A Deacetylase CqSIRT1 Promotes WSSV Infection by Binding to Viral Envelope Proteins in Cherax quadricarinatus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals, Hpt Cell Cultures and WSSV
2.2. RNA Extraction and Reverse Transcription
2.3. Gene Cloning and Molecular Characteristics Analysis
2.4. Semi-Quantitative RT-PCR and Real-Time Fluorescent Quantitative PCR
2.5. Double-Strand RNA Production and RNA Interference
2.6. Pharmacological Treatment and Cytotoxicity Test
2.7. Prokaryotic Expression and Purification of Recombinant Protein
2.8. Pull-Down Assay
2.9. Transfections and Co-Immunoprecipitation
2.10. SDS-PAGE and Western Blot Analysis
2.11. Immunofluorescence Assay
2.12. Statistical Analysis
3. Results
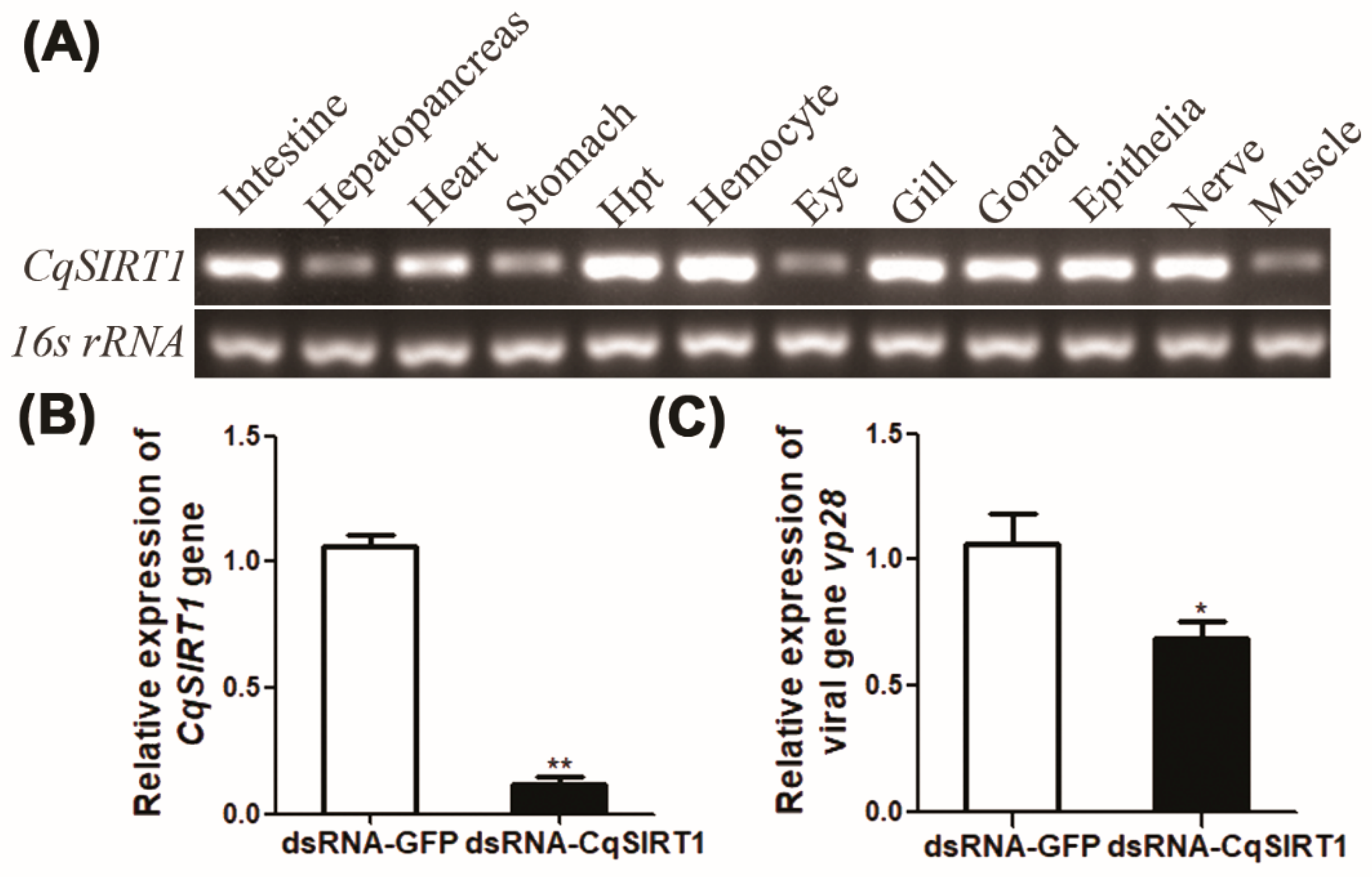
3.1. Molecular Characterization of CqSIRT1 from Red Claw Crayfish C. quadricarinatus
3.2. Tissue Distribution and the Effect of CqSIRT1 on WSSV Gene Transcription
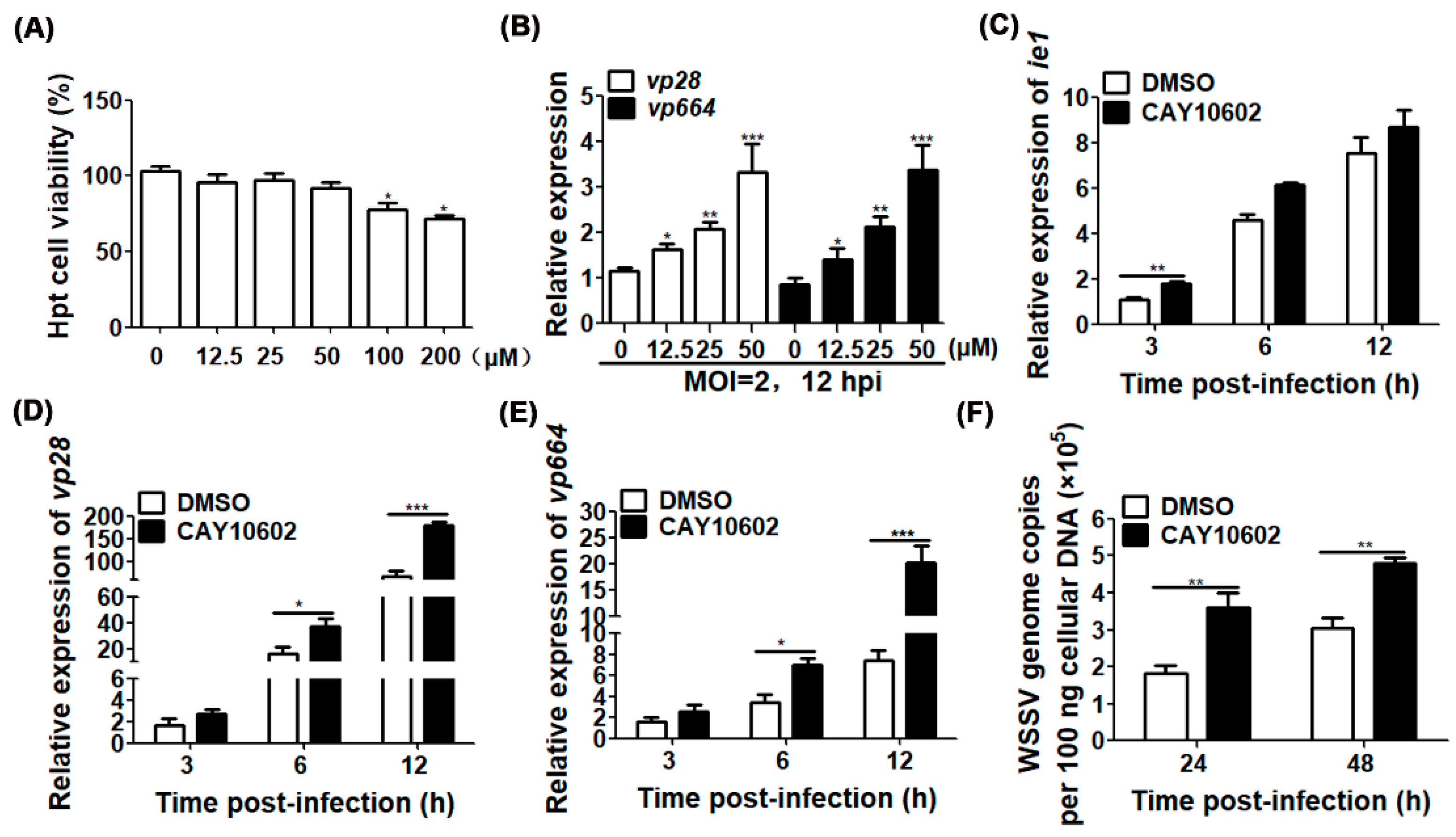
3.3. Enhancement of Deacetylase Activity of CqSIRT1 Promotes WSSV Replication
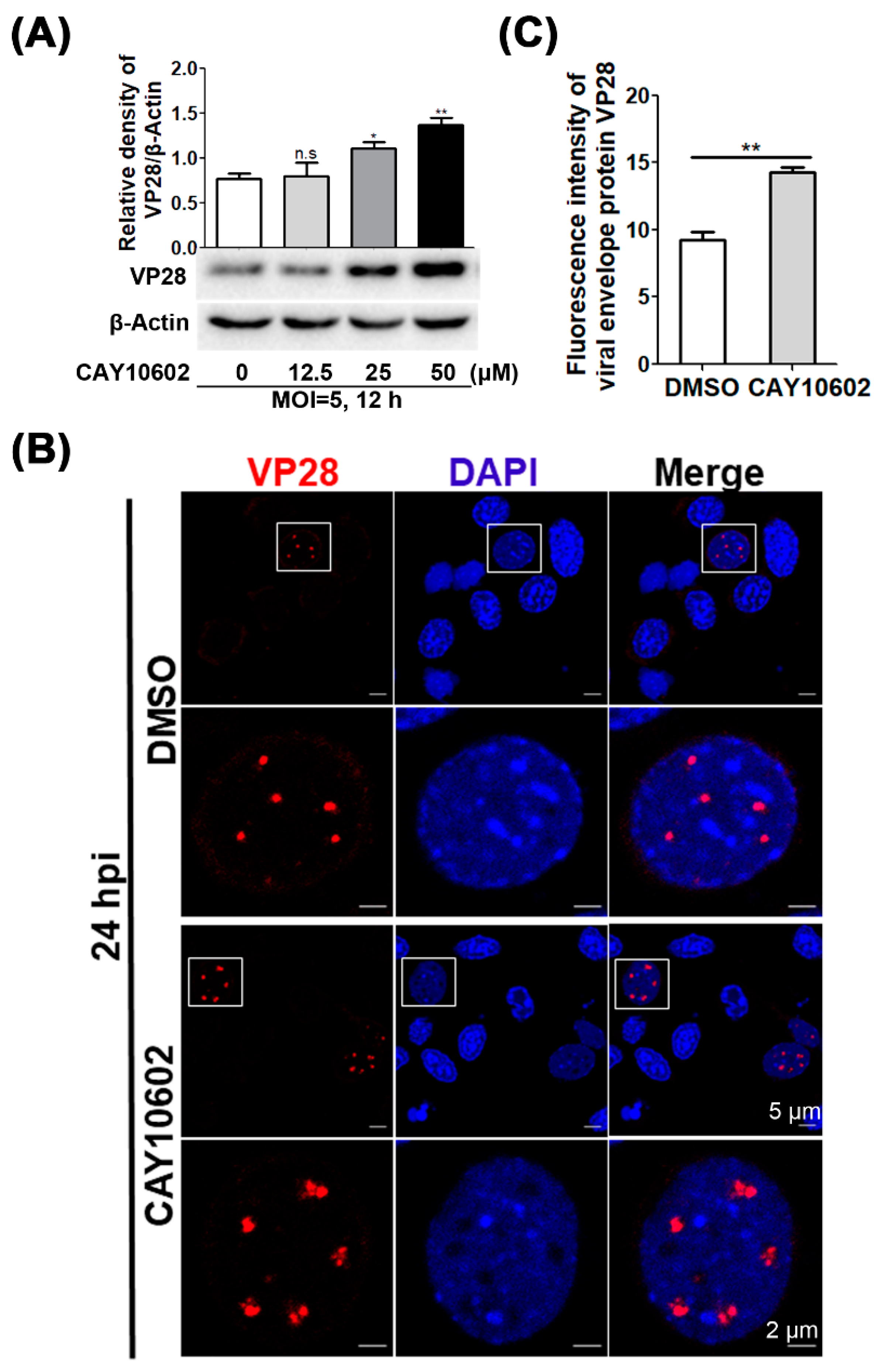
3.4. Enhancement of Deacetylase Activity of CqSIRT1 Facilitates VP28 Protein Expression
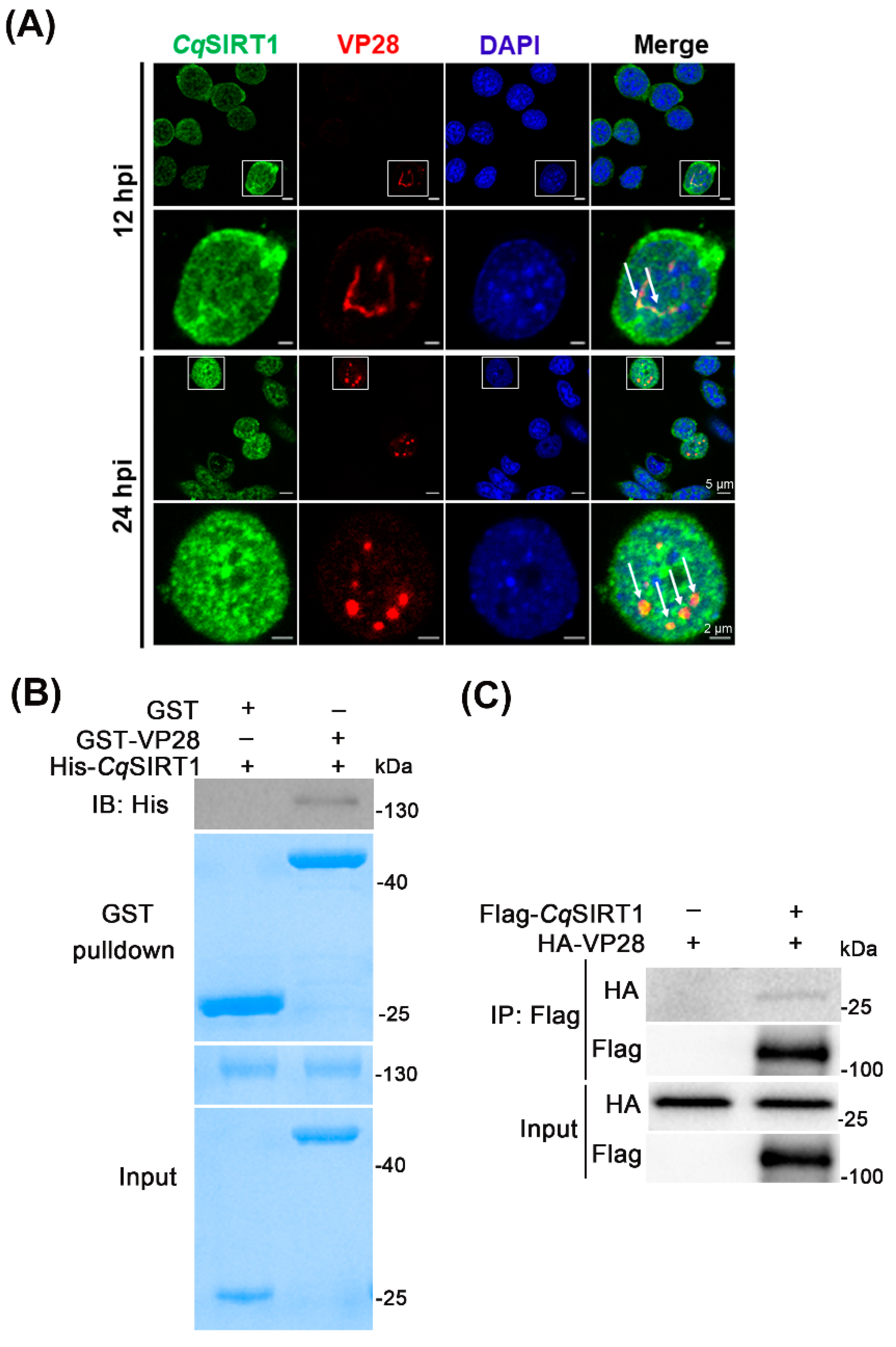
3.5. CqSIRT1 Co-localizes with VP28 in the Nuclei of Hpt Cells and Binds with VP28
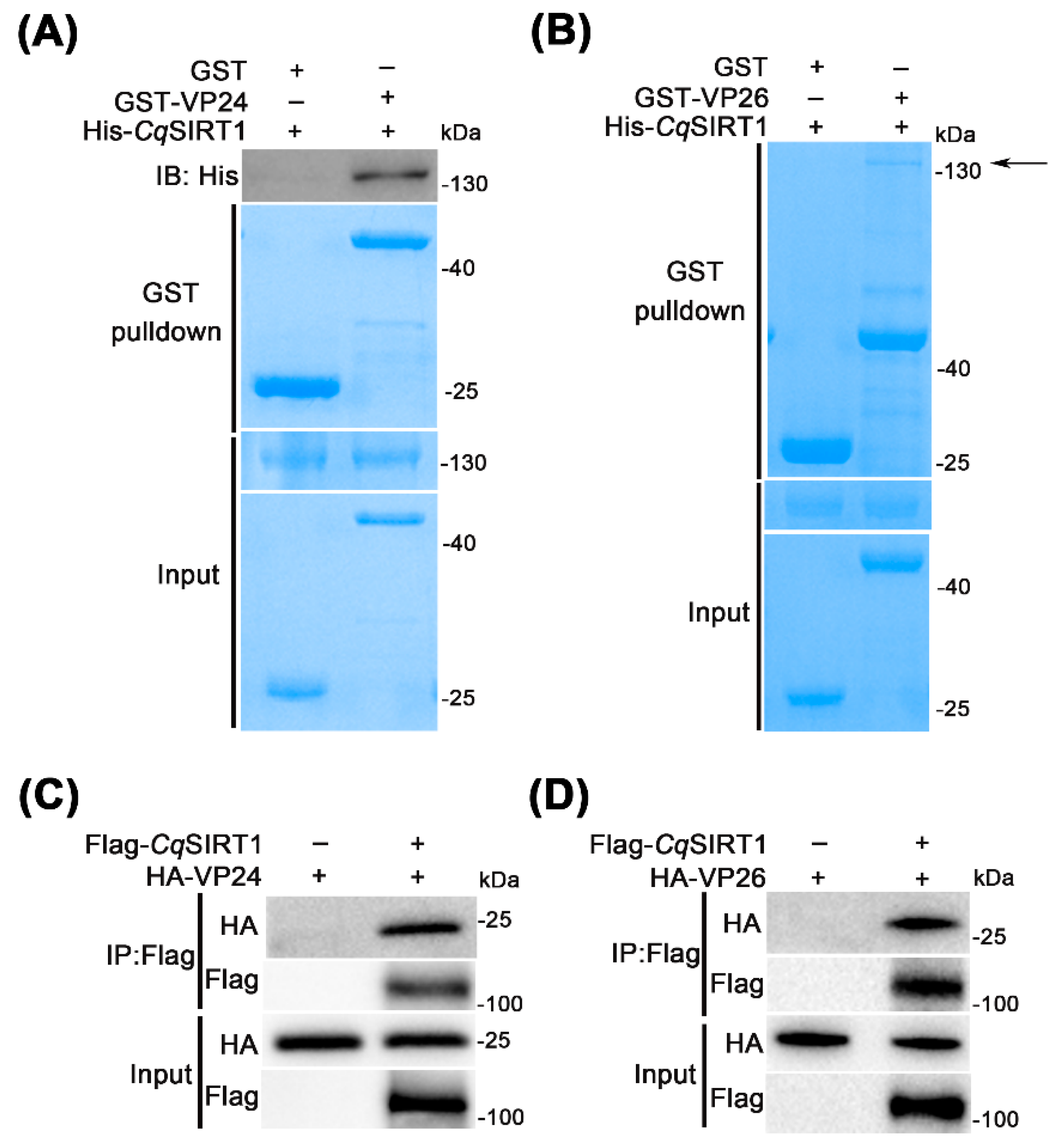
3.6. CqSIRT1 also Binds with Viral Envelope Proteins VP24 and VP26
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ali, I.; Conrad, R.J.; Verdin, E.; Ott, M. Lysine Acetylation Goes Global: From Epigenetics to Metabolism and Therapeutics. Chem. Rev. 2018, 118, 1216–1252. [Google Scholar] [CrossRef] [PubMed]
- Houtkooper, R.H.; Pirinen, E.; Auwerx, J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol. 2012, 13, 225–238. [Google Scholar] [CrossRef] [PubMed]
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. [Google Scholar] [CrossRef] [PubMed]
- Koyuncu, E.; Budayeva, H.G.; Miteva, Y.V.; Ricci, D.P.; Silhavy, T.J.; Shenk, T.; Cristea, I.M. Sirtuins Are Evolutionarily Conserved Viral Restriction Factors. Mbio 2014, 5, e02249-14. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; He, M.; Zhou, F.; Ye, F.; Gao, S.J. Activation of Kaposi’s sarcoma-associated herpesvirus (KSHV) by inhibitors of class III histone deacetylases: Identification of sirtuin 1 as a regulator of the KSHV life cycle. J. Virol. 2014, 88, 6355–6367. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.J.; Kong, K.E.; Gao, W.W.; Tang, H.V.; Chaudhary, V.; Cheng, Y.; Zhou, J.; Chan, C.P.; Wong, D.K.; Yuen, M.F.; et al. Interplay between SIRT1 and hepatitis B virus X protein in the activation of viral transcription. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 491–501. [Google Scholar] [CrossRef]
- Ren, J.H.; Tao, Y.; Zhang, Z.Z.; Chen, W.X.; Cai, X.F.; Chen, K.; Ko, B.C.; Song, C.L.; Ran, L.K.; Li, W.Y.; et al. Sirtuin 1 regulates hepatitis B virus transcription and replication by targeting transcription factor AP-1. J. Virol. 2014, 88, 2442–2451. [Google Scholar] [CrossRef]
- Das, D.; Smith, N.; Wang, X.; Morgan, I.M. The Deacetylase SIRT1 Regulates the Replication Properties of Human Papillomavirus 16 E1 and E2. J. Virol. 2017, 91, e00102-17. [Google Scholar] [CrossRef]
- Langsfeld, E.S.; Bodily, J.M.; Laimins, L.A. The Deacetylase Sirtuin 1 Regulates Human Papillomavirus Replication by Modulating Histone Acetylation and Recruitment of DNA Damage Factors NBS1 and Rad51 to Viral Genomes. PLoS Pathog. 2015, 11, e1005181. [Google Scholar] [CrossRef]
- Zheng, S.C.; Xu, J.Y.; Liu, H.P. Cellular entry of white spot syndrome virus and antiviral immunity mediated by cellular receptors in crustaceans. Fish Shellfish Immunol. 2019, 93, 580–588. [Google Scholar] [CrossRef]
- Li, D.L.; Yang, M.H.; Liu, L.K.; Meng, C.; Li, M.Q.; Liu, H.P. Invasion and Propagation of White Spot Syndrome Virus: Hijacking of the Cytoskeleton, Intracellular Transport Machinery, and Nuclear Import Transporters. J. Virol. 2022, 96, e0220521. [Google Scholar] [CrossRef] [PubMed]
- Meng, C.; Liu, L.K.; Li, D.L.; Gao, R.L.; Fan, W.W.; Wang, K.J.; Wang, H.C.; Liu, H.P. White Spot Syndrome Virus Benefits from Endosomal Trafficking, Substantially Facilitated by a Valosin-Containing Protein, to Escape Autophagic Elimination and Propagate in the Crustacean Cherax quadricarinatus. J. Virol. 2020, 94, e01570-20. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.Y.; Shen, K.L.; Chen, Z.; Fan, W.W.; Xie, X.L.; Meng, C.; Chang, X.J.; Zheng, L.B.; Jeswin, J.; Li, C.H.; et al. White spot syndrome virus entry is dependent on multiple endocytic routes and strongly facilitated by Cq-GABARAP in a CME-dependent manner. Sci. Rep. 2016, 6, 28694. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.C.; Chang, X.J.; Li, W.D.; Wang, H.; Guo, L.M.; Wang, K.J.; Liu, H.P. A novel RING finger protein CqRNF152-like with self-ubiquitination activity inhibits white spot syndrome virus infection in a crustacean Cherax quadricarinatus. Fish Shellfish Immunol. 2019, 94, 934–943. [Google Scholar] [CrossRef]
- Kao, Z.N.; Liu, C.H.; Liu, W.J.; Kumar, R.; Leu, J.H.; Wang, H.C. Shrimp SIRT1 activates of the WSSV IE1 promoter independently of the NF-κB binding site. Fish Shellfish Immunol. 2020, 106, 910–919. [Google Scholar] [CrossRef]
- Soderhall, I.; Bangyeekhun, E.; Mayo, S.; Soderhall, K. Hemocyte production and maturation in an invertebrate animal; proliferation and gene expression in hematopoietic stem cells of Pacifastacus leniusculus. Dev. Comp. Immunol. 2003, 27, 661–672. [Google Scholar] [CrossRef]
- Xie, X.X.; Li, H.Y.; Xu, L.M.; Yang, F. A simple and efficient method for purification of intact white spot syndrome virus (WSSV) viral particles. Virus Res. 2005, 108, 63–67. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Drazic, A.; Myklebust, L.M.; Ree, R.; Arnesen, T. The world of protein acetylation. Biochim. Biophys. Acta 2016, 1864, 1372–1401. [Google Scholar] [CrossRef]
- Bordone, L.; Guarente, L. Calorie restriction, SIRT1 and metabolism: Understanding longevity. Nat. Rev. Mol. Cell Biol. 2005, 6, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.X.; Xu, L.M.; Yang, F. Proteomic analysis of the major envelope and nucleocapsid proteins of white spot syndrome virus. J. Virol. 2006, 80, 10615–10623. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Yang, F. White spot syndrome virus VP24 interacts with VP28 and is involved in virus infection. J. Gen. Virol. 2006, 87 Pt 7, 1903–1908. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Xu, L.; Li, H.; Qi, Y.P.; Yang, F. Four major envelope proteins of white spot syndrome virus bind to form a complex. J. Virol. 2009, 83, 4709–4712. [Google Scholar] [CrossRef]
- Tang, X.; Wu, J.; Sivaraman, J.; Hew, C.L. Crystal structures of major envelope proteins VP26 and VP28 from white spot syndrome virus shed light on their evolutionary relationship. J. Virol. 2007, 81, 6709–6717. [Google Scholar] [CrossRef]
- Das, D.; Bristol, M.L.; Smith, N.W.; James, C.D.; Wang, X.; Pichierri, P.; Morgan, I.M. Werner Helicase Control of Human Papillomavirus 16 E1-E2 DNA Replication Is Regulated by SIRT1 Deacetylation. Mbio 2019, 10, e00263-19. [Google Scholar] [CrossRef]
- Gu, H.; Roizman, B. Engagement of the lysine-specific demethylase/HDAC1/CoREST/REST complex by herpes simplex virus 1. J. Virol. 2009, 83, 4376–4385. [Google Scholar] [CrossRef]
- Poon, A.P.W.; Gu, H.D.; Roizman, B. ICP0 and the U(S)3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 9993–9998. [Google Scholar] [CrossRef]
- Gu, H.D.; Roizman, B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc. Natl. Acad. Sci. USA 2007, 104, 17134–17139. [Google Scholar] [CrossRef] [PubMed]
- Moorman, N.J.; Cristea, I.M.; Terhune, S.S.; Rout, M.P.; Chait, B.T.; Shenk, T. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 2008, 3, 253–262. [Google Scholar] [CrossRef]
- Terhune, S.S.; Moorman, N.J.; Cristea, I.M.; Savaryn, J.P.; Cuevas-Bennett, C.; Rout, M.P.; Chait, B.T.; Shenk, T. Human Cytomegalovirus UL29/28 Protein Interacts with Components of the NuRD Complex Which Promote Accumulation of Immediate-Early RNA. PLoS Pathog. 2010, 6, e1000965. [Google Scholar] [CrossRef] [PubMed]

| Primer | Sequence (5′–3′) | Usage |
|---|---|---|
| SIRT1-F | ATGGCGGACGCAGCATACGGG | ORF |
| SIRT1-R | TCATAGGTCAGATGAAGAAC | |
| qSIRT1-F | CCCAGACCCCTTCCATCAAC | qRT-PCR |
| qSIRT1-R | GAGGCAACAGTCTCCCGAAT | |
| 16s rRNA-F | AATGGTTGGACGAGAAGGAA | qRT-PCR |
| 16s rRNA-R | CCAACTAAACACCCTGCTGATA | |
| qvp28-F | AAACCTCCGCATTCCTGT | qRT-PCR |
| qvp28-R | GTGCCAACTTCATCCTCATC | |
| qIE1-F | CTGGCACAACAACAGACCCTACC | qRT-PCR |
| qIE1-R | GGCTAGCGAAGTAAAATATCCCCC | |
| VP664(13785+) | TTCTACTGTTGTCGGTCGCC | qRT-PCR |
| VP664(13940−) | GCGTCTCTATTGATGCGGGA | |
| dsSIRT1-F | AGGAAGAGGAAGAGGATG | RNAi |
| dsSIRT1-R | ATTTGAAGAAGGGACGAG | |
| dsGFP-F | CGACGTAAACGGCCACAAGT | RNAi |
| dsGFP-R | TTCTTGTACAGCTCGTCCATGC | |
| SIRT1(32a)-F | CGGGATCCATGGCGGACGCAGCATACGG | Prokaryoticexpression |
| SIRT1(32a)-R | CCGCTCGAGTAGGTCAGATGAAGAACATT | |
| VP24-F | GAGAGGATCCACCAACATAGAACTTAAC | |
| VP24-R | GAGAGAATTCTTTTTCCCCAACCTTAAAC | |
| VP26-F | GAGAGGATCCATGACACGTGTTGGAAG | |
| VP26-F | GAGAGAATTCCTTCTTCTTGATTTCGTC | |
| Flag-S1-F | CGGGATCCATGGCGGACGCAGCATACGG | Eukaryotic expression |
| Flag-S1-R | CCGCTCGAGTCATAGGTCAGATGAAGAACATT | |
| HA-VP24-F | CCGGAATTCGGATGCACATGTGGGGGGTTTAC | |
| HA-VP24-R | CCGCTCGAGTTATTTTTCCCCAACCTTAAAC | |
| HA-VP26-F | CCGGAATTCGGATGGAATTTGGCAACCTAAC | |
| HA-VP26-R | CCGCTCGAGTTACTTCTTCTTGATTTCGT | |
| HA-VP28-F | CCGGAATTCGGATGGATCTTTCTTTCACTC | |
| HA-VP28-R | CCGCTCGAGTTA CGCGGATCCCTCGGTCTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, S.; Meng, F.; Li, D.; Liu, L.; Ge, D.; Wang, Q.; Liu, H. A Deacetylase CqSIRT1 Promotes WSSV Infection by Binding to Viral Envelope Proteins in Cherax quadricarinatus. Viruses 2022, 14, 1733. https://doi.org/10.3390/v14081733
Zheng S, Meng F, Li D, Liu L, Ge D, Wang Q, Liu H. A Deacetylase CqSIRT1 Promotes WSSV Infection by Binding to Viral Envelope Proteins in Cherax quadricarinatus. Viruses. 2022; 14(8):1733. https://doi.org/10.3390/v14081733
Chicago/Turabian StyleZheng, Shucheng, Fanjuan Meng, Dongli Li, Lingke Liu, Di Ge, Qing Wang, and Haipeng Liu. 2022. "A Deacetylase CqSIRT1 Promotes WSSV Infection by Binding to Viral Envelope Proteins in Cherax quadricarinatus" Viruses 14, no. 8: 1733. https://doi.org/10.3390/v14081733





