Preparation of the RIPK3 Polyclonal Antibody and Its Application in Immunoassays of Nephropathogenic Infectious Bronchitis Virus-Infected Chickens
Abstract
:1. Introduction
2. Results
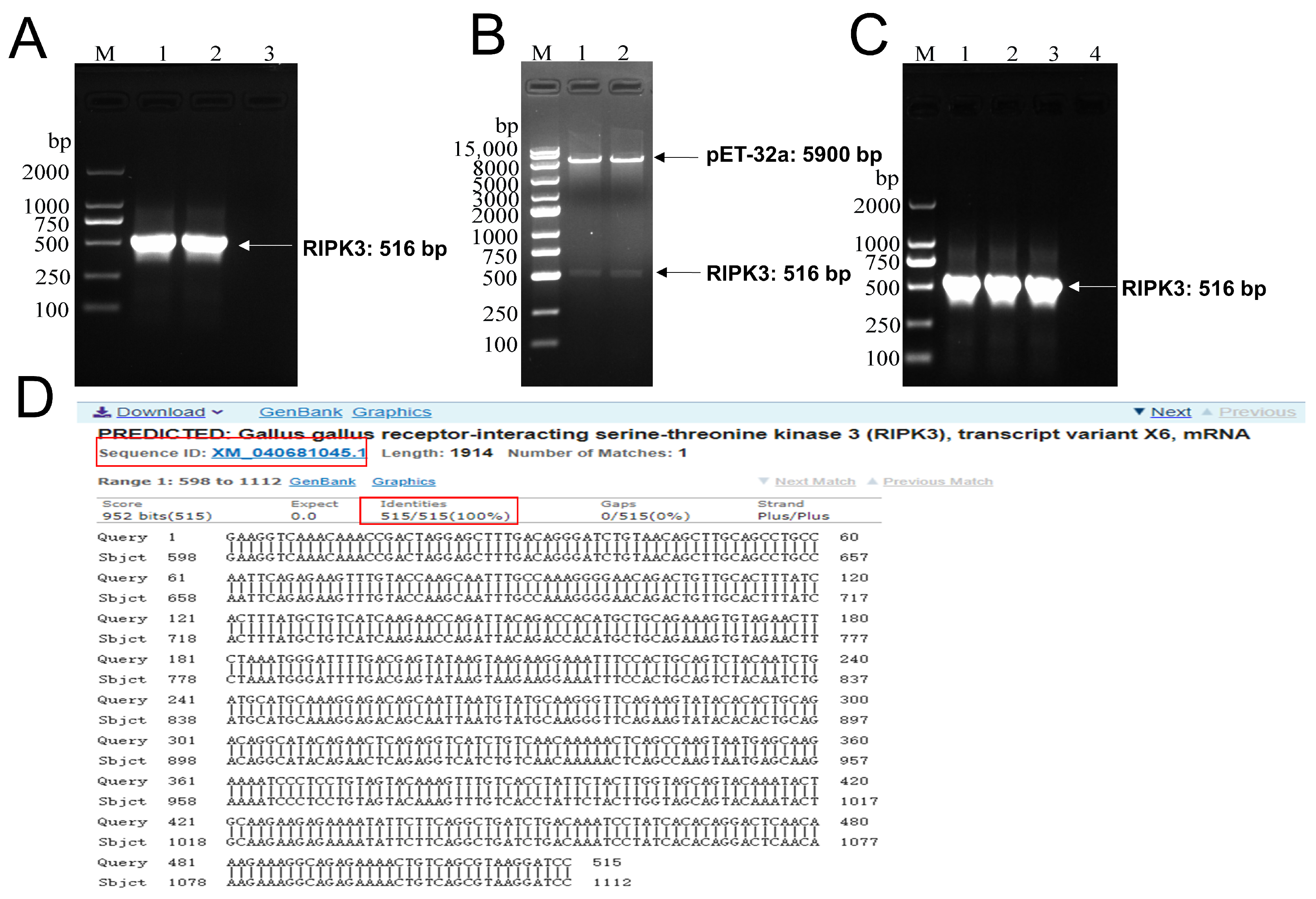
2.1. Construction of pET-32a-RIPK3 Prokaryotic Expression Vector
2.2. Induced Expression, Identification, Purification, and Renaturation of RIPK3 Truncated Protein
2.3. Antibody Titers Were Determined by Indirect ELISA
2.4. Histopathological Observation
2.5. Differential Expression of RIPK3 in Tissues
2.6. Effects of NIBV on RIPK3 Levels
2.7. NIBV Infection Induces Necroptosis
3. Discussion
4. Materials and Methods
4.1. Experimental Model and Subject Details
4.2. Construction of pET-32a-RIPK3 Expression Vector
4.3. Induction and Purification of pET-32a-RIPK3 Truncated Protein
4.4. Preparation of RIPK3 Truncated Protein Polyclonal Antibody
4.5. Histopathology
4.6. Western Blotting
4.7. Quantitative PCR Analysis
4.8. Immunofluorescence Assay
4.9. Coimmunoprecipitation
4.10. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahmood, Z.H.; Sleman, R.R.; Uthman, A.U. Isolation and molecular characterization of Sul/01/09 avian infectious bronchitis virus, indicates the emergence of a new genotype in the Middle East. Vet. Microbiol. 2011, 150, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Zheng, Y.; Yang, Y.; Liu, C.; Geng, Q.; Luo, C.; Zhang, W.; Li, F. Cryo-EM structure of infectious bronchitis coronavirus spike protein reveals structural and functional evolution of coronavirus spike proteins. PLoS Pathog. 2018, 14, e1007009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Zhai, X.; Lai, Y.; Zuo, L.; Zhang, Y.; Mei, X.; Xiang, R.; Kang, Z.; Zhou, L.; Wang, H. Construction and Immunogenicity of Novel Chimeric Virus-Like Particles Bearing Antigens of Infectious Bronchitis Virus and Newcastle Disease Virus. Viruses 2019, 11, 254. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.; Li, L.; Pan, L.; Wang, Z.; Chen, H.; Shao, C.; Yu, J.; Ren, Y.; Wang, X.; Huang, X.; et al. Infectious bronchitis virus: Identification of Gallus gallus APN high-affinity ligands with antiviral effects. Antivir. Res. 2021, 186, 104998. [Google Scholar] [CrossRef]
- Ahmed, Z.; Naeem, K.; Hameed, A. Detection and seroprevalence of infectious bronchitis virus strains in commercial poultry in Pakistan. Poult. Sci. 2007, 86, 1329–1335. [Google Scholar] [CrossRef]
- Cook, J.K.; Jackwood, M.; Jones, R.C. The long view: 40 years of infectious bronchitis research. Avian Pathol. 2012, 41, 239–250. [Google Scholar] [CrossRef]
- Ziegler, A.F.; Ladman, B.S.; Dunn, P.A.; Schneider, A.; Davison, S.; Miller, P.G.; Lu, H.; Weinstock, D.; Salem, M.; Eckroade, R.J.; et al. Nephropathogenic infectious bronchitis in Pennsylvania chickens 1997–2000. Avian Dis. 2002, 46, 847–858. [Google Scholar] [CrossRef]
- Xu, P.; Liu, P.; Zhou, C.; Shi, Y.; Wu, Q.; Yang, Y.; Li, G.; Hu, G.; Guo, X. A Multi-Omics Study of Chicken Infected by Nephropathogenic Infectious Bronchitis Virus. Viruses 2019, 11, 1070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Huang, C.; Chen, W.; Li, Z.; Hu, G.; Li, G.; Liu, P.; Hu, R.; Zhuang, Y.; Luo, J.; et al. Nephropathogenic Infectious Bronchitis Virus Mediates Kidney Injury in Chickens via the TLR7/NF-κB Signaling Axis. Front. Cell. Infect. Microbiol. 2022, 12, 865283. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.B.; June, C.H. Cytokine release syndrome in severe COVID-19. Science 2020, 368, 473–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, Y.; Nabar, N.R.; Shi, C.-S.; Kamenyeva, O.; Xiao, X.; Hwang, I.-Y.; Wang, M.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018, 9, 904. [Google Scholar] [CrossRef]
- Najjar, M.; Saleh, D.; Zelic, M.; Nogusa, S.; Shah, S.; Tai, A.; Finger, J.N.; Polykratis, A.; Gough, P.J.; Bertin, J.; et al. RIPK1 and RIPK3 Kinases Promote Cell-Death-Independent Inflammation by Toll-like Receptor 4. Immunity 2016, 45, 46–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, P.; Wang, C.; Li, J.; Su, H.; Yang, L.; Wu, P.; Lewno, M.T.; Liu, J.; Wang, X. COP9 Signalosome Suppresses RIPK1-RIPK3-Mediated Cardiomyocyte Necroptosis in Mice. Circ. Heart Fail. 2020, 13, e006996. [Google Scholar] [CrossRef]
- Devos, M.; Tanghe, G.; Gilbert, B.; Dierick, E.; Verheirstraeten, M.; Nemegeer, J.; de Reuver, R.; Lefebvre, S.; De Munck, J.; Rehwinkel, J.; et al. Sensing of endogenous nucleic acids by ZBP1 induces keratinocyte necroptosis and skin inflammation. J. Exp. Med. 2020, 217, e20191913. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, J.M.; Czabotar, P.E.; Hildebrand, J.M.; Lucet, I.S.; Zhang, J.-G.; Alvarez-Diaz, S.; Lewis, R.; Lalaoui, N.; Metcalf, D.; Webb, A.I.; et al. The pseudokinase MLKL mediates necroptosis via a molecular switch mechanism. Immunity 2013, 39, 443–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upton, J.W.; Shubina, M.; Balachandran, S. RIPK3-driven cell death during virus infections. Immunol. Rev. 2017, 277, 90–101. [Google Scholar] [CrossRef]
- Orzalli, M.H.; Kagan, J.C. Apoptosis and Necroptosis as Host Defense Strategies to Prevent Viral Infection. Trends Cell Biol. 2017, 27, 800–809. [Google Scholar] [CrossRef]
- Upton, J.W.; Kaiser, W.J.; Mocarski, E.S. Cytomegalovirus M45 cell death suppression requires receptor-interacting protein (RIP) homotypic interaction motif (RHIM)-dependent interaction with RIP1. J. Biol. Chem. 2008, 283, 16966–16970. [Google Scholar] [CrossRef] [Green Version]
- Thapa, R.J.; Ingram, J.P.; Ragan, K.B.; Nogusa, S.; Boyd, D.F.; Benitez, A.A.; Sridharan, H.; Kosoff, R.; Shubina, M.; Landsteiner, V.J.; et al. DAI Senses Influenza A Virus Genomic RNA and Activates RIPK3-Dependent Cell Death. Cell Host Microbe 2016, 20, 674–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuriakose, T.; Man, S.M.; Subbarao Malireddi, R.K.; Karki, R.; Kesavardhana, S.; Place, D.E.; Neale, G.; Vogel, P.; Kanneganti, T.-D. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 2016, 1, aag2045. [Google Scholar] [CrossRef] [Green Version]
- Shlomovitz, I.; Zargrian, S.; Gerlic, M. Mechanisms of RIPK3-induced inflammation. Immunol. Cell Biol. 2017, 95, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Christgen, S.; Zheng, M.; Kesavardhana, S.; Karki, R.; Malireddi, R.K.S.; Banoth, B.; Place, D.E.; Briard, B.; Sharma, B.R.; Tuladhar, S.; et al. Identification of the PANoptosome: A Molecular Platform Triggering Pyroptosis, Apoptosis, and Necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 2020, 10, 237. [Google Scholar] [CrossRef] [PubMed]
- Samir, P.; Malireddi, R.K.S.; Kanneganti, T.D. The PANoptosome: A Deadly Protein Complex Driving Pyroptosis, Apoptosis, and Necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 2020, 10, 238. [Google Scholar] [CrossRef]
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.S.; et al. Synergism of TNF-α and IFN-γ Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell 2021, 184, 149–168.e17. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jiang, H.; Chen, S.; Du, F.; Wang, X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell 2012, 148, 228–243. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Zhang, Y.; Cui, M.; Jin, L.; Wang, Y.; Lv, F.; Liu, Y.; Zheng, W.; Shang, H.; Zhang, J.; et al. CaMKII is a RIP3 substrate mediating ischemia- and oxidative stress-induced myocardial necroptosis. Nat. Med. 2016, 22, 175–182. [Google Scholar] [CrossRef]
- Sadeghi, H.M.M.; Rabbani, M.; Rismani, E.; Moazen, F.; Khodabakhsh, F.; Dormiani, K.; Khazaei, Y. Optimization of the expression of reteplase in Escherichia coli. Res. Pharm. Sci. 2011, 6, 87–92. [Google Scholar]
- Soman, M.; Mini, M.; Joseph, S.; Thomas, J.; Chacko, N.; Sumithra, T.G.; Ambily, R.; Mani, B.K.; Balan, R. Cloning and sequence analysis of a partial CDS of leptospiral ligA gene in pET-32a - Escherichia coli DH5α system. Vet. World 2018, 11, 557–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, H.; Chen, Y.; Huang, Q.; Guo, X.; Liu, P.; Liu, W.; Zhang, C.; Cao, H.; Hu, G. Prokaryotic expression of the chicken xanthine oxidase (XOD) subunit and its localization in liver and kidney. Int. J. Biol. Macromol. 2016, 87, 341–347. [Google Scholar] [CrossRef]
- Kim, S.; Jeong, H.; Kim, E.Y.; Kim, J.F.; Lee, S.Y.; Yoon, S.H. Genomic and transcriptomic landscape of Escherichia coli BL21(DE3). Nucleic Acids Res. 2017, 45, 5285–5293. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Lin, Z.; Qian, K.; Shao, H.; Ye, J.; Qin, A. Peptides with 16R in S2 protein showed broad reactions with sera against different types of infectious bronchitis viruses. Vet. Microbiol. 2019, 236, 108391. [Google Scholar] [CrossRef]
- Bande, F.; Arshad, S.S.; Omar, A.R.; Hair-Bejo, M.; Mahmuda, A.; Nair, V. Global distributions and strain diversity of avian infectious bronchitis virus: A review. Anim. Health Res. Rev. 2017, 18, 70–83. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Huang, C.; Shi, Y.; Li, N.; Wang, E.; Hu, R.; Li, G.; Yang, F.; Zhuang, Y.; Liu, P.; et al. Investigation of the crosstalk between GRP78/PERK/ATF-4 signaling pathway and renal apoptosis induced by nephropathogenic infectious bronchitis virus infection. J. Virol. 2021, 96, Jvi0142921. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.N.; Kanneganti, T.D. PANoptosis in Viral Infection: The Missing Puzzle Piece in the Cell Death Field. J. Mol. Biol. 2021, 434, 167249. [Google Scholar] [CrossRef] [PubMed]
- Carty, M.; Guy, C.; Bowie, A.G. Detection of Viral Infections by Innate Immunity. Biochem. Pharm. 2021, 183, 114316. [Google Scholar] [CrossRef]
- Zheng, M.; Williams, E.P.; Malireddi, R.S.; Karki, R.; Banoth, B.; Burton, A.; Webby, R.; Channappanavar, R.; Jonsson, C.B.; Kanneganti, T.D. Impaired NLRP3 inflammasome activation/pyroptosis leads to robust inflammatory cell death via caspase-8/RIPK3 during coronavirus infection. J. Biol. Chem. 2020, 295, 14040–14052. [Google Scholar] [CrossRef] [PubMed]
- Kuang, J.; Xu, P.; Shi, Y.; Yang, Y.; Liu, P.; Chen, S.; Zhou, C.; Li, G.; Zhuang, Y.; Hu, R.; et al. Nephropathogenic Infectious Bronchitis Virus Infection Altered the Metabolome Profile and Immune Function of the Bursa of Fabricius in Chicken. Front. Vet. Sci. 2020, 7, 628270. [Google Scholar] [CrossRef]
- Vucur, M.; Reisinger, F.; Gautheron, J.; Janssen, J.; Roderburg, C.; Cardenas, D.V.; Kreggenwinkel, K.; Koppe, C.; Hammerich, L.; Hakem, R.; et al. RIP3 inhibits inflammatory hepatocarcinogenesis but promotes cholestasis by controlling caspase-8- and JNK-dependent compensatory cell proliferation. Cell Rep. 2013, 4, 776–790. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Wang, H.; Wang, Z.; He, S.; Chen, S.; Liao, D.; Wang, L.; Yan, J.; Liu, W.; Lei, X.; et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012, 148, 213–227. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Shi, Y.; Li, G.; Huang, C.; Zhuang, Y.; Shu, B.; Cao, X.; Li, Z.; Hu, G.; Liu, P.; et al. Preparation of the peroxisome proliferator-activated receptor α polyclonal antibody: Its application in fatty liver hemorrhagic syndrome. Int. J. Biol. Macromol. 2021, 182, 179–186. [Google Scholar] [CrossRef]
- Lin, J.; Redies, C. Histological evidence: Housekeeping genes beta-actin and GAPDH are of limited value for normalization of gene expression. Dev. Genes Evol. 2012, 222, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.L.; Tsai, M.M.; Tsai, C.Y.; Huang, Y.H.; Chen, C.Y.; Chi, H.C.; Tseng, Y.H.; Chao, I.W.; Lin, W.C.; Wu, S.M.; et al. Glyoxalase-I is a novel prognosis factor associated with gastric cancer progression. PLoS ONE 2012, 7, e34352. [Google Scholar] [CrossRef]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, G.; Shi, Y.; Cao, X.; Chen, W.; Gu, Y.; Li, N.; Huang, C.; Zhuang, Y.; Li, G.; Liu, P.; et al. Preparation of the RIPK3 Polyclonal Antibody and Its Application in Immunoassays of Nephropathogenic Infectious Bronchitis Virus-Infected Chickens. Viruses 2022, 14, 1747. https://doi.org/10.3390/v14081747
Tian G, Shi Y, Cao X, Chen W, Gu Y, Li N, Huang C, Zhuang Y, Li G, Liu P, et al. Preparation of the RIPK3 Polyclonal Antibody and Its Application in Immunoassays of Nephropathogenic Infectious Bronchitis Virus-Infected Chickens. Viruses. 2022; 14(8):1747. https://doi.org/10.3390/v14081747
Chicago/Turabian StyleTian, Guanming, Yan Shi, Xianhong Cao, Wei Chen, Yueming Gu, Ning Li, Cheng Huang, Yu Zhuang, Guyue Li, Ping Liu, and et al. 2022. "Preparation of the RIPK3 Polyclonal Antibody and Its Application in Immunoassays of Nephropathogenic Infectious Bronchitis Virus-Infected Chickens" Viruses 14, no. 8: 1747. https://doi.org/10.3390/v14081747





