SERINC5-Mediated Restriction of HIV-1 Infectivity Correlates with Resistance to Cholesterol Extraction but Not with Lipid Order of Viral Membrane
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids, Cells, Lipids, and Chemicals
2.2. Virus Production and Purification
2.3. Large Unilamellar Vesicles Preparation
2.4. Lipid Probe Staining
2.5. Infectivity Measurements of Pseudoviruses Subjected to Lipid Manipulation Treatments
2.6. Lipid Order Imaging and Analysis
2.7. Immunofluorescence Imaging and Analysis
2.8. Statistical Analysis
3. Results
3.1. Laurdan and Nile Red Are Reliable Probes for Measuring Lipid Order of Large Unilamellar Vesicles
3.2. Lipophilic Probes Reveal a Highly Ordered Phase Structure in HIV-1 Pseudovirus Membranes
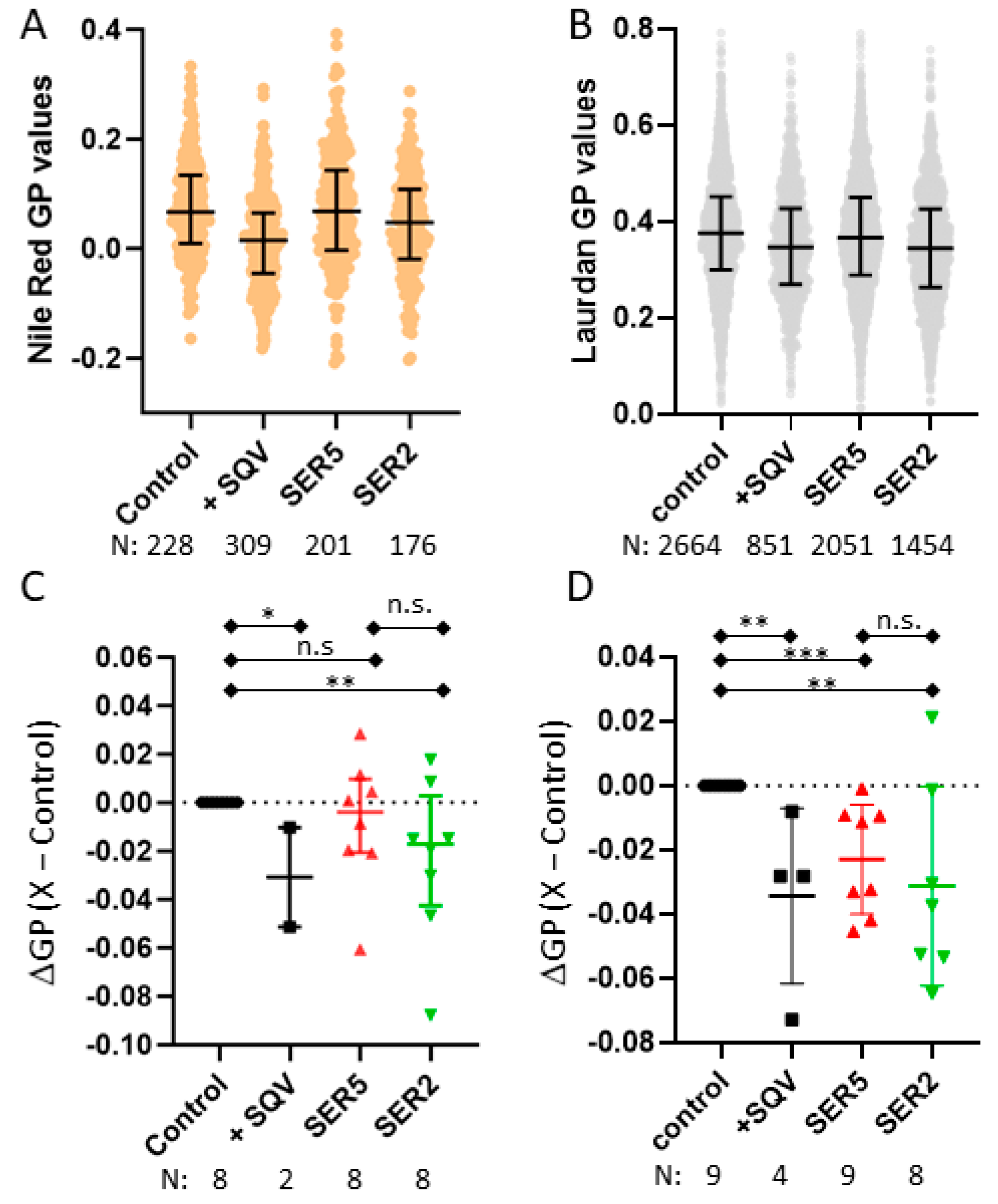
3.3. SER5 Incorporation Has Minimal Impact on the Global Lipid Order of the Viral Membrane
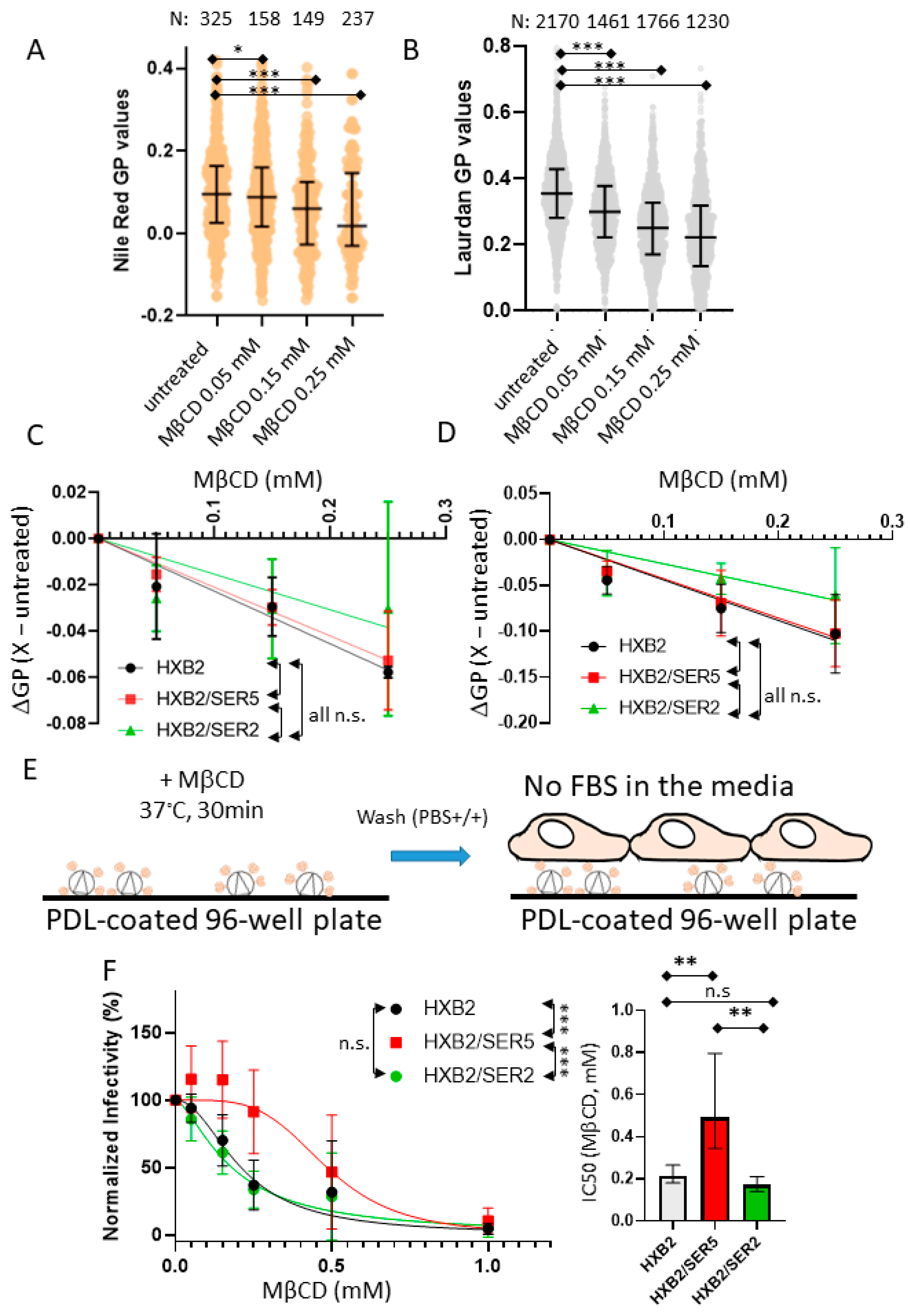
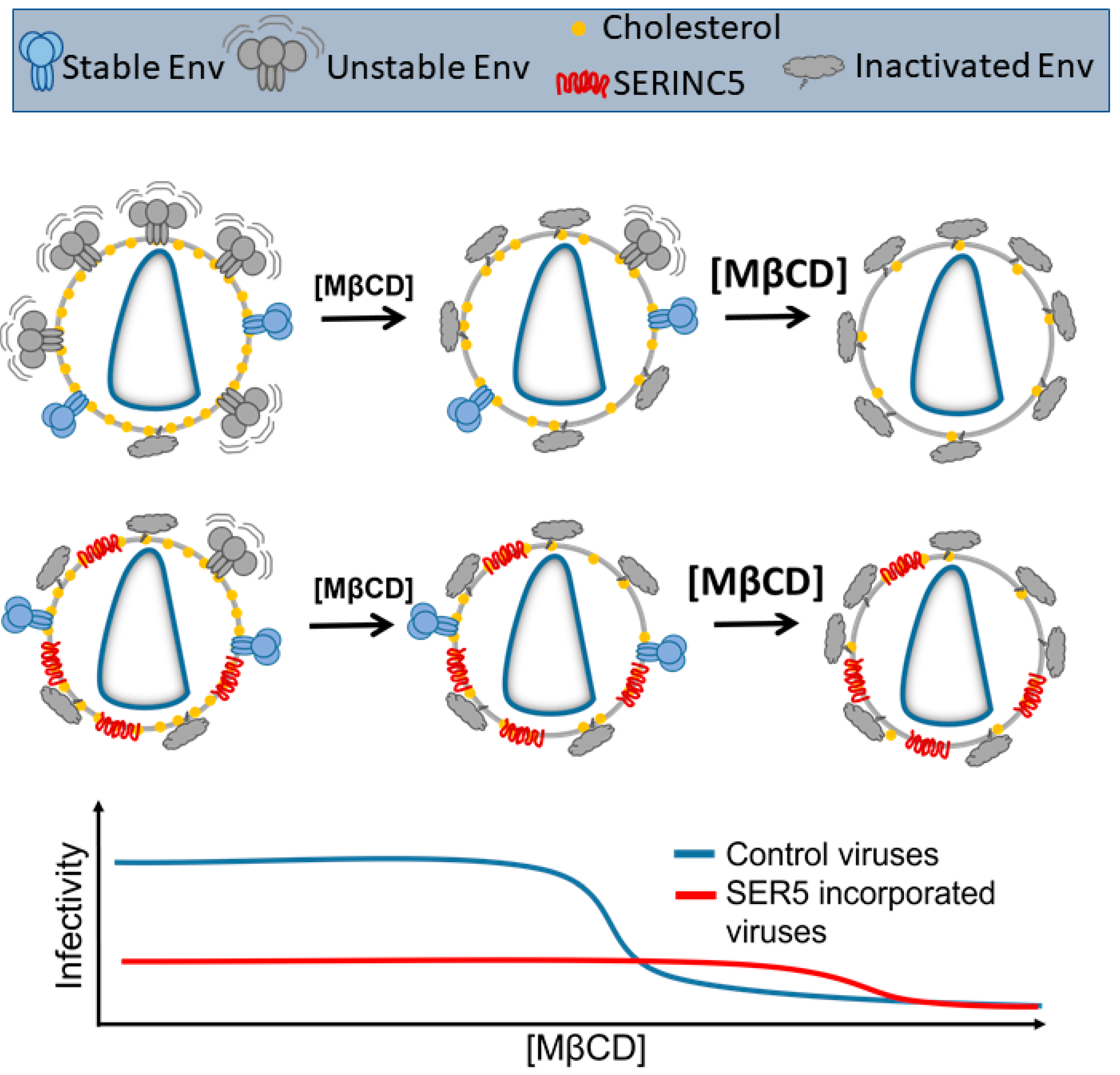
3.4. SER5 Incorporation Renders Pseudovirus Infectivity Less Sensitive to Cholesterol Extraction by MβCD
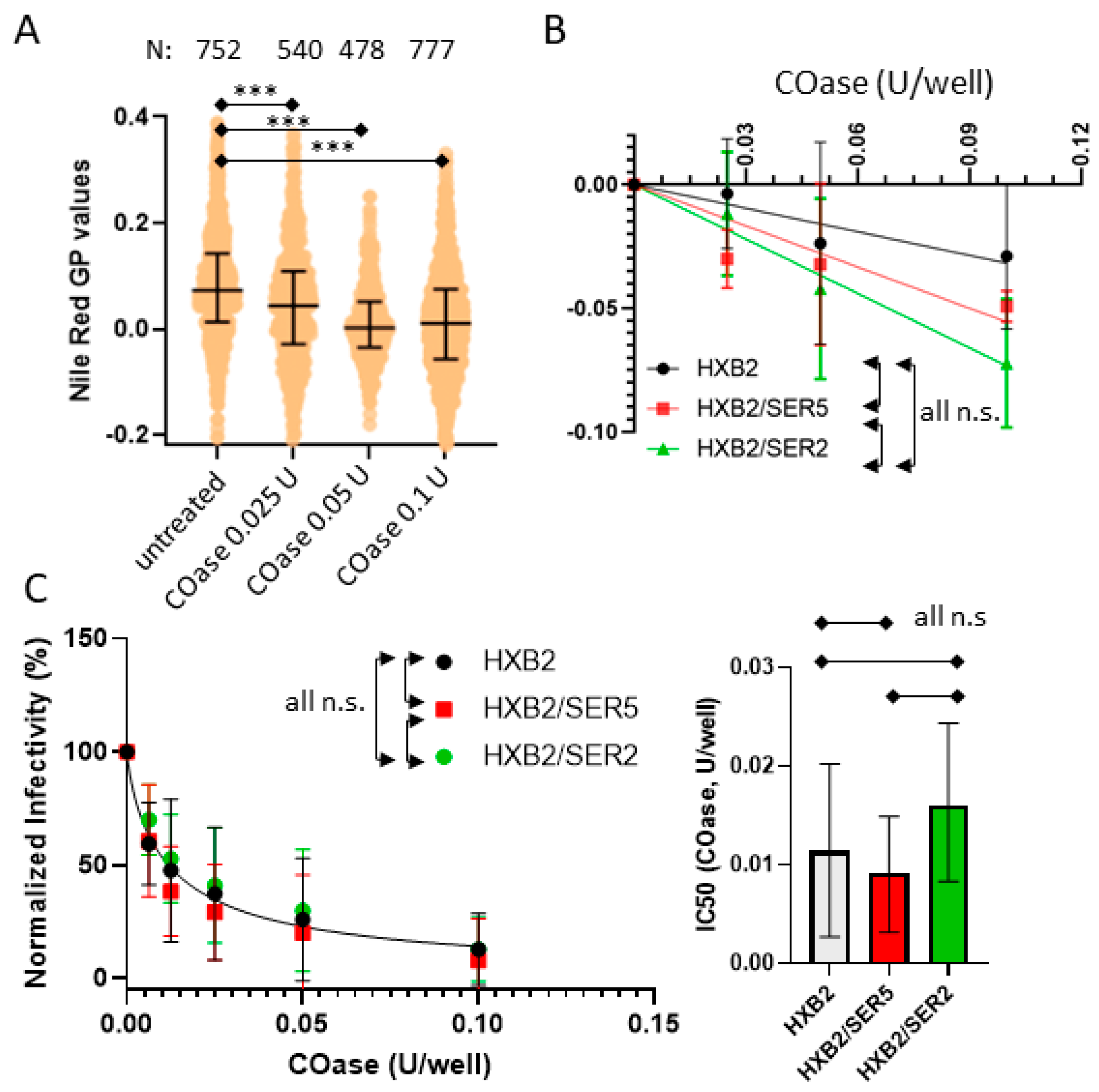
3.5. SER5 Incorporation Does Not Affect Pseudovirus Sensitivity to Cholesterol Oxidation
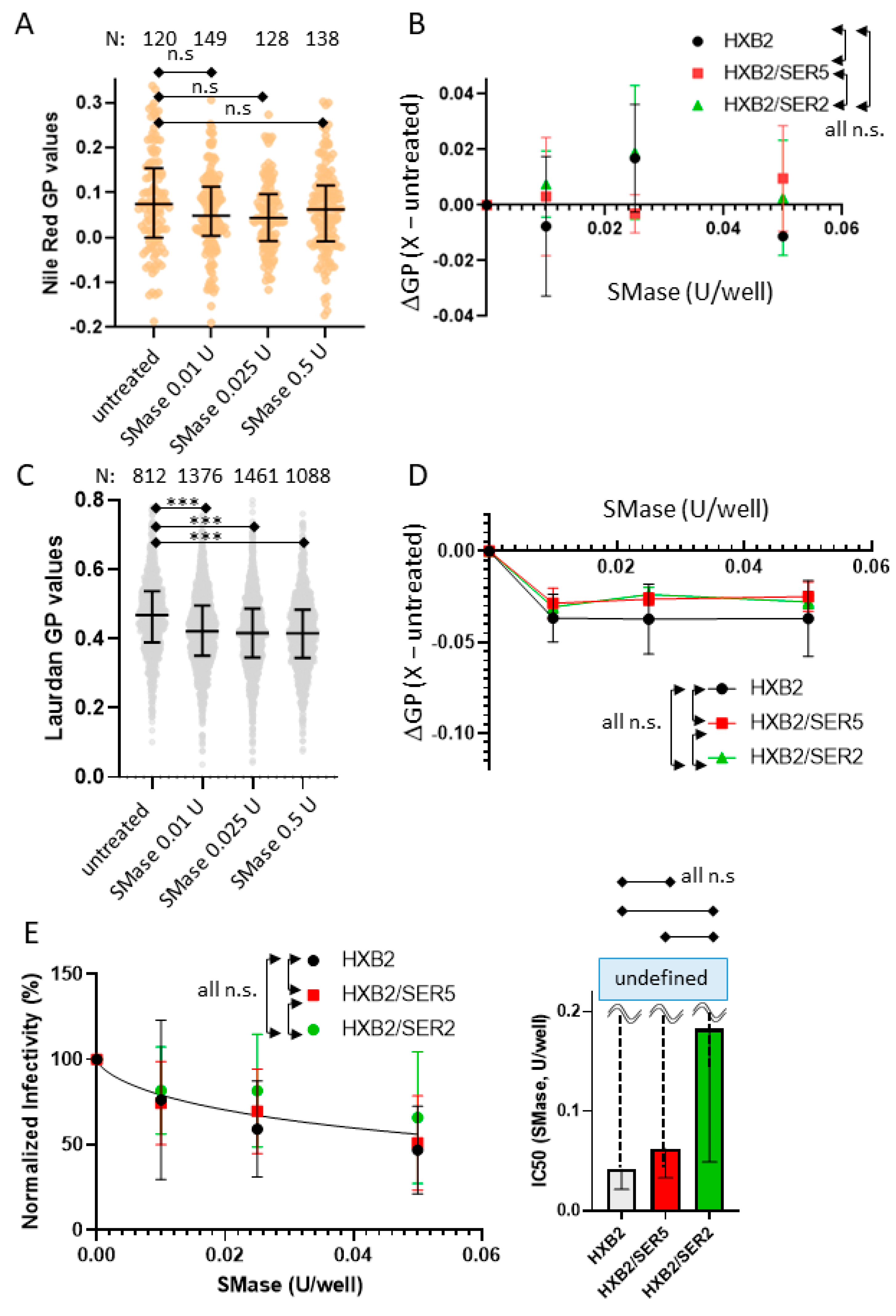
3.6. SER5 Incorporation Does Not Affect Pseudovirus Sensitivity to Sphingomyelinase Treatment
3.7. SER5-Containing Viruses Retain Resistance to Cholesterol Extraction despite Sphingomyelinase Treatment
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Freed, E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015, 13, 484–496. [Google Scholar] [CrossRef]
- Wilen, C.B.; Tilton, J.C.; Doms, R.W. HIV: Cell binding and entry. Cold Spring Harb. Perspect. Med. 2012, 2, a006866. [Google Scholar] [CrossRef]
- McManus, C.M.; Doms, R.W. Fusion mediated by the HIV-1 envelope protein. Subcell Biochem. 2000, 34, 457–481. [Google Scholar]
- Checkley, M.A.; Luttge, B.G.; Freed, E.O. HIV-1 envelope glycoprotein biosynthesis, trafficking, and incorporation. J. Mol. Biol. 2011, 410, 582–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, P.L.; Doms, R.W.; Moss, B. Oligomeric structure of the human immunodeficiency virus type 1 envelope glycoprotein. Proc. Natl. Acad. Sci. USA 1990, 87, 648–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, P.; Winkler, H.; Chertova, E.; Taylor, K.A.; Roux, K.H. Cryoelectron tomography of HIV-1 envelope spikes: Further evidence for tripod-like legs. PLoS Pathog. 2008, 4, e1000203. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, J.; Staudt, T.; Glass, B.; Bingen, P.; Engelhardt, J.; Anders, M.; Schneider, J.; Muller, B.; Hell, S.W.; Krausslich, H.G. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 2012, 338, 524–528. [Google Scholar] [CrossRef] [Green Version]
- Muranyi, W.; Malkusch, S.; Muller, B.; Heilemann, M.; Krausslich, H.G. Super-resolution microscopy reveals specific recruitment of HIV-1 envelope proteins to viral assembly sites dependent on the envelope C-terminal tail. PLoS Pathog. 2013, 9, e1003198. [Google Scholar] [CrossRef]
- Zhu, P.; Liu, J.; Bess, J., Jr.; Chertova, E.; Lifson, J.D.; Grise, H.; Ofek, G.A.; Taylor, K.A.; Roux, K.H. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 2006, 441, 847–852. [Google Scholar] [CrossRef]
- Brandenberg, O.F.; Magnus, C.; Rusert, P.; Regoes, R.R.; Trkola, A. Different infectivity of HIV-1 strains is linked to number of envelope trimers required for entry. PLoS Pathog. 2015, 11, e1004595. [Google Scholar] [CrossRef]
- Ladinsky, M.S.; Gnanapragasam, P.N.; Yang, Z.; West, A.P.; Kay, M.S.; Bjorkman, P.J. Electron tomography visualization of HIV-1 fusion with target cells using fusion inhibitors to trap the pre-hairpin intermediate. ELife 2020, 9, e58411. [Google Scholar] [CrossRef]
- Ono, A.; Freed, E.O. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. USA 2001, 98, 13925–13930. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.H.; Plemenitas, A.; Linnemann, T.; Fackler, O.T.; Peterlin, B.M. Nef increases infectivity of HIV via lipid rafts. Curr. Biol. CB 2001, 11, 875–879. [Google Scholar] [CrossRef] [Green Version]
- Brugger, B.; Glass, B.; Haberkant, P.; Leibrecht, I.; Wieland, F.T.; Krausslich, H.G. The HIV lipidome: A raft with an unusual composition. Proc. Natl. Acad. Sci. USA 2006, 103, 2641–2646. [Google Scholar] [CrossRef] [Green Version]
- Waheed, A.A.; Freed, E.O. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 2009, 143, 162–176. [Google Scholar] [CrossRef] [Green Version]
- Nieto-Garai, J.A.; Contreras, F.-X.; Arboleya, A.; Lorizate, M. Role of Protein–Lipid Interactions in Viral Entry. Adv. Biol. 2022, 6, 2101264. [Google Scholar] [CrossRef]
- Chan, R.; Uchil, P.D.; Jin, J.; Shui, G.; Ott, D.E.; Mothes, W.; Wenk, M.R. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J. Virol. 2008, 82, 11228–11238. [Google Scholar] [CrossRef] [Green Version]
- Mucksch, F.; Citir, M.; Luchtenborg, C.; Glass, B.; Traynor-Kaplan, A.; Schultz, C.; Brugger, B.; Krausslich, H.G. Quantification of phosphoinositides reveals strong enrichment of PIP2 in HIV-1 compared to producer cell membranes. Sci. Rep. 2019, 9, 17661. [Google Scholar] [CrossRef]
- Lorizate, M.; Krausslich, H.G. Role of lipids in virus replication. Cold Spring Harb. Perspect. Biol. 2011, 3, a004820. [Google Scholar] [CrossRef] [Green Version]
- Kalyana Sundaram, R.V.; Li, H.; Bailey, L.; Rashad, A.A.; Aneja, R.; Weiss, K.; Huynh, J.; Bastian, A.R.; Papazoglou, E.; Abrams, C.; et al. Impact of HIV-1 Membrane Cholesterol on Cell-Independent Lytic Inactivation and Cellular Infectivity. Biochemistry 2016, 55, 447–458. [Google Scholar] [CrossRef]
- Campbell, S.; Gaus, K.; Bittman, R.; Jessup, W.; Crowe, S.; Mak, J. The raft-promoting property of virion-associated cholesterol, but not the presence of virion-associated Brij 98 rafts, is a determinant of human immunodeficiency virus type 1 infectivity. J. Virol. 2004, 78, 10556–10565. [Google Scholar] [CrossRef] [Green Version]
- Waheed, A.A.; Ablan, S.D.; Roser, J.D.; Sowder, R.C.; Schaffner, C.P.; Chertova, E.; Freed, E.O. HIV-1 escape from the entry-inhibiting effects of a cholesterol-binding compound via cleavage of gp41 by the viral protease. Proc. Natl. Acad. Sci. USA 2007, 104, 8467–8471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barklis, E.; Alfadhli, A.; Kyle, J.E.; Bramer, L.M.; Bloodsworth, K.J.; Barklis, R.L.; Leier, H.C.; Petty, R.M.; Zelnik, I.D.; Metz, T.O.; et al. Ceramide synthase 2 deletion decreases the infectivity of HIV-1. J. Biol. Chem. 2021, 296, 100340. [Google Scholar] [CrossRef]
- Schwarzer, R.; Levental, I.; Gramatica, A.; Scolari, S.; Buschmann, V.; Veit, M.; Herrmann, A. The cholesterol-binding motif of the HIV-1 glycoprotein gp41 regulates lateral sorting and oligomerization. Cell Microbiol. 2014, 16, 1565–1581. [Google Scholar] [CrossRef]
- Schroeder, C. Cholesterol-binding viral proteins in virus entry and morphogenesis. Subcell Biochem. 2010, 51, 77–108. [Google Scholar]
- Chen, S.S.; Yang, P.; Ke, P.Y.; Li, H.F.; Chan, W.E.; Chang, D.K.; Chuang, C.K.; Tsai, Y.; Huang, S.C. Identification of the LWYIK motif located in the human immunodeficiency virus type 1 transmembrane gp41 protein as a distinct determinant for viral infection. J. Virol. 2009, 83, 870–883. [Google Scholar] [CrossRef] [Green Version]
- Nieto-Garai, J.A.; Arboleya, A.; Otaegi, S.; Chojnacki, J.; Casas, J.; Fabriàs, G.; Contreras, F.X.; Kräusslich, H.-G.; Lorizate, M. Cholesterol in the Viral Membrane is a Molecular Switch Governing HIV-1 Env Clustering. Adv. Sci. 2020, 8, 2003468. [Google Scholar] [CrossRef]
- Rosa, A.; Chande, A.; Ziglio, S.; De Sanctis, V.; Bertorelli, R.; Goh, S.L.; McCauley, S.M.; Nowosielska, A.; Antonarakis, S.E.; Luban, J.; et al. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 2015, 526, 212–217. [Google Scholar] [CrossRef] [Green Version]
- Usami, Y.; Wu, Y.; Gottlinger, H.G. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 2015, 526, 218–223. [Google Scholar] [CrossRef] [Green Version]
- Sood, C.; Marin, M.; Chande, A.; Pizzato, M.; Melikyan, G.B. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J. Biol. Chem. 2017, 292, 6014–6026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Featherstone, A.; Aiken, C. SERINC5 Inhibits HIV-1 Infectivity by Altering the Conformation of gp120 on HIV-1 Particles. J. Virol. 2020, 94, e00594-20. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.E.; Kiessling, V.; Pornillos, O.; White, J.M.; Ganser-Pornillos, B.K.; Tamm, L.K. HIV-cell membrane fusion intermediates are restricted by Serincs as revealed by cryo-electron and TIRF microscopy. J. Biol. Chem. 2020, 295, 15183–15195. [Google Scholar] [CrossRef] [PubMed]
- Schulte, B.; Selyutina, A.; Opp, S.; Herschhorn, A.; Sodroski, J.G.; Pizzato, M.; Diaz-Griffero, F. Localization to detergent-resistant membranes and HIV-1 core entry inhibition correlate with HIV-1 restriction by SERINC5. Virology 2018, 515, 52–65. [Google Scholar] [CrossRef]
- Chen, Y.C.; Sood, C.; Marin, M.; Aaron, J.; Gratton, E.; Salaita, K.; Melikyan, G.B. Super-Resolution Fluorescence Imaging Reveals That Serine Incorporator Protein 5 Inhibits Human Immunodeficiency Virus Fusion by Disrupting Envelope Glycoprotein Clusters. ACS Nano 2020, 14, 10929–10943. [Google Scholar] [CrossRef]
- Pye, V.E.; Rosa, A.; Bertelli, C.; Struwe, W.B.; Maslen, S.L.; Corey, R.; Liko, I.; Hassall, M.; Mattiuzzo, G.; Ballandras-Colas, A.; et al. A bipartite structural organization defines the SERINC family of HIV-1 restriction factors. Nat. Struct. Mol. Biol. 2020, 27, 78–83. [Google Scholar] [CrossRef]
- Leonhardt, S.; Purdy, M.; Grover, J.; Yang, Z.; Poulos, S.; McIntire, W.; Tatham, E.; Erramilli, S.; Nosol, K.; Lai, K.; et al. CryoEM Structures of the Human HIV-1 Restriction Factor SERINC3 and Function as a Lipid Transporter. bioRxiv 2022. [Google Scholar] [CrossRef]
- Trautz, B.; Wiedemann, H.; Luchtenborg, C.; Pierini, V.; Kranich, J.; Glass, B.; Krausslich, H.G.; Brocker, T.; Pizzato, M.; Ruggieri, A.; et al. The host-cell restriction factor SERINC5 restricts HIV-1 infectivity without altering the lipid composition and organization of viral particles. J. Biol. Chem. 2017, 292, 13702–13713. [Google Scholar] [CrossRef] [Green Version]
- Salimi, H.; Johnson, J.; Flores, M.G.; Zhang, M.S.; O’Malley, Y.; Houtman, J.C.; Schlievert, P.M.; Haim, H. The lipid membrane of HIV-1 stabilizes the viral envelope glycoproteins and modulates their sensitivity to antibody neutralization. J. Biol. Chem. 2020, 295, 348–362. [Google Scholar] [CrossRef]
- Malim, M.H.; Hauber, J.; Fenrick, R.; Cullen, B.R. Immunodeficiency virus rev trans-activator modulates the expression of the viral regulatory genes. Nature 1988, 335, 181–183. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowska, K.; Hordejuk, R.; Szymczyk, P.; Kulma, M.; Abdel-Shakor, A.B.; Plucienniczak, A.; Dolowy, K.; Szewczyk, A.; Sobota, A. Lysenin-His, a sphingomyelin-recognizing toxin, requires tryptophan 20 for cation-selective channel assembly but not for membrane binding. Mol. Membr. Biol. 2007, 24, 121–134. [Google Scholar] [CrossRef]
- Kondo, N.; Marin, M.; Kim, J.H.; Desai, T.M.; Melikyan, G.B. Distinct requirements for HIV-cell fusion and HIV-mediated cell-cell fusion. J. Biol. Chem. 2015, 290, 6558–6573. [Google Scholar] [CrossRef] [Green Version]
- Hammonds, J.; Chen, X.; Zhang, X.; Lee, F.; Spearman, P. Advances in methods for the production, purification, and characterization of HIV-1 Gag-Env pseudovirion vaccines. Vaccine 2007, 25, 8036–8048. [Google Scholar] [CrossRef]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matos, P.M.; Marin, M.; Ahn, B.; Lam, W.; Santos, N.C.; Melikyan, G.B. Anionic lipids are required for vesicular stomatitis virus G protein-mediated single particle fusion with supported lipid bilayers. J. Biol. Chem. 2013, 288, 12416–12425. [Google Scholar] [CrossRef] [Green Version]
- Bhatia, V.K.; Madsen, K.L.; Bolinger, P.Y.; Kunding, A.; Hedegard, P.; Gether, U.; Stamou, D. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. EMBO J. 2009, 28, 3303–3314. [Google Scholar] [CrossRef]
- Chojnacki, J.; Waithe, D.; Carravilla, P.; Huarte, N.; Galiani, S.; Enderlein, J.; Eggeling, C. Envelope glycoprotein mobility on HIV-1 particles depends on the virus maturation state. Nat. Commun. 2017, 8, 545. [Google Scholar] [CrossRef] [Green Version]
- De Chaumont, F.; Dallongeville, S.; Chenouard, N.; Herve, N.; Pop, S.; Provoost, T.; Meas-Yedid, V.; Pankajakshan, P.; Lecomte, T.; Le Montagner, Y.; et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods 2012, 9, 690–696. [Google Scholar] [CrossRef]
- Parasassi, T.; Di Stefano, M.; Loiero, M.; Ravagnan, G.; Gratton, E. Influence of cholesterol on phospholipid bilayers phase domains as detected by Laurdan fluorescence. Biophys. J. 1994, 66, 120–132. [Google Scholar] [CrossRef] [Green Version]
- Parasassi, T.; Gratton, E. Membrane lipid domains and dynamics as detected by Laurdan fluorescence. J. Fluoresc. 1995, 5, 59–69. [Google Scholar] [CrossRef]
- Bagatolli, L.A.; Gratton, E. Two-Photon Fluorescence Microscopy Observation of Shape Changes at the Phase Transition in Phospholipid Giant Unilamellar Vesicles. Biophys. J. 1999, 77, 2090–2101. [Google Scholar] [CrossRef] [Green Version]
- Saxena, R.; Shrivastava, S.; Haldar, S.; Klymchenko, A.S.; Chattopadhyay, A. Location, dynamics and solvent relaxation of a Nile Red-based phase-sensitive fluorescent membrane probe. Chem. Phys. Lipids 2014, 183, 1–8. [Google Scholar] [CrossRef]
- Kucherak, O.A.; Oncul, S.; Darwich, Z.; Yushchenko, D.A.; Arntz, Y.; Didier, P.; Mely, Y.; Klymchenko, A.S. Switchable nile red-based probe for cholesterol and lipid order at the outer leaflet of biomembranes. J. Am. Chem. Soc. 2010, 132, 4907–4916. [Google Scholar] [CrossRef] [PubMed]
- Zhanghao, K.; Liu, W.; Li, M.; Wu, Z.; Wang, X.; Chen, X.; Shan, C.; Wang, H.; Chen, X.; Dai, Q.; et al. High-dimensional super-resolution imaging reveals heterogeneity and dynamics of subcellular lipid membranes. Nat. Commun. 2020, 11, 5890. [Google Scholar] [CrossRef]
- Guido, C.A.; Mennucci, B.; Jacquemin, D.; Adamo, C. Planar vs. twisted intramolecular charge transfer mechanism in Nile Red: New hints from theory. Phys. Chem. Chem. Phys. 2010, 12, 8016–8023. [Google Scholar] [CrossRef]
- Kreder, R.; Pyrshev, K.A.; Darwich, Z.; Kucherak, O.A.; Mely, Y.; Klymchenko, A.S. Solvatochromic Nile Red probes with FRET quencher reveal lipid order heterogeneity in living and apoptotic cells. ACS Chem. Biol. 2015, 10, 1435–1442. [Google Scholar] [CrossRef]
- Carravilla, P.; Dasgupta, A.; Zhurgenbayeva, G.; Danylchuk, D.I.; Klymchenko, A.S.; Sezgin, E.; Eggeling, C. Long-term STED imaging of membrane packing and dynamics by exchangeable polarity-sensitive dyes. Biophys. Rep. 2021, 1, 100023. [Google Scholar] [CrossRef]
- Lorizate, M.; Brugger, B.; Akiyama, H.; Glass, B.; Muller, B.; Anderluh, G.; Wieland, F.T.; Krausslich, H.G. Probing HIV-1 membrane liquid order by Laurdan staining reveals producer cell-dependent differences. J. Biol. Chem. 2009, 284, 22238–22247. [Google Scholar] [CrossRef] [Green Version]
- Zidovetzki, R.; Levitan, I. Use of cyclodextrins to manipulate plasma membrane cholesterol content: Evidence, misconceptions and control strategies. Biochim. Biophys. Acta 2007, 1768, 1311–1324. [Google Scholar] [CrossRef] [Green Version]
- Rodal, S.K.; Skretting, G.; Garred, O.; Vilhardt, F.; van Deurs, B.; Sandvig, K. Extraction of cholesterol with methyl-beta-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol. Biol. Cell 1999, 10, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Montefiori, D.C. Measuring HIV neutralization in a luciferase reporter gene assay. Methods Mol. Biol. 2009, 485, 395–405. [Google Scholar]
- Neuvonen, M.; Manna, M.; Mokkila, S.; Javanainen, M.; Rog, T.; Liu, Z.; Bittman, R.; Vattulainen, I.; Ikonen, E. Enzymatic oxidation of cholesterol: Properties and functional effects of cholestenone in cell membranes. PLoS ONE 2014, 9, e103743. [Google Scholar] [CrossRef] [Green Version]
- Silvius, J.R. Role of cholesterol in lipid raft formation: Lessons from lipid model systems. Biochim. Biophys. Acta 2003, 1610, 174–183. [Google Scholar] [CrossRef] [Green Version]
- Das, A.; Brown, M.S.; Anderson, D.D.; Goldstein, J.L.; Radhakrishnan, A. Three pools of plasma membrane cholesterol and their relation to cholesterol homeostasis. Elife 2014, 3, e02882. [Google Scholar] [CrossRef]
- Johnson, K.A.; Radhakrishnan, A. Accessibility of cholesterol at cell surfaces. J. Lipid Res. 2020, 61, 1307. [Google Scholar] [CrossRef] [Green Version]
- Holopainen, J.M.; Subramanian, M.; Kinnunen, P.K. Sphingomyelinase induces lipid microdomain formation in a fluid phosphatidylcholine/sphingomyelin membrane. Biochemistry 1998, 37, 17562–17570. [Google Scholar] [CrossRef]
- Yamaji, A.; Sekizawa, Y.; Emoto, K.; Sakuraba, H.; Inoue, K.; Kobayashi, H.; Umeda, M. Lysenin, a novel sphingomyelin-specific binding protein. J. Biol. Chem. 1998, 273, 5300–5306. [Google Scholar] [CrossRef] [Green Version]
- Audi, A.; Soudani, N.; Dbaibo, G.; Zaraket, H. Depletion of Host and Viral Sphingomyelin Impairs Influenza Virus Infection. Front. Microbiol. 2020, 11, 612. [Google Scholar] [CrossRef]
- Kinnebrew, M.; Iverson, E.J.; Patel, B.B.; Pusapati, G.V.; Kong, J.H.; Johnson, K.A.; Luchetti, G.; Eckert, K.M.; McDonald, J.G.; Covey, D.F.; et al. Cholesterol accessibility at the ciliary membrane controls hedgehog signaling. Elife 2019, 8, e50051. [Google Scholar] [CrossRef]
- Spahn, C.; Grimm, J.B.; Lavis, L.D.; Lampe, M.; Heilemann, M. Whole-Cell, 3D, and Multicolor STED Imaging with Exchangeable Fluorophores. Nano Lett. 2019, 19, 500–505. [Google Scholar] [CrossRef] [Green Version]
- Slotte, J.P. Direct observation of the action of cholesterol oxidase in monolayers. Biochim. Biophys. Acta 1995, 1259, 180–186. [Google Scholar] [CrossRef]
- Han, Q.; Jones, J.A.; Nicely, N.I.; Reed, R.K.; Shen, X.; Mansouri, K.; Louder, M.; Trama, A.M.; Alam, S.M.; Edwards, R.J.; et al. Difficult-to-neutralize global HIV-1 isolates are neutralized by antibodies targeting open envelope conformations. Nat. Commun. 2019, 10, 2898. [Google Scholar] [CrossRef] [PubMed]
- Pezeshkian, N.; Groves, N.S.; van Engelenburg, S.B. Single-molecule imaging of HIV-1 envelope glycoprotein dynamics and Gag lattice association exposes determinants responsible for virus incorporation. Proc. Natl. Acad. Sci. USA 2019, 116, 25269–25277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, M.; Ma, X.; Castillo-Menendez, L.R.; Gorman, J.; Alsahafi, N.; Ermel, U.; Terry, D.S.; Chambers, M.; Peng, D.; Zhang, B.; et al. Associating HIV-1 envelope glycoprotein structures with states on the virus observed by smFRET. Nature 2019, 568, 415–419. [Google Scholar] [CrossRef]
- Kirschman, J.; Marin, M.; Chen, Y.-C.; Chen, J.; Herschhorn, A.; Smith, A.B.I.; Melikyan, G.B. SERINC5 Restricts HIV-1 Infectivity by Promoting Conformational Changes and Accelerating Functional Inactivation of Env. Viruses 2022, 14, 1388. [Google Scholar] [CrossRef]
- Zhang, X.; Shi, J.; Qiu, X.; Chai, Q.; Frabutt, D.A.; Schwartz, R.C.; Zheng, Y.H. CD4 Expression and Env Conformation Are Critical for HIV-1 Restriction by SERINC5. J. Virol. 2019, 93, e00544-19. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Raghunath, G.; Chen, Y.-C.; Marin, M.; Wu, H.; Melikyan, G.B. SERINC5-Mediated Restriction of HIV-1 Infectivity Correlates with Resistance to Cholesterol Extraction but Not with Lipid Order of Viral Membrane. Viruses 2022, 14, 1636. https://doi.org/10.3390/v14081636
Raghunath G, Chen Y-C, Marin M, Wu H, Melikyan GB. SERINC5-Mediated Restriction of HIV-1 Infectivity Correlates with Resistance to Cholesterol Extraction but Not with Lipid Order of Viral Membrane. Viruses. 2022; 14(8):1636. https://doi.org/10.3390/v14081636
Chicago/Turabian StyleRaghunath, Gokul, Yen-Cheng Chen, Mariana Marin, Hui Wu, and Gregory B. Melikyan. 2022. "SERINC5-Mediated Restriction of HIV-1 Infectivity Correlates with Resistance to Cholesterol Extraction but Not with Lipid Order of Viral Membrane" Viruses 14, no. 8: 1636. https://doi.org/10.3390/v14081636




