Central Role of Ubiquitination in Wheat Response to CWMV Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Sample Preparation
2.2. Protein Extraction and Trypsin Digestion
2.3. Affinity Enrichment of the Ubiquitinated Peptides
2.4. LC-MS/MS Analysis
2.5. Database Search and Bioinformatic Analysis
2.6. Construction of Recombinant Plasmids
2.7. Transient Expression and Virus Inoculation
2.8. RNA Extraction and Quantitative Reverse Transcription-PCR (qRT-PCR) Assays
2.9. Western Blot Analysis of the Ubiquitinated Proteins
3. Results
3.1. Large-Scale Profiling of Ubiquitination Sites in Wheat Infected with CWMV
3.2. Analysis of Ubiquitination Sites and Peptide Characteristics
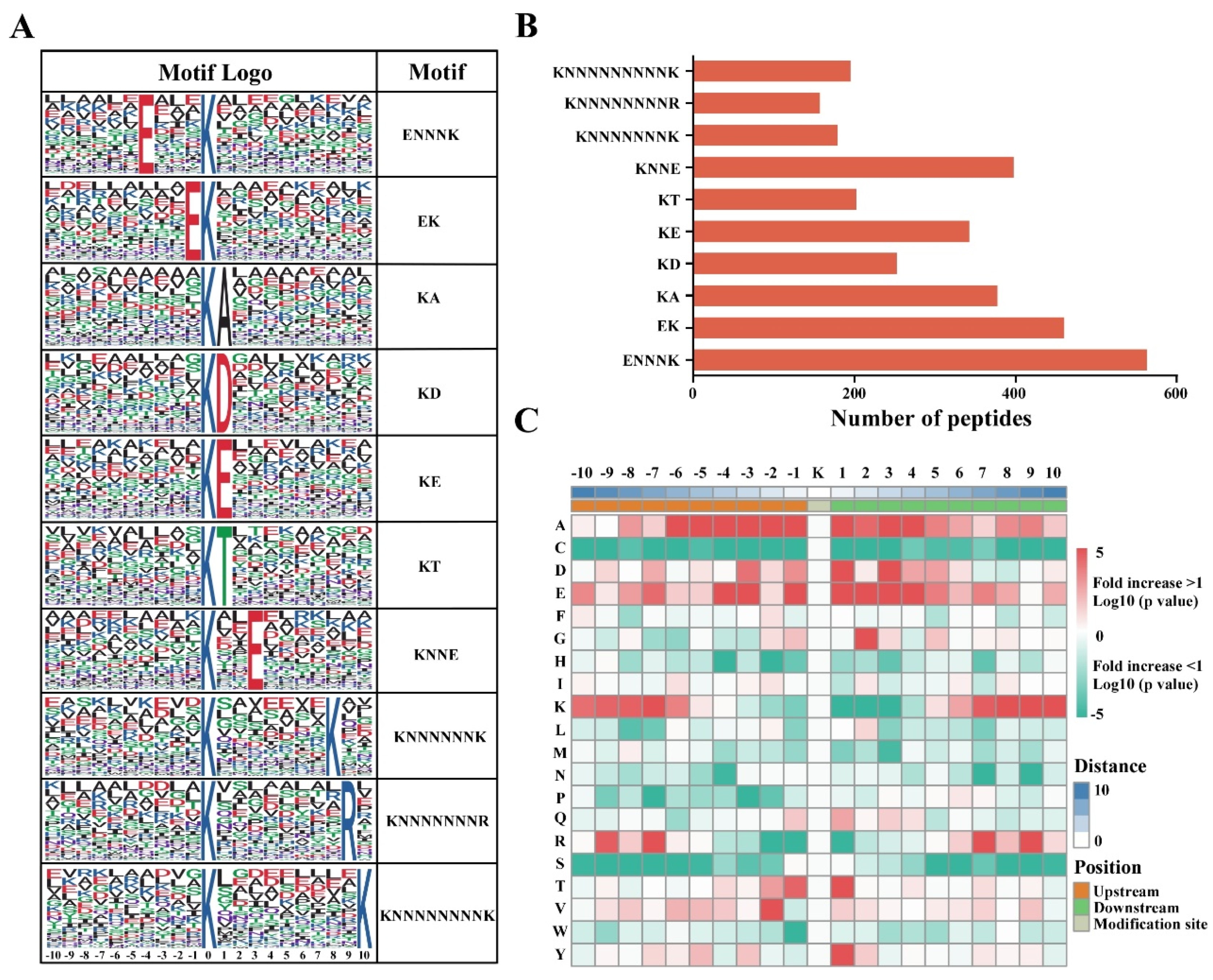
3.3. Analysis of Ubiquitination Motifs and the Conservation of Ubiquitination Site Characteristics
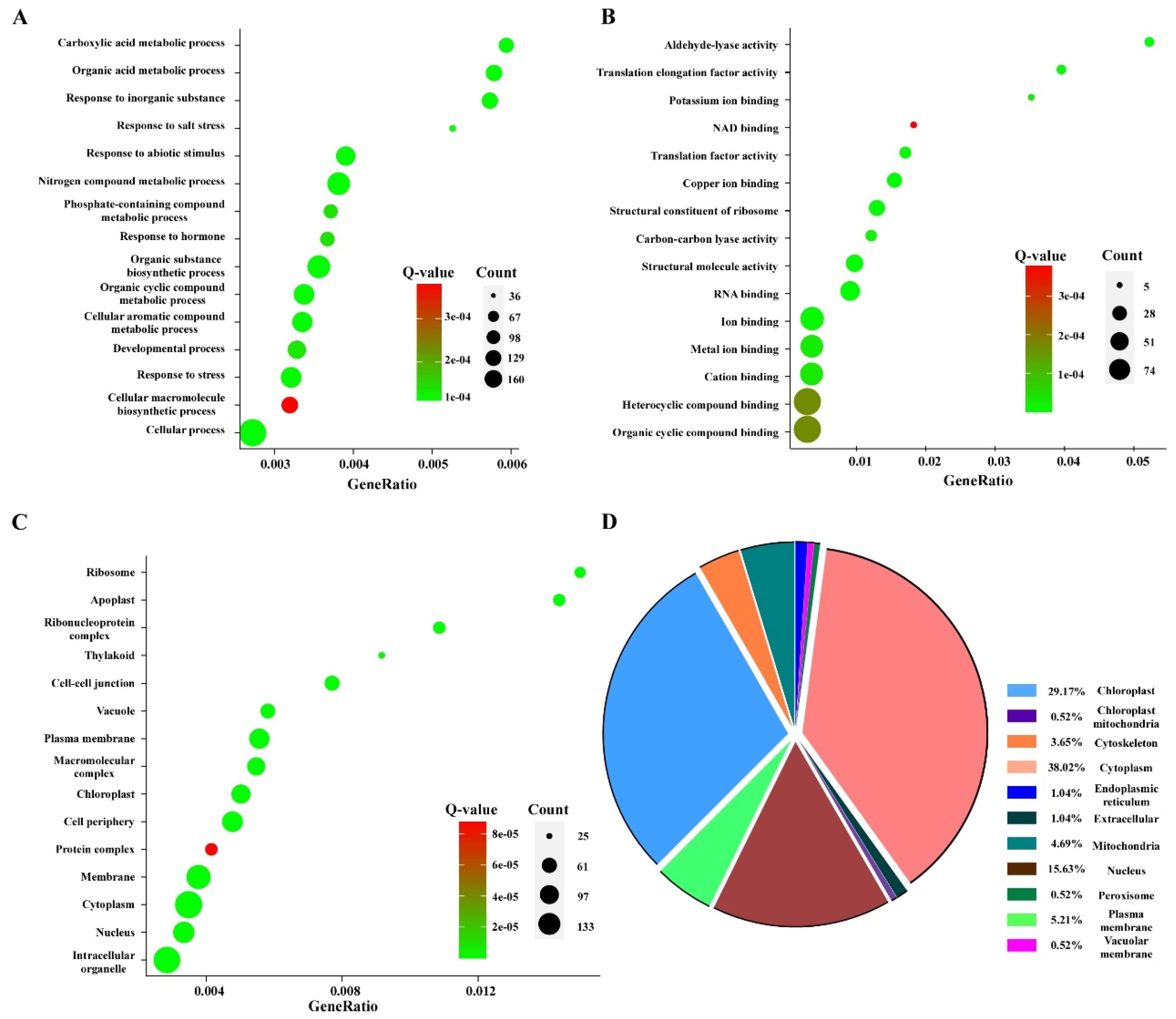
3.4. Gene Ontology Pathway Enrichment and Subcellular Localization Analyses of the Differentially Expressed Ubiquitinated Proteins
3.5. Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Enrichment and Protein–Protein Interaction (PPI) Network Analyses of Differentially Expressed Ubiquitinated Proteins
3.6. Ubiquitylation Patterns and Dynamics of Immunity-Related Proteins under CWMV Infection
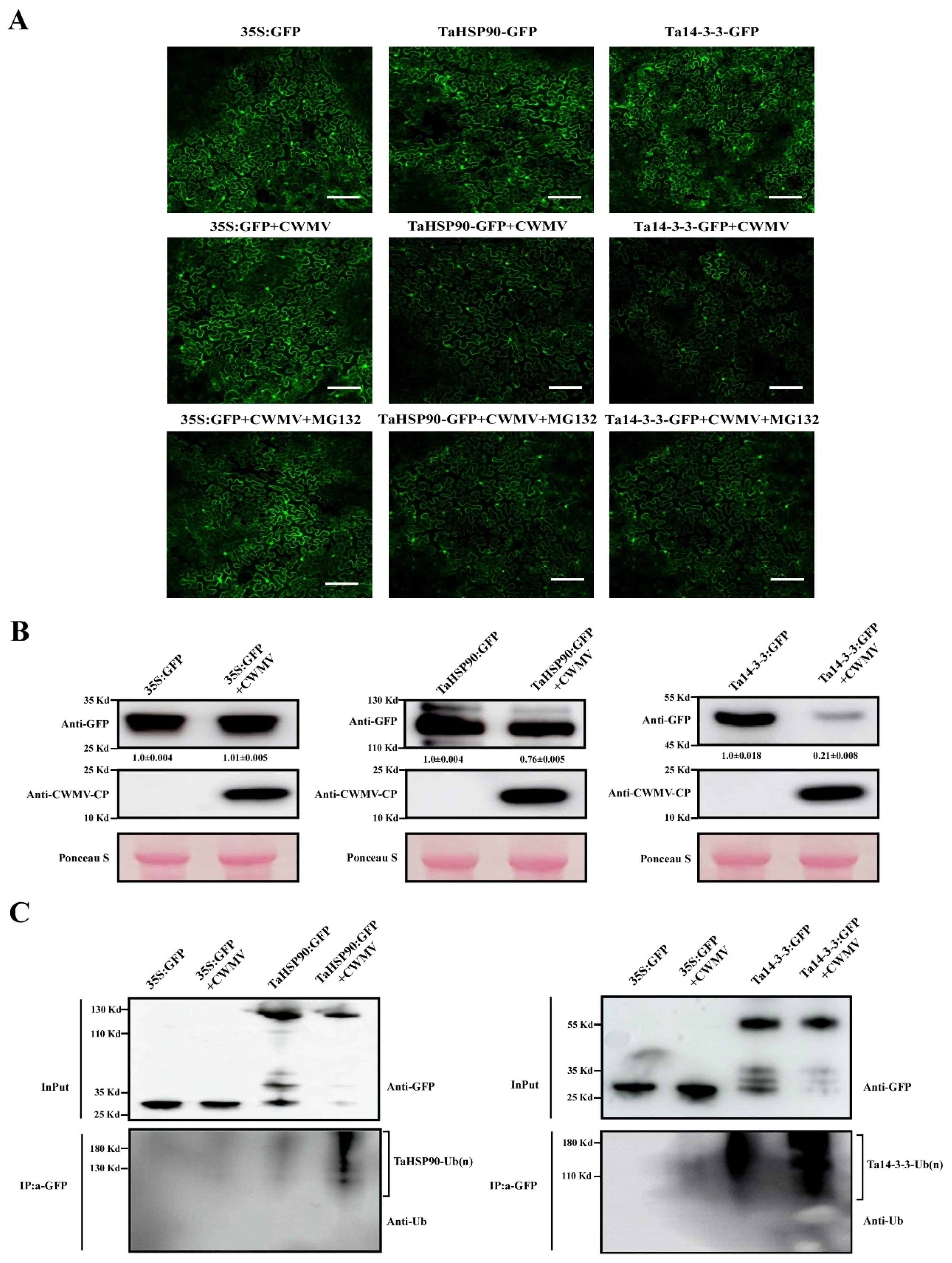
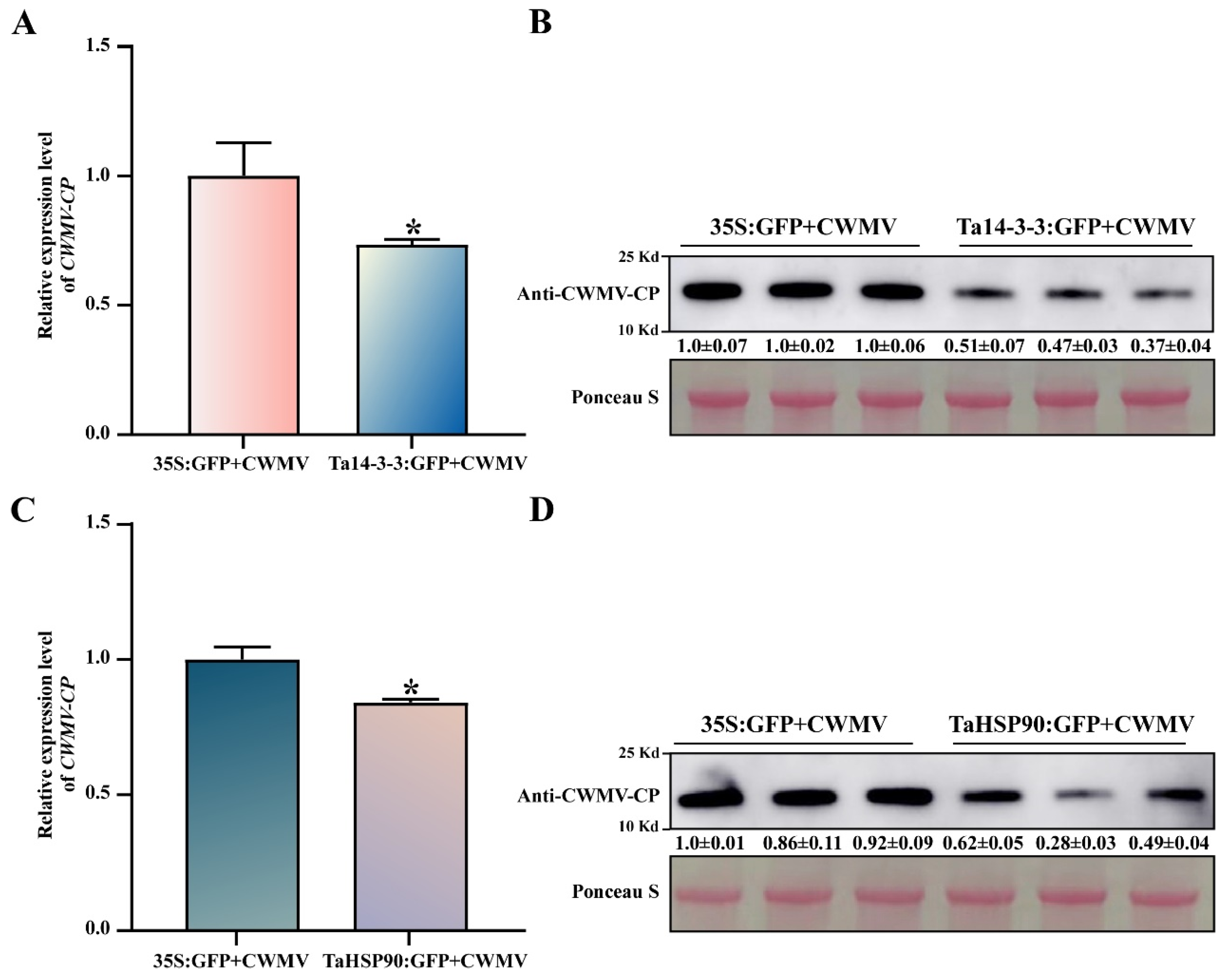
3.7. Ta14-3-3 and TaHSP90 Negatively Regulate CWMV Infection in Nicotiana benthamiana
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- He, D.; Damaris, R.; Li, M.; Khan, I.; Yang, P. Advances on Plant Ubiquitylome—From Mechanism to Application. Int. J. Mol. Sci. 2020, 21, 7909. [Google Scholar] [CrossRef] [PubMed]
- Millar, A.H.; Heazlewood, J.L.; Giglione, C.; Holdsworth, M.J.; Bachmair, A.; Schulze, W.X. The Scope, Functions, and Dynamics of Posttranslational Protein Modifications. Annu. Rev. Plant Biol. 2019, 70, 119–151. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Zhou, B.; Zeng, L. Conventional and unconventional ubiquitination in plant immunity. Mol. Plant Pathol. 2017, 18, 1313–1330. [Google Scholar] [CrossRef]
- Shu, K.; Yang, W. E3 Ubiquitin Ligases: Ubiquitous Actors in Plant Development and Abiotic Stress Responses. Plant Cell Physiol. 2017, 58, 1461–1476. [Google Scholar] [CrossRef]
- Wilson, R.S.; Swatek, K.N.; Thelen, J.J. Regulation of the Regulators: Post-Translational Modifications, Subcellular, and Spatiotemporal Distribution of Plant 14-3-3 Proteins. Front. Plant Sci. 2016, 7, 611. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhao, J.; Cai, J.; Patil, S.B. Ubiquitin-specific proteases function in plant development and stress responses. Plant Mol. Biol. 2017, 94, 565–576. [Google Scholar] [CrossRef]
- Stone, S.L. Role of the Ubiquitin Proteasome System in Plant Response to Abiotic Stress. Cell Mol. Biol. 2019, 343, 65–110. [Google Scholar] [CrossRef]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Hu, M.; Pei, B.-L.; Zhang, L.-F.; Li, Y.-Z. Histone H2B Monoubiquitination Is Involved in Regulating the Dynamics of Microtubules during the Defense Response to Verticillium dahliae Toxins in Arabidopsis. Plant Physiol. 2014, 164, 1857–1865. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, Y.; Fei, F.; Wang, Z.; Wang, W.; Cao, A.; Liu, Y.; Han, S.; Xing, L.; Wang, H.; et al. E3 ubiquitin ligase gene CMPG1-V from Haynaldia villosa L. contributes to powdery mildew resistance in common wheat (Triticum aestivum L.). Plant J. 2015, 84, 154–168. [Google Scholar] [CrossRef]
- Danielsen, J.M.R.; Sylvestersen, K.B.; Bekker-Jensen, S.; Szklarczyk, D.; Poulsen, J.W.; Horn, H.; Jensen, L.J.; Mailand, N.; Nielsen, M.L. Mass Spectrometric Analysis of Lysine Ubiquitylation Reveals Promiscuity at Site Level. Mol. Cell. Proteom. 2011, 10, M110.003590. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Schwartz, D.; Elias, J.E.; Thoreen, C.C.; Cheng, D.; Marsischky, G.; Roelofs, J.; Finley, D.; Gygi, S.P. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 2003, 21, 921–926. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Paige, J.S.; Jaffrey, S.R. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 2010, 28, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Meierhofer, D.; Wang, X.; Huang, L.; Kaiser, P. Quantitative Analysis of global Ubiquitination in HeLa Cells by Mass Spectrometry. J. Proteome Res. 2008, 7, 4566–4576. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Bennett, E.J.; Huttlin, E.L.; Guo, A.; Li, J.; Possemato, A.; Sowa, M.E.; Rad, R.; Rush, J.; Comb, M.J.; et al. Systematic and Quantitative Assessment of the Ubiquitin-Modified Proteome. Mol. Cell 2011, 44, 325–340. [Google Scholar] [CrossRef]
- Zhu, L.; Cheng, H.; Peng, G.; Wang, S.; Zhang, Z.; Ni, E.; Fu, X.; Zhuang, C.; Liu, Z.; Zhou, H. Ubiquitinome Profiling Reveals the Landscape of Ubiquitination Regulation in Rice Young Panicles. Genom. Proteom. Bioinform. 2020, 18, 305–320. [Google Scholar] [CrossRef]
- Song, Y.; Shi, X.; Zou, Y.; Guo, J.; Huo, N.; Chen, S.; Zhao, C.; Li, H.; Wu, G.; Peng, Y. Proteome-wide identification and functional analysis of ubiquitinated proteins in peach leaves. Sci. Rep. 2020, 10, 2447. [Google Scholar] [CrossRef]
- Zhang, P.Y.; Liu, F.L.; Chen, S.Q.; Liang, Z.R.; Wang, W.J.; Sun, X.T. Comparative Ubiquitome Analysis under Heat Stress Reveals Diverse Functions of Ubiquitination in Saccharina japonica. Int. J. Mol. Sci. 2020, 21, 8210. [Google Scholar]
- Chen, X.-L.; Xie, X.; Wu, L.; Liu, C.; Zeng, L.; Zhou, X.; Luo, F.; Wang, G.-L.; Liu, W. Proteomic Analysis of Ubiquitinated Proteins in Rice (Oryza sativa) After Treatment With Pathogen-Associated Molecular Pattern (PAMP) Elicitors. Front. Plant Sci. 2018, 9, 1064. [Google Scholar] [CrossRef]
- Zhang, P.; Liu, Y.; Liu, W.; Cao, M.; Massart, S.; Wang, X. Identification, Characterization and Full-Length Sequence Analysis of a Novel Polerovirus Associated with Wheat Leaf Yellowing Disease. Front. Microbiol. 2017, 8, 1689. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.-M.; He, J.; Li, J.; Chen, J.-P.; Zhang, H.-M. Chinese wheat mosaic virus: A long-term threat to wheat in China. J. Integr. Agric. 2019, 18, 821–829. [Google Scholar] [CrossRef]
- Adams, M.J.; Antoniw, J.F.; Kreuze, J. Virgaviridae: A new family of rod-shaped plant viruses. Arch. Virol. 2009, 154, 1967–1972. [Google Scholar] [CrossRef] [PubMed]
- Diao, A.; Chen, J.; Gitton, F.; Antoniw, J.F.; Mullins, J.; Hall, A.M.; Adams, M.J. Sequences of European wheat mosaic virus and oat golden stripe virus and genome analysis of the genus furovirus. Virology 1999, 261, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Andika, I.B.; Zheng, S.; Tan, Z.; Sun, L.; Kondo, H.; Zhou, X.; Chen, J. Endoplasmic reticulum export and vesicle formation of the movement protein of Chinese wheat mosaic virus are regulated by two transmembrane domains and depend on the secretory pathway. Virology 2013, 435, 493–503. [Google Scholar] [CrossRef]
- Sun, L.; Andika, I.B.; Kondo, H.; Chen, J. Identification of the amino acid residues and domains in the cysteine-rich protein of Chinese wheat mosaic virus that are important for RNA silencing suppression and subcellular localization. Mol. Plant Pathol. 2013, 14, 265–278. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, T.Y.; Liao, Q.S.; He, L.; Li, J.; Zhang, H.M.; Chen, X.; Li, J.; Yang, J.; Li, J.B.; et al. Chinese Wheat Mosaic Virus-Induced Gene Silencing in Monocots and Dicots at Low Temperature. Front. Plant Sci. 2018, 9, 1627. [Google Scholar] [CrossRef]
- Zheng, W.; Hu, H.; Lu, Q.; Jin, P.; Cai, L.; Hu, C.; Yang, J.; Dai, L.; Chen, J. Genome-Wide Identification and Characterization of Long Noncoding RNAs Involved in Chinese Wheat Mosaic Virus Infection of Nicotiana benthamiana. Biology 2021, 10, 232. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, F.; Xie, L.; Song, X.-J.; Li, J.; Chen, J.-P.; Zhang, H.-M. Functional identification of two minor capsid proteins from Chinese wheat mosaic virus using its infectious full-length cDNA clones. J. Gen. Virol. 2016, 97, 2441–2450. [Google Scholar] [CrossRef]
- Verchot, J. Plant Virus Infection and the Ubiquitin Proteasome Machinery: Arms Race along the Endoplasmic Reticulum. Viruses 2016, 8, 314. [Google Scholar] [CrossRef]
- Lu, J.; Xu, Y.; Fan, Y.; Wang, Y.; Zhang, G.; Liang, Y.; Jiang, C.; Hong, B.; Gao, J.; Ma, C. Proteome and Ubiquitome Changes during Rose Petal Senescence. Int. J. Mol. Sci. 2019, 20, 6108. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2018, 18, 623–632. [Google Scholar] [CrossRef]
- Zhang, Y.; Zeng, L. Crosstalk between Ubiquitination and Other Post-translational Protein Modifications in Plant Immunity. Plant Commun. 2020, 1, 100041. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Kang, H.; Liu, W.; Wang, G.-L. Comprehensive Profiling of the Rice Ubiquitome Reveals the Significance of Lysine Ubiquitination in Young Leaves. J. Proteome Res. 2015, 14, 2017–2025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhang, L.; Shi, C.; Tian, Q.; Lv, G.; Wang, Y.; Cui, D.; Chen, F. Comprehensive profiling of lysine ubiquitome reveals diverse functions of lysine ubiquitination in common wheat. Sci. Rep. 2017, 7, 13601. [Google Scholar] [CrossRef]
- van Butselaar, T.; Van den Ackerveken, G. Salicylic Acid Steers the Growth–Immunity Tradeoff. Trends Plant Sci. 2020, 25, 566–576. [Google Scholar] [CrossRef]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–Defense Tradeoffs in Plants: A Balancing Act to Optimize Fitness. Mol. Plant 2014, 7, 1267–1287. [Google Scholar] [CrossRef]
- Gao, C.; Tang, D.Z.; Wang, W. The Role of Ubiquitination in Plant Immunity: Fine-Tuning Immune Signaling and Beyond. Plant and Cell Physiol. 2022, pcac105. [Google Scholar] [CrossRef]
- Kraft, C.; Deplazes, A.; Sohrmann, M.; Peter, M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 2008, 10, 602–610. [Google Scholar] [CrossRef]
- Baba, A.I.; Rigó, G.; Ayaydin, F.; Rehman, A.U.; Andrási, N.; Zsigmond, L.; Valkai, I.; Urbancsok, J.; Vass, I.; Pasternak, T.; et al. Functional Analysis of the Arabidopsis thaliana CDPK-Related Kinase Family: AtCRK1 Regulates Responses to Continuous Light. Int. J. Mol. Sci. 2018, 19, 1282. [Google Scholar] [CrossRef]
- Cui, L.H.; Min, H.J.; Yu, S.G.; Byun, M.Y.; Oh, T.R.; Lee, A.; Yang, H.W.; Kim, W.T. OsATL38 mediates mono-ubiquitination of the 14-3-3 protein OsGF14d and negatively regulates the cold stress response in rice. J. Exp. Bot. 2021, 73, 307–323. [Google Scholar] [CrossRef]
- Oh, C.-S. Characteristics of 14-3-3 Proteins and Their Role in Plant Immunity. Plant Pathol. J. 2010, 26, 1–7. [Google Scholar] [CrossRef]
- Yang, X.; Wang, W.-M.; Coleman, M.; Orgil, U.; Feng, J.; Ma, X.; Ferl, R.; Turner, J.G.; Xiao, S. Arabidopsis 14-3-3 lambda is a positive regulator of RPW8-mediated disease resistance. Plant J. 2009, 60, 539–550. [Google Scholar] [CrossRef]
- Oh, C.-S.; Pedley, K.F.; Martin, G.B. Tomato 14-3-3 Protein 7 Positively Regulates Immunity-Associated Programmed Cell Death by Enhancing Protein Abundance and Signaling Ability of MAPKKK α. Plant Cell 2010, 22, 260–272. [Google Scholar] [CrossRef]
- Margaritopoulou, T.; Kryovrysanaki, N.; Megkoula, P.; Prassinos, C.; Samakovli, D.; Milioni, D.; Hatzopoulos, P. HSP90 canonical content organizes a molecular scaffold mechanism to progress flowering. Plant J. 2016, 87, 174–187. [Google Scholar] [CrossRef]
- Tillmann, B.; Roth, S.; Bublak, D.; Sommer, M.; Stelzer, E.H.K.; Scharf, K.-D.; Schleiff, E. Hsp90 is involved in the regulation of cytosolic precursor protein abundance in tomato. Mol. Plant 2015, 8, 228–241. [Google Scholar] [CrossRef]
- Wei, Y.; Zhu, B.; Liu, W.; Cheng, X.; Lin, D.; He, C.; Shi, H. Heat shock protein 90 co-chaperone modules fine-tune the antagonistic interaction between salicylic acid and auxin biosynthesis in cassava. Cell Rep. 2021, 34, 108717. [Google Scholar] [CrossRef]
- Wei, Y.; Zeng, H.; Liu, W.; Cheng, X.; Zhu, B.; Guo, J.; Shi, H. Autophagy-related genes serve as heat shock protein 90 co-chaperones in disease resistance against cassava bacterial blight. Plant J. 2021, 107, 925–937. [Google Scholar] [CrossRef]

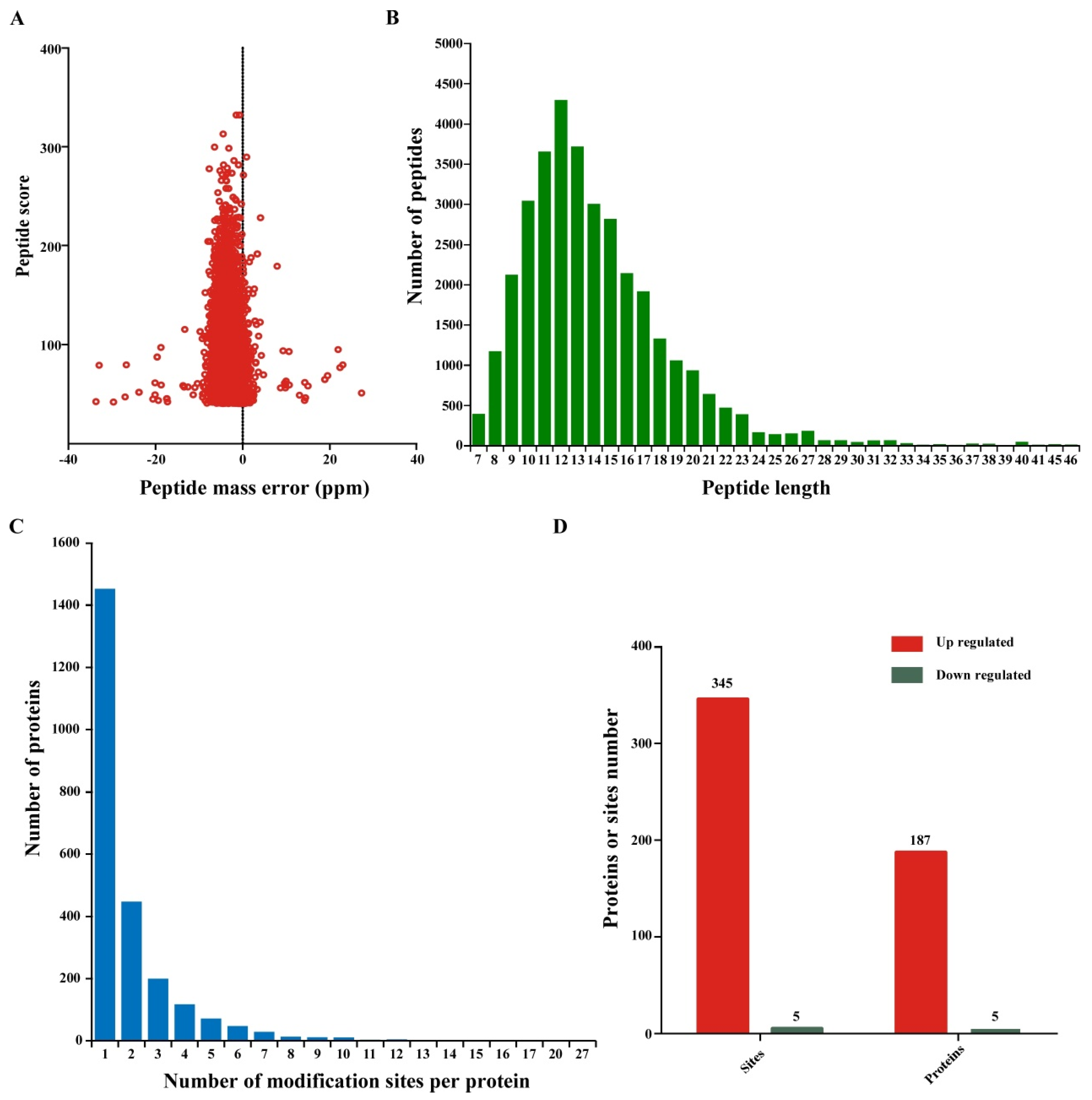

| Total Spectrum | Matched Spectrum | Peptides | Modified Peptides | Identified Proteins | Quantifiable Proteins | Identified Sites | Quantifiable Sites |
|---|---|---|---|---|---|---|---|
| 193,615 | 30,518 | 7643 | 4684 | 2413 | 1255 | 4813 | 2297 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, H.; Cai, L.; Zhang, T.; Liu, T.; Jiang, Y.; Liu, H.; Lu, Q.; Yang, J.; Chen, J. Central Role of Ubiquitination in Wheat Response to CWMV Infection. Viruses 2022, 14, 1789. https://doi.org/10.3390/v14081789
Hu H, Cai L, Zhang T, Liu T, Jiang Y, Liu H, Lu Q, Yang J, Chen J. Central Role of Ubiquitination in Wheat Response to CWMV Infection. Viruses. 2022; 14(8):1789. https://doi.org/10.3390/v14081789
Chicago/Turabian StyleHu, Haichao, Linna Cai, Tianye Zhang, Tingting Liu, Yaoyao Jiang, Hanhong Liu, Qisen Lu, Jian Yang, and Jianping Chen. 2022. "Central Role of Ubiquitination in Wheat Response to CWMV Infection" Viruses 14, no. 8: 1789. https://doi.org/10.3390/v14081789






