMed]
- Bayer, A.; Lennemann, N.J.; Ouyang, Y.; Bramley, J.C.; Morosky, S.; Marques, E.T.D.A.; Cherry, S.; Sadovsky, Y.; Coyne, C.B. Type III Interferons Produced by Human Placental Trophoblasts Confer Protection against Zika Virus Infection. Cell Host Microbe 2016, 19, 705–712. [Google Scholar] [CrossRef] [Green Version]
- Szaba, F.M.; Tighe, M.; Kummer, L.W.; Lanzer, K.G.; Ward, J.M.; Lanthier, P.; Kim, I.J.; Kuki, A.; Blackman, M.A.; Thomas, S.J.; et al. Zika virus infection in immunocompetent pregnant mice causes fetal damage and placental pathology in the absence of fetal infection. PLoS Pathog. 2018, 14, e1006994. [Google Scholar] [CrossRef]
- Bayer, A.; Lennemann, N.J.; Ouyang, Y.; Sadovsky, E.; Sheridan, M.A.; Roberts, R.M.; Coyne, C.B.; Sadovsky, Y. Chromosome 19 microRNAs exert antiviral activity independent from type III interferon signaling. Placenta 2018, 61, 33–38. [Google Scholar] [CrossRef]
- Ouyang, Y.; Bayer, A.; Chu, T.; Tyurin, V.; Kagan, V.; Morelli, A.E.; Coyne, C.B.; Sadovsky, Y. Isolation of human trophoblastic extracellular vesicles and characterization of their cargo and antiviral activity. Placenta 2016, 47, 86–95. [Google Scholar] [CrossRef] [Green Version]
- Malnou, E.C.; Umlauf, D.; Mouysset, M.; Cavaillé, J. Imprinted MicroRNA Gene Clusters in the Evolution, Development, and Functions of Mammalian Placenta. Front. Genet. 2019, 9, 706. [Google Scholar] [CrossRef]


| Week of Pregnancy | Type of Birth | % of RT-PCR Positive Brain | % of RT-PCR Positive Blood | % of Death/Birth Defect |
|---|---|---|---|---|
| First | Natural delivery | 12% | 16% (2 weeks after delivery) | 15% (11/73) * |
| Cesarean (2° week) | 6% | ND | 3% (2/52) # | |
| Second | Natural delivery | 0% | 0% | 0% (0/41) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martin, H.; Barthelemy, J.; Chin, Y.; Bergamelli, M.; Moinard, N.; Cartron, G.; Tanguy Le Gac, Y.; Malnou, C.E.; Simonin, Y. Usutu Virus Infects Human Placental Explants and Induces Congenital Defects in Mice. Viruses 2022, 14, 1619. https://doi.org/10.3390/v14081619
Martin H, Barthelemy J, Chin Y, Bergamelli M, Moinard N, Cartron G, Tanguy Le Gac Y, Malnou CE, Simonin Y. Usutu Virus Infects Human Placental Explants and Induces Congenital Defects in Mice. Viruses. 2022; 14(8):1619. https://doi.org/10.3390/v14081619
Chicago/Turabian StyleMartin, Hélène, Jonathan Barthelemy, Yamileth Chin, Mathilde Bergamelli, Nathalie Moinard, Géraldine Cartron, Yann Tanguy Le Gac, Cécile E. Malnou, and Yannick Simonin. 2022. "Usutu Virus Infects Human Placental Explants and Induces Congenital Defects in Mice" Viruses 14, no. 8: 1619. https://doi.org/10.3390/v14081619
APA StyleMartin, H., Barthelemy, J., Chin, Y., Bergamelli, M., Moinard, N., Cartron, G., Tanguy Le Gac, Y., Malnou, C. E., & Simonin, Y. (2022). Usutu Virus Infects Human Placental Explants and Induces Congenital Defects in Mice. Viruses, 14(8), 1619. https://doi.org/10.3390/v14081619







