Harnessing Human Papillomavirus’ Natural Tropism to Target Tumors
Abstract
1. Introduction
2. HPV Background
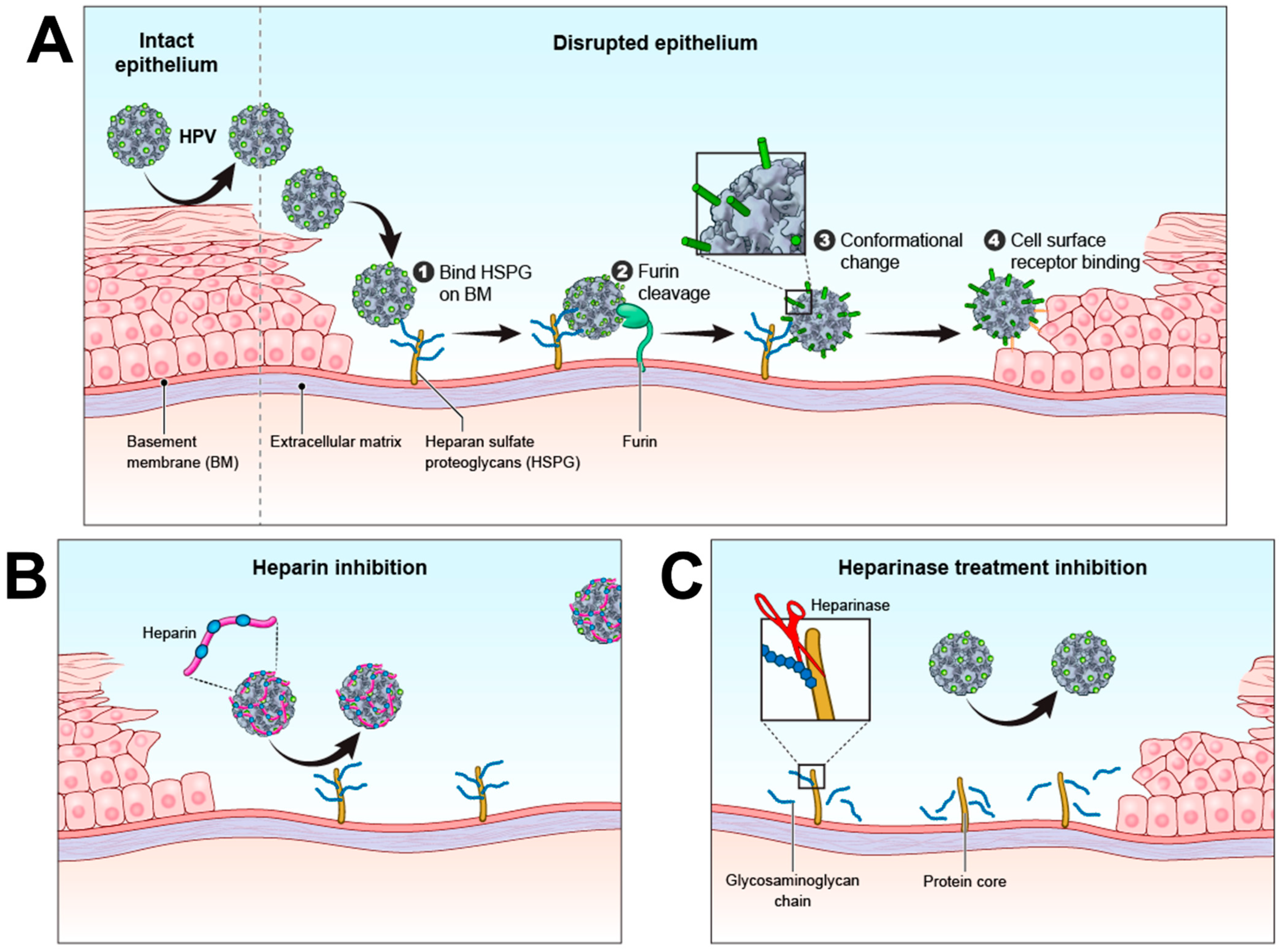
3. HPV Infection Mechanism
4. Proteoglycans
5. HSPGs and Cancer
6. Proteoglycans as Targets for Tumor Therapy
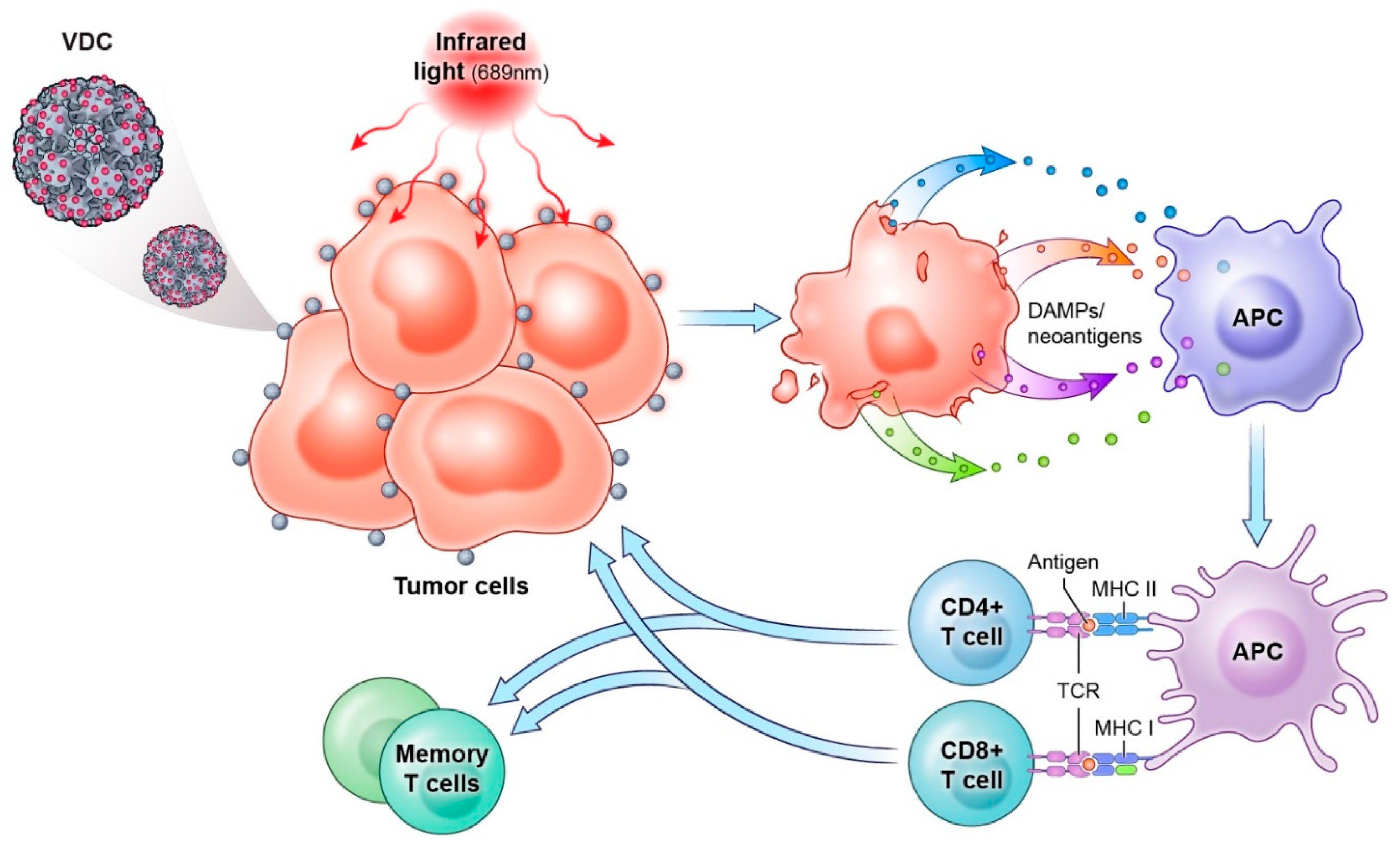
7. HPV Capsids as a Tumor Therapeutic
7.1. Gene Delivery
7.2. HPV Virus-like Particles as Drug Conjugates
8. Conclusions
9. Patents
Author Contributions
Funding
Conflicts of Interest
References
- Bernard, H.-U.; Burk, R.D.; Chen, Z.; van Doorslaer, K.; Hausen, H.Z.; de Villiers, E.-M. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology 2010, 401, 70–79. [Google Scholar] [CrossRef] [PubMed]
- de Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 2017, 141, 664–670. [Google Scholar] [CrossRef] [PubMed]
- Burd, E.M. Human Papillomavirus and Cervical Cancer. Clin. Microbiol. Rev. 2003, 16, 637–646. [Google Scholar] [CrossRef] [PubMed]
- Gillison, M.L.; Alemany, L.; Snijders, P.J.; Chaturvedi, A.; Steinberg, B.M.; Schwartz, S.; Castellsagué, X. Human papillomavirus and diseases of the upper airway: Head and neck cancer and respiratory papillomatosis. Vaccine 2012, 30 (Suppl. 5), F34–F54. [Google Scholar] [CrossRef] [PubMed]
- Egawa, N.; Doorbar, J. The low-risk papillomaviruses. Virus Res. 2017, 231, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Fortes, H.R.; von Ranke, F.M.; Escuissato, D.L.; Araujo Neto, C.A.; Zanetti, G.; Hochhegger, B.; Souza, C.A.; Marchiori, E. Recurrent respiratory papillomatosis: A state-of-the-art review. Respir. Med. 2017, 126, 116–121. [Google Scholar] [CrossRef]
- Dyson, N.; Howley, P.M.; Munger, K.; Harlow, E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science 1989, 243, 934–937. [Google Scholar] [CrossRef]
- Hawley-Nelson, P.; Vousden, K.H.; Hubbert, N.L.; Lowy, D.R.; Schiller, J.T. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989, 8, 3905–3910. [Google Scholar] [CrossRef]
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; and Howley, P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990, 63, 1129–1136. [Google Scholar] [CrossRef]
- Conde-Ferráez, L.; Chan May Ade, A.; Carrillo-Martínez, J.R.; Ayora-Talavera, G.; González-Losa Mdel, R. Human papillomavirus infection and spontaneous abortion: A case-control study performed in Mexico. Eur. J. Obstet. Gynecol. Reprod. Biol. 2013, 170, 468–473. [Google Scholar] [CrossRef]
- Knör, M.; Tziridis, K.; Agaimy, A.; Zenk, J.; Wendler, O. Human papillomavirus (HPV) prevalence in nasal and antrochoanal polyps and association with clinical data. PLoS ONE 2015, 10, e0141722. [Google Scholar] [CrossRef] [PubMed]
- Ambühl, L.M.; Baandrup, U.; Dybkær, K.; Blaakær, J.; Uldbjerg, N.; Sørensen, S. Human papillomavirus infection as a possible cause of spontaneous abortion and spontaneous preterm Delivery. Infect. Dis. Obstet. Gynecol. 2016, 2016, 3086036. [Google Scholar] [CrossRef]
- Ambühl, L.M.M.; Leonhard, A.K.; Widen Zakhary, C.; Jørgensen, A.; Blaakaer, J.; Dybkaer, K.; Baandrup, U.; Uldbjerg, N.; Sørensen, S. Human papillomavirus infects placental trophoblast and Hofbauer cells, but appears not to play a causal role in miscarriage and preterm labor. Acta Obstet. Gynecol. Scand. 2017, 96, 1188–1196. [Google Scholar] [CrossRef]
- Xiong, Y.Q.; Mo, Y.; Luo, Q.M.; Huo, S.T.; He, W.Q.; Chen, Q. The risk of human papillomavirus infection for spontaneous abortion, spontaneous preterm birth, and pregnancy rate of assisted reproductive technologies: A systematic review and meta-analysis. Gynecol. Obstet. Investig. 2018, 83, 417–427. [Google Scholar] [CrossRef]
- Jeršovienė, V.; Gudlevičienė, Ž.; Rimienė, J.; Butkauskas, D. Human papillomavirus and infertility. Medicina 2019, 55, 377. [Google Scholar] [CrossRef]
- Tognon, M.; Tagliapietra, A.; Magagnoli, F.; Mazziotta, C.; Oton-Gonzalez, L.; Lanzillotti, C.; Vesce, F.; Contini, C.; Rotondo, J.C.; Martini, F. Investigation on spontaneous abortion and human papillomavirus infection. Vaccines 2020, 8, 473. [Google Scholar] [CrossRef] [PubMed]
- Muscianisi, F.; De Toni, L.; Giorato, G.; Carosso, A.; Foresta, C.; Garolla, A. Is HPV the novel target in male idiopathic infertility? A systematic review of the literature. Front. Endocrinol. 2021, 12, 643539. [Google Scholar] [CrossRef]
- Oton-Gonzalez, L.; Rotondo, J.C.; Cerritelli, L.; Malagutti, N.; Lanzillotti, C.; Bononi, I.; Ciorba, A.; Bianchini, C.; Mazziotta, C.; De Mattei, M.; et al. Association between oncogenic human papillomavirus type 16 and Killian polyp. Infect. Agent Cancer 2021, 16, 3. [Google Scholar] [CrossRef]
- Zhou, J.; Sun, X.Y.; Stenzel, D.J.; Frazer, I.H. Expression of vaccinia recombinant HPV 16 L1 and L2 ORF proteins in epithelial cells is sufficient for assembly of HPV virion-like particles. Virology 1991, 185, 251–257. [Google Scholar] [CrossRef]
- Kirnbauer, R.; Booy, F.; Cheng, N.; Lowy, D.R.; Schiller, J.T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 1992, 89, 12180–12184. [Google Scholar] [CrossRef]
- Kirnbauer, R.; Taub, J.; Greenstone, H.; Roden, R.; Dürst, M.; Gissmann, L.; Lowy, D.R.; Schiller, J.T. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J. Virol. 1993, 67, 6929–6936. [Google Scholar] [CrossRef] [PubMed]
- Rose, R.C.; Bonnez, W.; Reichman, R.C.; Garcea, R.L. Expression of human papillomavirus type 11 L1 protein in insect cells: In vivo and in vitro assembly of viruslike particles. J. Virol. 1993, 67, 1936–1944. [Google Scholar] [CrossRef]
- Buck, C.B.; Pastrana, D.V.; Lowy, D.R.; Schiller, J.T. Efficient intracellular assembly of papillomaviral vectors. J. Virol. 2004, 78, 751–757. [Google Scholar] [CrossRef] [PubMed]
- Cerqueira, C.; Pang, Y.Y.; Day, P.M.; Thompson, C.D.; Buck, C.B.; Lowy, D.R.; Schiller, J.T. A Cell-Free Assembly System for Generating Infectious Human Papillomavirus 16 Capsids Implicates a Size Discrimination Mechanism for Preferential Viral Genome Packaging. J. Virol. 2016, 90, 1096–1107. [Google Scholar] [CrossRef]
- Day, P.M.; Lowy, D.R.; Schiller, J.T. Heparan sulfate-independent cell binding and infection with furin-precleaved papillomavirus capsids. J. Virol. 2008, 82, 12565–12568. [Google Scholar] [CrossRef] [PubMed]
- Roberts, J.N.; Buck, C.B.; Thompson, C.D.; Kines, R.; Bernardo, M.; Choyke, P.L.; Lowy, D.R.; Schiller, J.T. Genital transmission of HPV in a mouse model is potentiated by nonoxynol-9 and inhibited by carrageenan. Nat. Med. 2007, 13, 857–861. [Google Scholar] [CrossRef]
- Griffin, L.M.; Cicchini, L.; Pyeon, D. Human papillomavirus infection is inhibited by host autophagy in primary human keratinocytes. Virology 2013, 437, 12–19. [Google Scholar] [CrossRef]
- Joyce, J.G.; Tung, J.S.; Przysiecki, C.T.; Cook, J.C.; Lehman, E.D.; Sands, J.A.; Jansen, K.U.; Keller, P.M. The L1 major capsid protein of human papillomavirus type 11 recombinant virus-like particles interacts with heparin and cell-surface glycosaminoglycans on human keratinocytes. J. Biol. Chem. 1999, 274, 5810–5822. [Google Scholar] [CrossRef]
- Giroglou, T.; Florin, L.; Schäfer, F.; Streeck, R.E.; Sapp, M. Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 2001, 75, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Knappe, M.; Bodevin, S.; Selinka, H.C.; Spillmann, D.; Streeck, R.E.; Chen, X.S.; Lindahl, U.; Sapp, M. Surface-exposed amino acid residues of HPV16 L1 protein mediating interaction with cell surface heparan sulfate. J. Biol. Chem. 2007, 282, 27913–27922. [Google Scholar] [CrossRef] [PubMed]
- Cladel, N.M.; Hu, J.; Balogh, K.; Mejia, A.; Christensen, N.D. Wounding prior to challenge substantially improves infectivity of cottontail rabbit papillomavirus and allows for standardization of infection. J. Virol. Methods 2008, 148, 34–39. [Google Scholar] [CrossRef][Green Version]
- Johnson, K.M.; Kines, R.C.; Roberts, J.N.; Lowy, D.R.; Schiller, J.T.; Day, P.M. Role of heparan sulfate in attachment to and infection of the murine female genital tract by human papillomavirus. J. Virol. 2009, 83, 2067–2074. [Google Scholar] [CrossRef]
- Olczyk, P.; Mencner, Ł.; Komosinska-Vassev, K. Diverse Roles of Heparan Sulfate and Heparin in Wound Repair. Biomed. Res. Int. 2015, 2015, 549417. [Google Scholar] [CrossRef]
- Richards, R.M.; Lowy, D.R.; Schiller, J.T.; Day, P.M. Cleavage of the papillomavirus minor capsid protein, L2, at a furin consensus site is necessary for infection. Proc. Natl. Acad. Sci. USA 2006, 103, 1522–1527. [Google Scholar] [CrossRef]
- Kines, R.C.; Thompson, C.D.; Lowy, D.R.; Schiller, J.T.; Day, P.M. The initial steps leading to papillomavirus infection occur on the basement membrane prior to cell surface binding. Proc. Natl. Acad. Sci. USA 2009, 106, 20458–20463. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, A.H.; Park, P.W. Heparan sulfate proteoglycans in infection. In Glycans in Diseases and Therapeutics; Springer: Berlin/Heidelberg, Germany, 2011; pp. 31–62. [Google Scholar] [CrossRef]
- Cagno, V.; Tseligka, E.D.; Jones, S.T.; Tapparel, C. Heparan sulfate proteoglycans and viral attachment: True receptors or adaptation bias? Viruses 2019, 11, 596. [Google Scholar] [CrossRef]
- De Pasquale, V.; Quiccione, M.S.; Tafuri, S.; Avallone, L.; Pavone, L.M. Heparan sulfate proteoglycans in viral infection and treatment: A special focus on SARS-CoV-2. Int. J. Mol. Sci. 2021, 22, 6574. [Google Scholar] [CrossRef]
- Dechecchi, M.C.; Tamanini, A.; Bonizzato, A.; Cabrini, G. Heparan sulfate glycosaminoglycans are involved in adenovirus type 5 and 2-host cell interactions. Virology 2000, 268, 382–390. [Google Scholar] [CrossRef]
- Dechecchi, M.C.; Melotti, P.; Bonizzato, A.; Santacatterina, M.; Chilosi, M.; Cabrini, G. Heparan sulfate glycosaminoglycans are receptors sufficient to mediate the initial binding of adenovirus types 2 and 5. J. Virol. 2001, 75, 8772–8780. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Maguire, T.; Hileman, R.E.; Fromm, J.R.; Esko, J.D.; Linhardt, R.J.; Marks, R.M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 1997, 3, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Gripon, P.; Urban, S. Hepatitis B virus infection initiates with a large surface protein–dependent binding to heparan sulfate proteoglycans. Hepatology 2007, 46, 1759–1768. [Google Scholar] [CrossRef]
- Leistner, C.M.; Gruen-Bernhard, S.; Glebe, D. Role of glycosaminoglycans for binding and infection of hepatitis B virus. Cell Microbiol. 2008, 10, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Barth, H.; Schafer, C.; Adah, M.I.; Zhang, F.; Linhardt, R.J.; Toyoda, H.; Kinoshita-Toyoda, A.; Toida, T.; Van Kuppevelt, T.H.; Depla, E.; et al. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 2003, 278, 41003–41012. [Google Scholar] [CrossRef] [PubMed]
- Barth, H.; Schnober, E.K.; Zhang, F.; Linhardt, R.J.; Depla, E.; Boson, B.; Cosset, F.L.; Patel, A.H.; Blum, H.E.; Baumert, T.F. Viral and cellular determinants of the hepatitis C virus envelope-heparan sulfate interaction. J. Virol. 2006, 80, 10579–10590. [Google Scholar] [CrossRef] [PubMed]
- Kalia, M.; Chandra, V.; Rahman, S.A.; Sehgal, D.; Jameel, S. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J. Virol. 2009, 83, 12714–12724. [Google Scholar] [CrossRef]
- Patel, M.; Yanagishita, M.; Roderiquez, G.; Bou-Habib, D.C.; Oravecz, T.; Hascall, V.C.; Norcross, M.A. Cell-surface heparan sulfate proteoglycan mediates HIV-1 infection of T-cell lines. AIDS Res. Hum. Retrovir. 1993, 9, 167–174. [Google Scholar] [CrossRef]
- Tyagi, M.; Rusnati, M.; Presta, M.; Giacca, M. Internalization of HIV-1 tat requires cell surface heparan sulfate proteoglycans. J. Biol. Chem. 2001, 276, 3254–3261. [Google Scholar] [CrossRef]
- Vivès, R.R.; Imberty, A.; Sattentau, Q.J.; Lortat-Jacob, H. Heparan sulfate targets the HIV-1 envelope glycoprotein gp120 coreceptor binding site. J. Biol. Chem. 2005, 280, 21353–21357. [Google Scholar] [CrossRef]
- Pomin, V.H.; Bezerra, F.F.; Soares, P.A.G. Sulfated Glycans in HIV Infection and Therapy. Curr. Pharm. Des. 2017, 23, 3405–3414. [Google Scholar] [CrossRef]
- Compton, T.; Nowlin, D.M.; Cooper, N.R. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology 1993, 193, 834–841. [Google Scholar] [CrossRef]
- Boyle, K.A.; Compton, T. Receptor-binding properties of a soluble form of human cytomegalovirus glycoprotein B. J. Virol. 1998, 72, 1826–1833. [Google Scholar] [CrossRef] [PubMed]
- Mitra, D.; Hasan, M.H.; Bates, J.T.; Bierdeman, M.A.; Ederer, D.R.; Parmar, R.C.; Fassero, L.A.; Liang, Q.; Qiu, H.; Tiwari, V.; et al. The degree of polymerization and sulfation patterns in heparan sulfate are critical determinants of cytomegalovirus entry into host cells. PLoS Pathog. 2021, 17, e1009803. [Google Scholar] [CrossRef] [PubMed]
- WuDunn, D.; Spear, P.G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J. Virol. 1989, 63, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Shieh, M.T.; WuDunn, D.; Montgomery, R.I.; Esko, J.D.; Spear, P.G. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J. Cell Biol. 1992, 116, 1273–1281. [Google Scholar] [CrossRef]
- Tal-Singer, R.; Peng, C.; Ponce De Leon, M.; Abrams, W.R.; Banfield, B.W.; Tufaro, F.; Cohen, G.H.; Eisenberg, R.J. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J. Virol. 1995, 69, 4471–4483. [Google Scholar] [CrossRef] [PubMed]
- Feyzi, E.; Trybala, E.; Bergström, T.; Lindahl, U.; Spillmann, D. Structural requirement of heparan sulfate for interaction with herpes simplex virus type 1 virions and isolated glycoprotein C. J. Biol. Chem. 1997, 272, 24850–24857. [Google Scholar] [CrossRef][Green Version]
- Shukla, D.; Liu, J.; Blaiklock, P.; Shworak, N.W.; Bai, X.; Esko, J.D.; Cohen, G.H.; Eisenberg, R.J.; Rosenberg, R.D.; Spear, P.G. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell 1999, 99, 13–22. [Google Scholar] [CrossRef]
- Shukla, D.; Spear, P.G. Herpesviruses and heparan sulfate: An intimate relationship in aid of viral entry. J. Clin. Investig. 2001, 108, 503–510. [Google Scholar] [CrossRef]
- Williams, R.K.; Straus, S.E. Specificity and affinity of binding of herpes simplex virus type 2 glycoprotein B to glycosaminoglycans. J. Virol. 1997, 71, 1375–1380. [Google Scholar] [CrossRef]
- Gerber, S.I.; Belval, B.J.; Herold, B.C. Differences in the role of glycoprotein C of HSV-1 and HSV-2 in viral binding may contribute to serotype differences in cell tropism. Virology 1995, 214, 29–39. [Google Scholar] [CrossRef]
- Schowalter, R.M.; Pastrana, D.V.; Buck, C.B. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLoS Pathog. 2011, 7, e1002161. [Google Scholar] [CrossRef] [PubMed]
- Bayer, N.J.; Januliene, D.; Zocher, G.; Stehle, T.; Moeller, A.; Blaum, B.S. Structure of merkel cell polyomavirus capsid and interaction with its glycosaminoglycan attachment receptor. J. Virol. 2020, 94, e01664-19. [Google Scholar] [CrossRef]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell 2020, 183, 1043–1057.e1015. [Google Scholar] [CrossRef]
- Liu, L.; Chopra, P.; Li, X.; Bouwman, K.M.; Tompkins, S.M.; Wolfert, M.A.; de Vries, R.P.; Boons, G.-J. Heparan sulfate proteoglycans as attachment factor for SARS-CoV-2. ACS Cent. Sci. 2021, 7, 1009–1018. [Google Scholar] [CrossRef]
- Jacquet, A.; Haumont, M.; Chellun, D.; Massaer, M.; Tufaro, F.; Bollen, A.; Jacobs, P. The varicella zoster virus glycoprotein B (gB) plays a role in virus binding to cell surface heparan sulfate proteoglycans. Virus Res. 1998, 53, 197–207. [Google Scholar] [CrossRef]
- Weiland, M.E.; Palm, J.E.; Griffiths, W.J.; McCaffery, J.M.; Svärd, S.G. Characterisation of alpha-1 giardin: An immunodominant Giardia lamblia annexin with glycosaminoglycan-binding activity. Int. J. Parasitol. 2003, 33, 1341–1351. [Google Scholar] [CrossRef]
- Frevert, U.; Sinnis, P.; Cerami, C.; Shreffler, W.; Takacs, B.; Nussenzweig, V. Malaria circumsporozoite protein binds to heparan sulfate proteoglycans associated with the surface membrane of hepatocytes. J. Exp. Med. 1993, 177, 1287–1298. [Google Scholar] [CrossRef] [PubMed]
- Fried, M.; Duffy, P.E. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science 1996, 272, 1502–1504. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Kato, K.; Sugi, T.; Takemae, H.; Pandey, K.; Gong, H.; Tohya, Y.; Akashi, H. Plasmodium falciparum BAEBL binds to heparan sulfate proteoglycans on the human erythrocyte surface. J. Biol. Chem. 2010, 285, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Barria, E.; Boothroyd, J.C. A Toxoplasma lectin-like activity specific for sulfated polysaccharides is involved in host cell infection. J. Biol. Chem. 1999, 274, 1267–1276. [Google Scholar] [CrossRef]
- Carruthers, V.B.; Håkansson, S.; Giddings, O.K.; Sibley, L.D. Toxoplasma gondii uses sulfated proteoglycans for substrate and host cell attachment. Infect. Immun. 2000, 68, 4005–4011. [Google Scholar] [CrossRef] [PubMed]
- Jacquet, A.; Coulon, L.; De Nève, J.; Daminet, V.; Haumont, M.; Garcia, L.; Bollen, A.; Jurado, M.; Biemans, R. The surface antigen SAG3 mediates the attachment of Toxoplasma gondii to cell-surface proteoglycans. Mol. Biochem. Parasitol. 2001, 116, 35–44. [Google Scholar] [CrossRef]
- Azzouz, N.; Kamena, F.; Laurino, P.; Kikkeri, R.; Mercier, C.; Cesbron-Delauw, M.-F.; Dubremetz, J.-F.; De Cola, L.; Seeberger, P.H. Toxoplasma gondii secretory proteins bind to sulfated heparin structures. Glycobiology 2012, 23, 106–120. [Google Scholar] [CrossRef]
- Ortega-Barria, E.; Pereira, M.E. A novel T. cruzi heparin-binding protein promotes fibroblast adhesion and penetration of engineered bacteria and trypanosomes into mammalian cells. Cell 1991, 67, 411–421. [Google Scholar] [CrossRef]
- Oliveira, F.O., Jr.; Alves, C.R.; Calvet, C.M.; Toma, L.; Bouças, R.I.; Nader, H.B.; Castro Côrtes, L.M.; Krieger, M.A.; Meirelles Mde, N.; Souza Pereira, M.C. Trypanosoma cruzi heparin-binding proteins and the nature of the host cell heparan sulfate-binding domain. Microb. Pathog. 2008, 44, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Bambino-Medeiros, R.; Oliveira, F.O.; Calvet, C.M.; Vicente, D.; Toma, L.; Krieger, M.A.; Meirelles, M.N.; Pereira, M.C. Involvement of host cell heparan sulfate proteoglycan in Trypanosoma cruzi amastigote attachment and invasion. Parasitology 2011, 138, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Hannah, J.H.; Menozzi, F.D.; Renauld, G.; Locht, C.; Brennan, M.J. Sulfated glycoconjugate receptors for the Bordetella pertussis adhesin filamentous hemagglutinin (FHA) and mapping of the heparin-binding domain on FHA. Infect. Immun. 1994, 62, 5010–5019. [Google Scholar] [CrossRef] [PubMed]
- Utt, M.; Wadström, T. Identification of heparan sulphate binding surface proteins of Helicobacter pylori: Inhibition of heparan sulphate binding with sulphated carbohydrate polymers. J. Med. Microbiol. 1997, 46, 541–546. [Google Scholar] [CrossRef]
- Utt, M.; Danielsson, B.; Wadström, T. Helicobacter pylori vacuolating cytotoxin binding to a putative cell surface receptor, heparan sulfate, studied by surface plasmon resonance. FEMS Immunol. Med. Microbiol. 2001, 30, 109–113. [Google Scholar] [CrossRef]
- Alvarez-Domínguez, C.; Vázquez-Boland, J.A.; Carrasco-Marín, E.; López-Mato, P.; Leyva-Cobián, F. Host cell heparan sulfate proteoglycans mediate attachment and entry of Listeria monocytogenes, and the listerial surface protein ActA is involved in heparan sulfate receptor recognition. Infect. Immun. 1997, 65, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Menozzi, F.D.; Rouse, J.H.; Alavi, M.; Laude-Sharp, M.; Muller, J.; Bischoff, R.; Brennan, M.J.; Locht, C. Identification of a heparin-binding hemagglutinin present in mycobacteria. J. Exp. Med. 1996, 184, 993–1001. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Belland, R.J.; Wilson, J.; Swanson, J. Adherence of pilus- Opa+ gonococci to epithelial cells in vitro involves heparan sulfate. J. Exp. Med. 1995, 182, 511–517. [Google Scholar] [CrossRef] [PubMed]
- van Putten, J.P.; Paul, S.M. Binding of syndecan-like cell surface proteoglycan receptors is required for Neisseria gonorrhoeae entry into human mucosal cells. EMBO J. 1995, 14, 2144–2154. [Google Scholar] [CrossRef]
- Klotz, S.A.; Smith, R.L. Glycosaminoglycans inhibit Candida albicans adherence to extracellular matrix proteins. FEMS Microbiol. Lett. 1992, 78, 205–208. [Google Scholar] [CrossRef]
- Hoffman, M.P.; Haidaris, C.G. Identification and characterization of a Candida albicans-binding proteoglycan secreted from rat submandibular salivary glands. Infect. Immun. 1994, 62, 828–836. [Google Scholar] [CrossRef]
- Ordiales, H.; Vázquez-López, F.; Pevida, M.; Vázquez-Losada, B.; Vázquez, F.; Quirós, L.M.; Martín, C. Glycosaminoglycans are involved in the adhesion of Candida albicans and Malassezia species to keratinocytes but not to dermal fibroblasts. Actas Dermo-Sifiliográficas 2021, 112, 619–624. [Google Scholar] [CrossRef]
- Caughey, B.; Raymond, G.J. Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J. Virol. 1993, 67, 643–650. [Google Scholar] [CrossRef]
- Hundt, C.; Peyrin, J.M.; Haïk, S.; Gauczynski, S.; Leucht, C.; Rieger, R.; Riley, M.L.; Deslys, J.P.; Dormont, D.; Lasmézas, C.I.; et al. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO J. 2001, 20, 5876–5886. [Google Scholar] [CrossRef]
- Wong, C.; Xiong, L.W.; Horiuchi, M.; Raymond, L.; Wehrly, K.; Chesebro, B.; Caughey, B. Sulfated glycans and elevated temperature stimulate PrP(Sc)-dependent cell-free formation of protease-resistant prion protein. EMBO J. 2001, 20, 377–386. [Google Scholar] [CrossRef]
- Ben-Zaken, O.; Tzaban, S.; Tal, Y.; Horonchik, L.; Esko, J.D.; Vlodavsky, I.; Taraboulos, A. Cellular heparan sulfate participates in the metabolism of prions. J. Biol. Chem. 2003, 278, 40041–40049. [Google Scholar] [CrossRef]
- Bishop, J.R.; Schuksz, M.; Esko, J.D. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 2007, 446, 1030–1037. [Google Scholar] [CrossRef]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. In Cold Spring Harb Perspect Biol; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2011; Volume 3, p. a004952. [Google Scholar] [CrossRef] [PubMed]
- Merry, C.L.R.; Lindahl, U.; Couchman, J.; Esko, J.D. Proteoglycans and sulfated glycosaminoglycans. In Essentials of Glycobiology; Varki, A., Cummings, R.D., Esko, J.D., Stanley, P., Hart, G.W., Aebi, M., Mohnen, D., Kinoshita, T., Packer, N.H., Prestegard, J.H., et al., Eds.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2022; pp. 217–232. [Google Scholar]
- Kaltenbach, D.D.; Jaishankar, D.; Hao, M.; Beer, J.C.; Volin, M.V.; Desai, U.R.; Tiwari, V. Sulfotransferase and Heparanase: Remodeling engines in promoting virus infection and disease development. Front. Pharmacol. 2018, 9, 1315. [Google Scholar] [CrossRef]
- De Pasquale, V.; Pavone, L.M. Heparan sulfate proteoglycans: The sweet side of development turns sour in mucopolysaccharidoses. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 165539. [Google Scholar] [CrossRef]
- Fons, N.R.; Kines, R.C.; Thompson, C.D.; Day, P.M.; Lowy, D.R.; Schiller, J.T. Chondroitin sulfate proteoglycans are de facto cellular receptors for human papillomavirus 16 under high serum conditions. J. Virol. 2022, 96, e0185721. [Google Scholar] [CrossRef]
- Fuster, M.M.; Esko, J.D. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat. Rev. Cancer 2005, 5, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Hammond, E.; Khurana, A.; Shridhar, V.; Dredge, K. The role of heparanase and sulfatases in the modification of heparan sulfate proteoglycans within the tumor microenvironment and opportunities for novel cancer therapeutics. Front. Oncol. 2014, 4, 195. [Google Scholar] [CrossRef]
- Nagarajan, A.; Malvi, P.; Wajapeyee, N. Heparan sulfate and heparan sulfate proteoglycans in cancer initiation and progression. Front. Endocrinol. 2018, 9, 483. [Google Scholar] [CrossRef]
- Jayson, G.C.; Lyon, M.; Paraskeva, C.; Turnbull, J.E.; Deakin, J.A.; Gallagher, J.T. Heparan sulfate undergoes specific structural changes during the progression from human colon adenoma to carcinoma in vitro. J. Biol. Chem. 1998, 273, 51–57. [Google Scholar] [CrossRef]
- Weyers, A.; Yang, B.; Yoon, D.S.; Park, J.H.; Zhang, F.; Lee, K.B.; Linhardt, R.J. A structural analysis of glycosaminoglycans from lethal and nonlethal breast cancer tissues: Toward a novel class of theragnostics for personalized medicine in oncology? Omics 2012, 16, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Vega, I.; García, O.; Crespo, A.; Castañón, S.; Menéndez, P.; Astudillo, A.; Quirós, L.M. Specific genes involved in synthesis and editing of heparan sulfate proteoglycans show altered expression patterns in breast cancer. BMC Cancer 2013, 13, 24. [Google Scholar] [CrossRef]
- Fernández-Vega, I.; García-Suárez, O.; García, B.; Crespo, A.; Astudillo, A.; Quirós, L.M. Heparan sulfate proteoglycans undergo differential expression alterations in right sided colorectal cancer, depending on their metastatic character. BMC Cancer 2015, 15, 742. [Google Scholar] [CrossRef]
- Suhovskih, A.V.; Domanitskaya, N.V.; Tsidulko, A.Y.; Prudnikova, T.Y.; Kashuba, V.I.; Grigorieva, E.V. Tissue-specificity of heparan sulfate biosynthetic machinery in cancer. Cell Adh. Migr. 2015, 9, 452–459. [Google Scholar] [CrossRef]
- Rangel, M.P.; de Sá, V.K.; Prieto, T.; Martins, J.R.M.; Olivieri, E.R.; Carraro, D.; Takagaki, T.; Capelozzi, V.L. Biomolecular analysis of matrix proteoglycans as biomarkers in non small cell lung cancer. Glycoconj. J. 2018, 35, 233–242. [Google Scholar] [CrossRef]
- Denys, A.; Allain, F. The emerging roles of heparan sulfate 3-O-sulfotransferases in cancer. Front. Oncol. 2019, 9, 507. [Google Scholar] [CrossRef]
- Elgundi, Z.; Papanicolaou, M.; Major, G.; Cox, T.R.; Melrose, J.; Whitelock, J.M.; Farrugia, B.L. Cancer Metastasis: The role of the extracellular matrix and the heparan sulfate proteoglycan perlecan. Front. Oncol. 2020, 9, 1482. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.; Reis, C.A.; Vivès, R.R.; Magalhães, A. Heparan sulfate biosynthesis and sulfation profiles as modulators of cancer signalling and progression. Front. Oncol. 2021, 11, 778752. [Google Scholar] [CrossRef]
- Vlodavsky, I.; Friedmann, Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J. Clin. Investig. 2001, 108, 341–347. [Google Scholar] [CrossRef]
- Ramani, V.C.; Purushothaman, A.; Stewart, M.D.; Thompson, C.A.; Vlodavsky, I.; Au, J.L.; Sanderson, R.D. The heparanase/syndecan-1 axis in cancer: Mechanisms and therapies. FEBS J. 2013, 280, 2294–2306. [Google Scholar] [CrossRef]
- Masola, V.; Zaza, G.; Secchi, M.F.; Gambaro, G.; Lupo, A.; Onisto, M. Heparanase is a key player in renal fibrosis by regulating TGF-β expression and activity. Biochim. Biophys. Acta 2014, 1843, 2122–2128. [Google Scholar] [CrossRef] [PubMed]
- Secchi, M.F.; Masola, V.; Zaza, G.; Lupo, A.; Gambaro, G.; Onisto, M. Recent data concerning heparanase: Focus on fibrosis, inflammation and cancer. Biomol. Concepts 2015, 6, 415–421. [Google Scholar] [CrossRef]
- Esko, J.D.; Rostand, K.S.; Weinke, J.L. Tumor Formation Dependent on Proteoglycan Biosynthesis. Science 1988, 241, 1092–1096. [Google Scholar] [CrossRef]
- Liu, D.; Shriver, Z.; Venkataraman, G.; Shabrawi, Y.E.; Sasisekharan, R. Tumor cell surface heparan sulfate as cryptic promoters or inhibitors of tumor growth and metastasis. Proc. Nat. Acad. Sci. USA 2002, 99, 568–573. [Google Scholar] [CrossRef]
- Liu, X.; Zhou, Z.-H.; Li, W.; Zhang, S.-K.; Li, J.; Zhou, M.-J.; Song, J.-W. Heparanase promotes tumor growth and liver metastasis of colorectal cancer cells by activating the p38/MMP1 axis. Front. Oncol. 2019, 9, 216. [Google Scholar] [CrossRef]
- Wang, X.; Zuo, D.; Chen, Y.; Li, W.; Liu, R.; He, Y.; Ren, L.; Zhou, L.; Deng, T.; Wang, X.; et al. Shed Syndecan-1 is involved in chemotherapy resistance via the EGFR pathway in colorectal cancer. Br. J. Cancer 2014, 111, 1965–1976. [Google Scholar] [CrossRef]
- Olah, C.; Tschirdewahn, S.; Hoffmann, M.J.; Krafft, U.; Hadaschik, B.; Nyirady, P.; Szendröi, A.; Módos, O.; Csizmarik, A.; Kovalszky, I.; et al. Soluble syndecan-1 levels are associated with survival in platinum-treated bladder cancer patients. Diagnostics 2020, 10, 864. [Google Scholar] [CrossRef]
- Rangarajan, S.; Richter, J.R.; Richter, R.P.; Bandari, S.K.; Tripathi, K.; Vlodavsky, I.; Sanderson, R.D. Heparanase-enhanced shedding of syndecan-1 and its role in driving disease pathogenesis and progression. J. Histochem. Cytochem. 2020, 68, 823–840. [Google Scholar] [CrossRef]
- Czarnowski, D. Syndecans in cancer: A review of function, expression, prognostic value, and therapeutic significance. Cancer Treat. Res. Commun. 2021, 27, 100312. [Google Scholar] [CrossRef]
- Hilgers, K.; Ibrahim, S.A.; Kiesel, L.; Greve, B.; Espinoza-Sánchez, N.A.; Götte, M. Differential impact of membrane-bound and soluble forms of the prognostic marker syndecan-1 on the invasiveness, migration, apoptosis, and proliferation of cervical cancer cells. Front. Oncol. 2022, 12, 803899. [Google Scholar] [CrossRef] [PubMed]
- Kalscheuer, S.; Khanna, V.; Kim, H.; Li, S.; Sachdev, D.; DeCarlo, A.; Yang, D.; Panyam, J. Discovery of HSPG2 (perlecan) as a therapeutic target in triple negative breast cancer. Sci. Rep. 2019, 9, 12492. [Google Scholar] [CrossRef]
- Folkman, J.; Langer, R.; Linhardt, R.J.; Haudenschild, C.; Taylor, S. Angiogenesis inhibition and tumor regression caused by heparin or a heparin fragment in the presence of cortisone. Science 1983, 221, 719–725. [Google Scholar] [CrossRef] [PubMed]
- Askari Rizvi, S.F.; Zhang, H. Emerging trends of receptor-mediated tumor targeting peptides: A review with perspective from molecular imaging modalities. Eur. J. Med. Chem. 2021, 221, 113538. [Google Scholar] [CrossRef]
- Berdiaki, A.; Neagu, M.; Giatagana, E.-M.; Kuskov, A.; Tsatsakis, A.M.; Tzanakakis, G.N.; Nikitovic, D. Glycosaminoglycans: Carriers and targets for tailored anti-cancer therapy. Biomolecules 2021, 11, 395. [Google Scholar] [CrossRef]
- Bloise, N.; Okkeh, M.; Restivo, E.; Della Pina, C.; Visai, L. Targeting the “Sweet Side” of tumor with glycan-binding molecules conjugated-nanoparticles: Implications in cancer therapy and diagnosis. Nanomaterials 2021, 11, 289. [Google Scholar] [CrossRef] [PubMed]
- McKee, T.D.; Grandi, P.; Mok, W.; Alexandrakis, G.; Insin, N.; Zimmer, J.P.; Bawendi, M.G.; Boucher, Y.; Breakefield, X.O.; Jain, R.K. Degradation of fibrillar collagen in a human melanoma xenograft improves the efficacy of an oncolytic herpes simplex virus vector. Cancer Res. 2006, 66, 2509–2513. [Google Scholar] [CrossRef]
- Farrera-Sal, M.; Moreno, R.; Mato-Berciano, A.; Maliandi, M.V.; Bazan-Peregrino, M.; Alemany, R. Hyaluronidase expression within tumors increases virotherapy efficacy and T cell accumulation. Mol. Ther. Oncolytics 2021, 22, 27–35. [Google Scholar] [CrossRef]
- Gao, W.; Tang, Z.; Zhang, Y.F.; Feng, M.; Qian, M.; Dimitrov, D.S.; Ho, M. Immunotoxin targeting glypican-3 regresses liver cancer via dual inhibition of Wnt signalling and protein synthesis. Nat. Commun. 2015, 6, 6536. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Gao, W.; Feng, M.; Pastan, I.; Ho, M. Construction of an immunotoxin, HN3-mPE24, targeting glypican-3 for liver cancer therapy. Oncotarget 2017, 8, 32450–32460. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.X.; Gold, P.J.; El-Khoueiry, A.B.; Abrams, T.A.; Morikawa, H.; Ohishi, N.; Ohtomo, T.; Philip, P.A. First-in-man phase I study of GC33, a novel recombinant humanized antibody against glypican-3, in patients with advanced hepatocellular carcinoma. Clin. Cancer Res. 2013, 19, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Abou-Alfa, G.K.; Puig, O.; Daniele, B.; Kudo, M.; Merle, P.; Park, J.W.; Ross, P.; Peron, J.M.; Ebert, O.; Chan, S.; et al. Randomized phase II placebo controlled study of codrituzumab in previously treated patients with advanced hepatocellular carcinoma. J. Hepatol. 2016, 65, 289–295. [Google Scholar] [CrossRef]
- Zhang, Y.-F.; Ho, M. Humanization of high-affinity antibodies targeting glypican-3 in hepatocellular carcinoma. Sci. Rep. 2016, 6, 33878. [Google Scholar] [CrossRef]
- Fu, Y.; Urban, D.J.; Nani, R.R.; Zhang, Y.-F.; Li, N.; Fu, H.; Shah, H.; Gorka, A.P.; Guha, R.; Chen, L.; et al. Glypican-3-Specific Antibody Drug Conjugates Targeting Hepatocellular Carcinoma. Hepatology 2019, 70, 563–576. [Google Scholar] [CrossRef]
- Hanaoka, H.; Nagaya, T.; Sato, K.; Nakamura, Y.; Watanabe, R.; Harada, T.; Gao, W.; Feng, M.; Phung, Y.; Kim, I.; et al. Glypican-3 targeted human heavy chain antibody as a drug carrier for hepatocellular carcinoma therapy. Mol. Pharm. 2015, 12, 2151–2157. [Google Scholar] [CrossRef]
- Liu, X.; Gao, F.; Jiang, L.; Jia, M.; Ao, L.; Lu, M.; Gou, L.; Ho, M.; Jia, S.; Chen, F.; et al. 32A9, a novel human antibody for designing an immunotoxin and CAR-T cells against glypican-3 in hepatocellular carcinoma. J. Transl. Med. 2020, 18, 295. [Google Scholar] [CrossRef]
- Ishiguro, T.; Sano, Y.; Komatsu, S.I.; Kamata-Sakurai, M.; Kaneko, A.; Kinoshita, Y.; Shiraiwa, H.; Azuma, Y.; Tsunenari, T.; Kayukawa, Y.; et al. An anti-glypican 3/CD3 bispecific T cell-redirecting antibody for treatment of solid tumors. Sci. Transl. Med. 2017, 9, eaal4291. [Google Scholar] [CrossRef]
- Pan, J.; Li, N.; Renn, A.; Zhu, H.; Chen, L.; Shen, M.; Hall, M.D.; Qian, M.; Pastan, I.; Ho, M. GPC1-targeted immunotoxins inhibit pancreatic tumor growth in mice via depletion of short-lived GPC1 and downregulation of Wnt signaling. Mol. Can. Ther. 2022, 21, 960–973. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Fu, H.; Hewitt, S.M.; Dimitrov, D.S.; Ho, M. Therapeutically targeting glypican-2 via single-domain antibody-based chimeric antigen receptors and immunotoxins in neuroblastoma. Proc. Nat. Acad. Sci. USA 2017, 114, E6623–E6631. [Google Scholar] [CrossRef]
- Gao, H.; Li, K.; Tu, H.; Pan, X.; Jiang, H.; Shi, B.; Kong, J.; Wang, H.; Yang, S.; Gu, J.; et al. Development of T cells redirected to glypican-3 for the treatment of hepatocellular carcinoma. Clin Cancer Res 2014, 20, 6418–6428. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Jiang, X.; Chen, S.; Lai, Y.; Wei, X.; Li, B.; Lin, S.; Wang, S.; Wu, Q.; Liang, Q.; et al. Anti-GPC3-CAR T cells suppress the growth of tumor cells in patient-derived xenografts of hepatocellular carcinoma. Front. Immunol. 2017, 7, 690. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Li, N.; Zhang, Y.F.; Fu, H.; Feng, M.; Schneider, D.; Su, L.; Wu, X.; Zhou, J.; Mackay, S.; et al. Persistent polyfunctional chimeric antigen receptor T cells that target glypican 3 eliminate orthotopic hepatocellular carcinomas in mice. Gastroenterology 2020, 158, 2250–2265.e2220. [Google Scholar] [CrossRef]
- Shi, D.; Shi, Y.; Kaseb, A.O.; Qi, X.; Zhang, Y.; Chi, J.; Lu, Q.; Gao, H.; Jiang, H.; Wang, H.; et al. Chimeric antigen receptor-glypican-3 T-Cell therapy for advanced hepatocellular carcinoma: Results of phase I trials. Clin. Cancer Res. 2020, 26, 3979–3989. [Google Scholar] [CrossRef]
- Elson-Schwab, L.; Garner, O.B.; Schuksz, M.; Crawford, B.E.; Esko, J.D.; Tor, Y. Guanidinylated neomycin delivers large, bioactive cargo into cells through a heparan sulfate-dependent pathway. J. Biol. Chem. 2007, 282, 13585–13591. [Google Scholar] [CrossRef]
- Rapraeger, A.C.; Ell, B.J.; Roy, M.; Li, X.; Morrison, O.R.; Thomas, G.M.; Beauvais, D.M. Vascular endothelial-cadherin stimulates syndecan-1-coupled insulin-like growth factor-1 receptor and cross-talk between αVβ3 integrin and vascular endothelial growth factor receptor 2 at the onset of endothelial cell dissemination during angiogenesis. FEBS J. 2013, 280, 2194–2206. [Google Scholar] [CrossRef]
- Melo, C.M.; Wang, H.; Fujimura, K.; Strnadel, J.; Meneghetti, M.C.Z.; Nader, H.B.; Klemke, R.L.; Pinhal, M.A.S. The heparan sulfate binding peptide in tumor progression of triple-negative breast cancer. Front. Oncol. 2021, 11, 697626. [Google Scholar] [CrossRef]
- Brunetti, J.; Pillozzi, S.; Falciani, C.; Depau, L.; Tenori, E.; Scali, S.; Lozzi, L.; Pini, A.; Arcangeli, A.; Menichetti, S.; et al. Tumor-selective peptide-carrier delivery of Paclitaxel increases in vivo activity of the drug. Sci. Rep. 2015, 5, 17736. [Google Scholar] [CrossRef][Green Version]
- Brunetti, J.; Depau, L.; Falciani, C.; Gentile, M.; Mandarini, E.; Riolo, G.; Lupetti, P.; Pini, A.; Bracci, L. Insights into the role of sulfated glycans in cancer cell adhesion and migration through use of branched peptide probe. Sci. Rep. 2016, 6, 27174. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, J.; Riolo, G.; Depau, L.; Mandarini, E.; Bernini, A.; Karousou, E.; Passi, A.; Pini, A.; Bracci, L.; Falciani, C. Unraveling heparan sulfate proteoglycan binding motif for cancer cell selectivity. Front. Oncol. 2019, 9, 843. [Google Scholar] [CrossRef]
- Zheng, X.; Gai, X.; Han, S.; Moser, C.D.; Hu, C.; Shire, A.M.; Floyd, R.A.; Roberts, L.R. The human sulfatase 2 inhibitor 2,4-disulfonylphenyl-tert-butylnitrone (OKN-007) has an antitumor effect in hepatocellular carcinoma mediated via suppression of TGFB1/SMAD2 and Hedgehog/GLI1 signaling. Genes Chromosomes Cancer 2013, 52, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.M.; Hosono-Fukao, T.; Tang, R.; Sugaya, N.; van Kuppevelt, T.H.; Jenniskens, G.J.; Kimata, K.; Rosen, S.D.; Uchimura, K. Direct detection of HSulf-1 and HSulf-2 activities on extracellular heparan sulfate and their inhibition by PI-88. Glycobiology 2010, 20, 175–186. [Google Scholar] [CrossRef]
- Chen, P.J.; Lee, P.H.; Han, K.H.; Fan, J.; Cheung, T.T.; Hu, R.H.; Paik, S.W.; Lee, W.C.; Chau, G.Y.; Jeng, L.B.; et al. A phase III trial of muparfostat (PI-88) as adjuvant therapy in patients with hepatitis virus related hepatocellular carcinoma (HV-HCC) after resection. Ann. Oncol. 2017, 28, v213. [Google Scholar] [CrossRef]
- Marchetti, D.; Reiland, J.; Erwin, B.; Roy, M. Inhibition of heparanase activity and heparanase-induced angiogenesis by suramin analogues. Int. J. Cancer 2003, 104, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Dredge, K.; Hammond, E.; Davis, K.; Li, C.P.; Liu, L.; Johnstone, K.; Handley, P.; Wimmer, N.; Gonda, T.J.; Gautam, A.; et al. The PG500 series: Novel heparan sulfate mimetics as potent angiogenesis and heparanase inhibitors for cancer therapy. Investig. New Drugs 2010, 28, 276–283. [Google Scholar] [CrossRef]
- Dredge, K.; Hammond, E.; Handley, P.; Gonda, T.J.; Smith, M.T.; Vincent, C.; Brandt, R.; Ferro, V.; Bytheway, I. PG545, a dual heparanase and angiogenesis inhibitor, induces potent anti-tumour and anti-metastatic efficacy in preclinical models. Br. J. Cancer 2011, 104, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Hammond, E.; Handley, P.; Dredge, K.; Bytheway, I. Mechanisms of heparanase inhibition by the heparan sulfate mimetic PG545 and three structural analogues. FEBS Open Bio. 2013, 3, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Roy, S.; Cochran, E.; Zouaoui, R.; Chu, C.L.; Duffner, J.; Zhao, G.; Smith, S.; Galcheva-Gargova, Z.; Karlgren, J.; et al. M402, a novel heparan sulfate mimetic, targets multiple pathways implicated in tumor progression and metastasis. PLoS ONE 2011, 6, e21106. [Google Scholar] [CrossRef]
- Ritchie, J.P.; Ramani, V.C.; Ren, Y.; Naggi, A.; Torri, G.; Casu, B.; Penco, S.; Pisano, C.; Carminati, P.; Tortoreto, M.; et al. SST0001, a chemically modified heparin, inhibits myeloma growth and angiogenesis via disruption of the heparanase/syndecan-1 axis. Clin. Cancer Res. 2011, 17, 1382–1393. [Google Scholar] [CrossRef]
- Raman, K.; Ninomiya, M.; Nguyen, T.K.N.; Tsuzuki, Y.; Koketsu, M.; Kuberan, B. Novel glycosaminoglycan biosynthetic inhibitors affect tumor-associated angiogenesis. Biochem. Biophys. Res. Commun. 2011, 404, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Chua, J.S.; Kuberan, B. Synthetic Xylosides: Probing the glycosaminoglycan biosynthetic machinery for biomedical applications. Acc. Chem. Res. 2017, 50, 2693–2705. [Google Scholar] [CrossRef]
- Ranki, T.; Kanerva, A.; Ristimäki, A.; Hakkarainen, T.; Särkioja, M.; Kangasniemi, L.; Raki, M.; Laakkonen, P.; Goodison, S.; Hemminki, A. A heparan sulfate-targeted conditionally replicative adenovirus, Ad5.pk7-Delta24, for the treatment of advanced breast cancer. Gene Ther. 2007, 14, 58–67. [Google Scholar] [CrossRef]
- Yu, T.; Li, Y.; Gu, X.; Li, Q. Development of a hyaluronic acid-based nanocarrier incorporating doxorubicin and cisplatin as a pH-sensitive and CD44-targeted anti-breast cancer drug delivery system. Front. Pharmacol. 2020, 11, 532457. [Google Scholar] [CrossRef]
- Salanti, A.; Clausen, T.M.; Agerbæk, M.; Al Nakouzi, N.; Dahlbäck, M.; Oo, H.Z.; Lee, S.; Gustavsson, T.; Rich, J.R.; Hedberg, B.J.; et al. Targeting human cancer by a glycosaminoglycan binding malaria protein. Cancer Cell 2015, 28, 500–514. [Google Scholar] [CrossRef]
- Seiler, R.; Oo, H.Z.; Tortora, D.; Clausen, T.M.; Wang, C.K.; Kumar, G.; Pereira, M.A.; Ørum-Madsen, M.S.; Agerbæk, M.; Gustavsson, T.; et al. An oncofetal glycosaminoglycan modification provides therapeutic access to cisplatin-resistant bladder cancer. Eur. Urol. 2017, 72, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Zhou, J.; Li, Z.; Appelman, H.D.; Zhao, L.; Zhu, J.; Wang, T.D. Sorafenib encapsulated in nanocarrier functionalized with glypican-3 specific peptide for targeted therapy of hepatocellular carcinoma. Colloids Surf. B Biointerfaces 2019, 184, 110498. [Google Scholar] [CrossRef] [PubMed]
- Kuo, P.H.; Teng, Y.H.; Cin, A.L.; Han, W.; Huang, P.W.; Wang, L.H.; Chou, Y.T.; Yang, J.L.; Tseng, Y.L.; Kao, M.; et al. Heparan sulfate targeting strategy for enhancing liposomal drug accumulation and facilitating deep distribution in tumors. Drug Deliv. 2020, 27, 542–555. [Google Scholar] [CrossRef]
- Park, J.O.; Stephen, Z.; Sun, C.; Veiseh, O.; Kievit, F.M.; Fang, C.; Leung, M.; Mok, H.; Zhang, M. Glypican-3 targeting of liver cancer cells using multifunctional nanoparticles. Mol. Imaging 2011, 10, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-J.; Tsai, K.-C.; Kuo, P.-H.; Chang, P.-L.; Wang, W.-C.; Chuang, Y.-J.; Chang, M.D.-T. A heparan sulfate-binding cell penetrating peptide for tumor targeting and migration inhibition. Biomed. Res. Int. 2015, 2015, 237969. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Fan, C.; Zhu, H.; Le, W.; Cui, S.; Chen, X.; Li, W.; Zhang, F.; Huang, Y.; Sh, D.; et al. Glypican-1-antibody-conjugated Gd-Au nanoclusters for FI/MRI dual-modal targeted detection of pancreatic cancer. Int. J. Nanomed. 2018, 13, 2585–2599. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Zhang, H.; Chen, X.; Song, L.; Cui, W.; Ren, S.; Wang, Y.; Guo, K.; Li, D.; Chen, R.; et al. A GPC1-targeted and gemcitabine-loaded biocompatible nanoplatform for pancreatic cancer multimodal imaging and therapy. Nanomedicine 2019, 14, 2339–2353. [Google Scholar] [CrossRef]
- Kines, R.C.; Cerio, R.J.; Roberts, J.N.; Thompson, C.D.; de Los Pinos, E.; Lowy, D.R.; Schiller, J.T. Human papillomavirus capsids preferentially bind and infect tumor cells. Int. J. Cancer 2016, 138, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.-F.; Chiang, A.J.; Tsai, H.-H.; Pomper, M.G.; Kang, T.H.; Roden, R.R.; Wu, T.C. Ovarian cancer gene therapy using HPV-16 pseudovirion carrying the HSV-TK gene. PLoS ONE 2012, 7, e40983. [Google Scholar] [CrossRef] [PubMed]
- Hojeij, R.; Domingos-Pereira, S.; Nkosi, M.; Gharbi, D.; Derré, L.; Schiller, J.T.; Jichlinski, P.; Nardelli-Haefliger, D. Immunogenic human papillomavirus pseudovirus-mediated suicide-gene therapy for bladder cancer. Int. J. Mol. Sci. 2016, 17, 1125. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, Y.; Maeda, H. A new concept for macromolecular therapeutics in cancer chemotherapy: Mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986, 46, 6387–6392. [Google Scholar] [PubMed]
- Tang, L.; Yang, X.; Yin, Q.; Cai, K.; Wang, H.; Chaudhury, I.; Yao, C.; Zhou, Q.; Kwon, M.; Hartman, J.A.; et al. Investigating the optimal size of anticancer nanomedicine. Proc. Nat. Acad. Sci. USA 2014, 111, 15344–15349. [Google Scholar] [CrossRef]
- Mitsunaga, M.; Ogawa, M.; Kosaka, N.; Rosenblum, L.T.; Choyke, P.L.; Kobayashi, H. Cancer cell-selective in vivo near infrared photoimmunotherapy targeting specific membrane molecules. Nat. Med. 2011, 17, 1685–1691. [Google Scholar] [CrossRef] [PubMed]
- Kines, R.C.; Varsavsky, I.; Choudhary, S.; Bhattacharya, D.; Spring, S.; McLaughlin, R.; Kang, S.J.; Grossniklaus, H.E.; Vavvas, D.; Monks, S.; et al. An infrared dye-conjugated virus-like particle for the treatment of primary uveal melanoma. Mol. Cancer Ther. 2018, 17, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Kines, R.C.; Thompson, C.D.; Spring, S.; Li, Z.; de Los Pinos, E.; Monks, S.; Schiller, J.T. Virus-like particle-drug conjugates induce protective, long-lasting adaptive antitumor immunity in the absence of specifically targeted tumor antigens. Cancer Immunol. Res. 2021, 9, 693–706. [Google Scholar] [CrossRef]
- Lenz, P.; Thompson, C.D.; Day, P.M.; Bacot, S.M.; Lowy, D.R.; Schiller, J.T. Interaction of papillomavirus virus-like particles with human myeloid antigen-presenting cells. Clin. Immunol. 2003, 106, 231–237. [Google Scholar] [CrossRef]
- Jager, M.J.; Shields, C.L.; Cebulla, C.M.; Abdel-Rahman, M.H.; Grossniklaus, H.E.; Stern, M.-H.; Carvajal, R.D.; Belfort, R.N.; Jia, R.; Shields, J.A.; et al. Uveal melanoma. Nat. Rev. Dis. Primers 2020, 6, 24. [Google Scholar] [CrossRef]
- Savinainen, A.; Grossniklaus, H.; Kang, S.; Rasmussen, C.; Bentley, E.; Krakova, Y.; Struble, C.B.; Rich, C. Ocular distribution and efficacy after suprachoroidal injection of AU-011 for treatment of ocular melanoma. Investig. Ophthalmol. Vis. Sci. 2020, 61, 3615. [Google Scholar]
- Narvekar, A.; Rich, C.; Savinainen, A.; Kim, I.K. Nanoparticles for the Treatment of Uveal Melanoma. In Uveal Melanoma: Biology and Management; Bernicker, E.H., Ed.; Springer Nature: Cham, Switzerland, 2021; pp. 135–149. [Google Scholar]

| Pathogen | Ligand | Reference |
|---|---|---|
| Viruses | ||
| Adenovirus (AdV) | fiber | [39,40] |
| Dengue virus | envelope | [41] |
| Hepatitis B virus (HBV) | L-envelope | [42,43] |
| Hepatitis C virus (HCV) | E2 envelope | [44,45] |
| Hepatitis E virus (HEV) | ORF2 capsid protein | [46] |
| Human immunodeficiency virus (HIV) | gp120, Tat | [47,48,49,50] |
| Human cytomegalovirus (HMCV) | gB | [51,52,53] |
| Human papillomavirus (HPV) | L1 capsid potein | [28,30,32] |
| Herpes simplex virus type 1 (HSV-1) | gB, gC, gD | [54,55,56,57,58,59] |
| Herpes simplex virus type 2 (HSV-2) | gB, gC | [54,59,60,61] |
| Merkel cell polyomavirus (MCPyV) | VP1 | [62,63] |
| Severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) | spike | [64,65] |
| Varicella zoster virus (VZV) | gB | [59,66] |
| Parasites | ||
| Giardia lamblia | Alpha-1 giardin | [67] |
| Plasmodium falciparum | BAEBL, VAR2CSA, CS | [68,69,70] |
| Toxoplasma gondii | SAG3, ROP2, ROP4, GRA2, SAG1 | [71,72,73,74] |
| Trypanosoma cruzi | Heparin binding proteins (HBP) | [75,76,77] |
| Bacteria | ||
| Bordatella pertussis | FHA | [78] |
| Helicobacter pylori | VacA | [79,80] |
| Listeria monocytogenes | ActA | [81] |
| Mycobacterium tuberculosis | HA | [82] |
| Neisseria gonorrhoaea | Opa | [83,84] |
| Other | ||
| Candida albicans | n.d. | [85,86,87] |
| Malassezia spp. | n.d. | [87] |
| Prion | PrP | [88,89,90,91] |
| Name | Target/Mechanism | Reference |
|---|---|---|
| Antibodies | ||
| HN3 | PE38, PE24 conjugates targeting glypican-3 | [129,130] |
| GC33 | ADCC; targeting glypican-3 | [131,132] |
| YP7 | PE38, Duocarmycin, IRdye700DX conjugates; pyrrolobenzodiazepine dimer; targeting glypican-3 | [129,133,134,135] |
| 32A9 | PE24 conjugates targeting glypican-3 | [136] |
| ERY974 | bi-specific antibody agains glypican-3 and CD3 | [137] |
| D4 (camel) | PE38-conjugated camelid nanobody targeting glypican-1 | [138] |
| LH7 | PE38-conjugated human single domain anti-glypican-2 | [139] |
| CAR-T | ||
| GC33 | Glypican-3 | [140,141] |
| hYP7 | Glypican-3 | [142] |
| 32A9 | Glypican-3 | [136] |
| Y035 | Glypican-3 | [143] |
| LH7 | Glypican-2 | [139] |
| Small molecule/peptide mimics/false substrates | ||
| Guanidinylated neomycin (Gneo) | LMW HS binding peptide carrying saporin | [144] |
| Synstatin (SSTN)92–119 | Peptide blocks syndecan-1/IGF1R complex-blocks integrin signaling and VEGFR2 activation | [145] |
| RGWRGEKIGN peptide | HS binding peptide blocks FGF2/HS binding | [146] |
| NT4 | General heparin, HSPG, CSPG mimetic (tetra-branched polypeptide); interferes with cell migration; delivers paclitaxel | [147,148,149] |
| OKN-007 | Sulfatase-2 inhibitor | [150] |
| PI-88 (muparfostat) | Heparanase inhibitor (heparin mimetic); interferes with VEGF, FGF1, FGF2 leading to reduction in angiogenesis and sulf1 and sulf2 activity | [151,152] |
| Suramin analogs | Heparanse inhibition; inhibits degradation of ECM and blocks angiogenic events by preventing release of FGF from ECM HS | [153] |
| PG545 | HS mimetic; blocks heparanase activity; prevents growth factor release and activation | [154,155,156] |
| M402 (neuparanib) | HS mimetic; inhibits HS interactions and activity of VEGF, FGF2, SDF-1α, P-selectin, and heparanase | [157] |
| SST0001 (roneparstat) | Split heparin; inhibits heparanase, downregulates HGF, VEGF, and MMP-9 expression and suppresses angiogenesis | [158] |
| Xylosides | Blocks GAG biosynthesis | [159,160] |
| Nanoparticles/Pathogens | ||
| Ad5 | Fiber modified to bind HSPG (bypass CAR) | [161] |
| hyaluronic acid micelle nanocarrier | Hyaluronic acid nanocarrier targeting CD44; incorporate doxorubicin and cisplatin | [162] |
| rVAR2CSA | Targets oncofetal CS (CD44, CDPG4; syndecan-1); conjugated with diptheria toxin or hemiasterlin | [163,164] |
| liposomes | Composed of glypican-3 targeting peptide incorporating sorafenib; GAG binding peptide incorporating doxorubicin | [165,166] |
| metal conjugates | HSPG targeted peptide and glypican-3 antibody delivering Fe3O4 for imaging; Gold nanocluster with gadolinum conjugated to anti-glypican-1; Gold nanocages incorporating gemcetabine conjugated to anti-glypican-1 for theranostics | [167,168,169,170] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kines, R.C.; Schiller, J.T. Harnessing Human Papillomavirus’ Natural Tropism to Target Tumors. Viruses 2022, 14, 1656. https://doi.org/10.3390/v14081656
Kines RC, Schiller JT. Harnessing Human Papillomavirus’ Natural Tropism to Target Tumors. Viruses. 2022; 14(8):1656. https://doi.org/10.3390/v14081656
Chicago/Turabian StyleKines, Rhonda C., and John T. Schiller. 2022. "Harnessing Human Papillomavirus’ Natural Tropism to Target Tumors" Viruses 14, no. 8: 1656. https://doi.org/10.3390/v14081656
APA StyleKines, R. C., & Schiller, J. T. (2022). Harnessing Human Papillomavirus’ Natural Tropism to Target Tumors. Viruses, 14(8), 1656. https://doi.org/10.3390/v14081656





