Connect to Protect: Dynamics and Genetic Connections of Highly Pathogenic Avian Influenza Outbreaks in Poultry from 2016 to 2021 in Germany
Abstract
1. Introduction
2. Materials and Methods
2.1. Definitions—Outbreaks, Cases, Clusters, and Hotspots
2.2. Sample Selection and RNA Extraction
2.3. IAV-End-RT-PCR
2.4. Next-Generation Sequencing and Consensus Generation
2.5. Phylogenetic and Phylogeographic Analysis
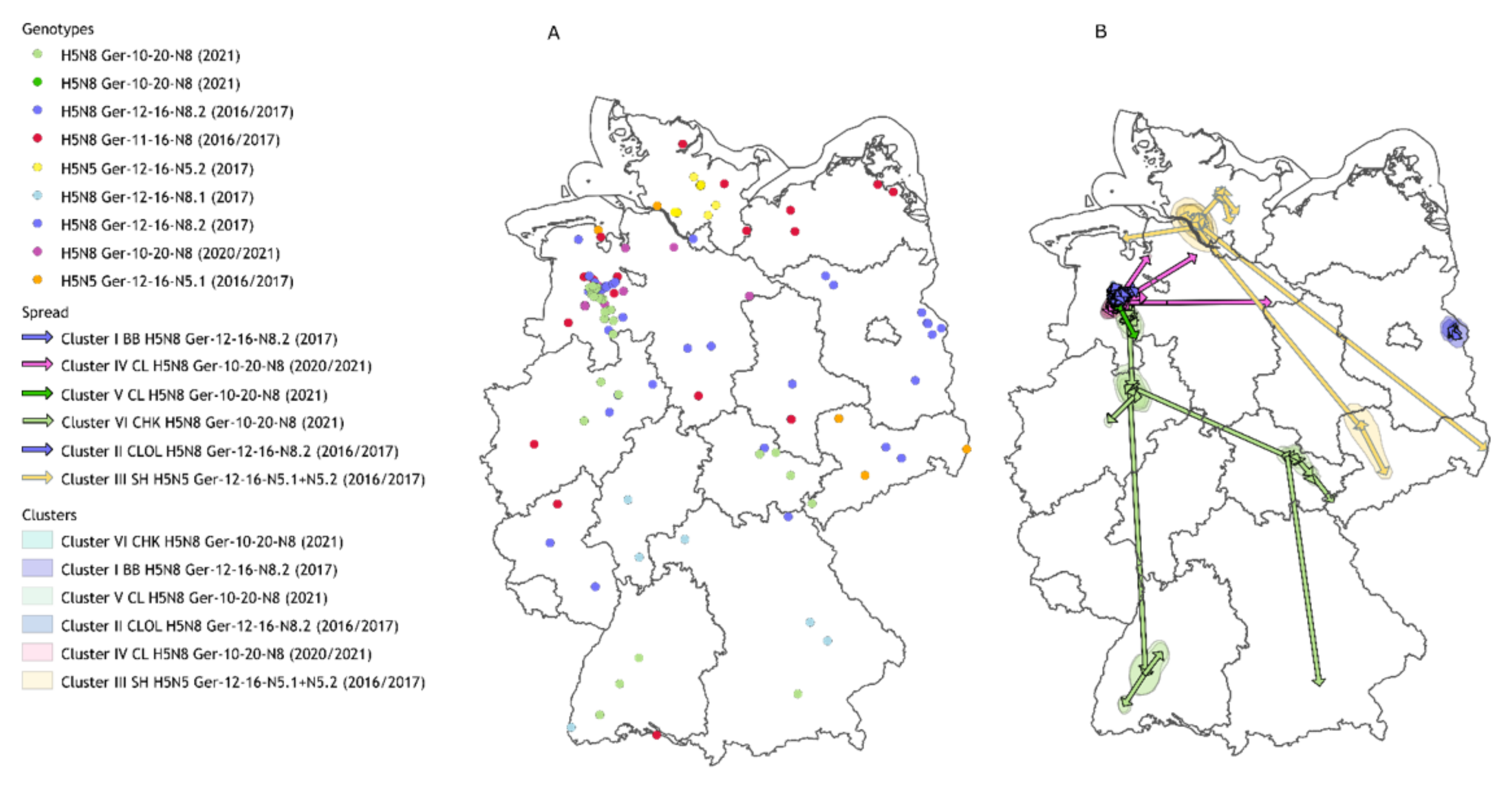
3. Results
3.1. Background Information
3.2. Direct Wild Bird Introductions and Spread—H5N8 Ger-11-16 and Ger-12-16.1
3.2.1. H5N8 Ger-11-16
Epidemiological Data
Phylogenetic Analysis of Ger-11-16
Wild Bird-Mediated Transmissions of Ger-11-16
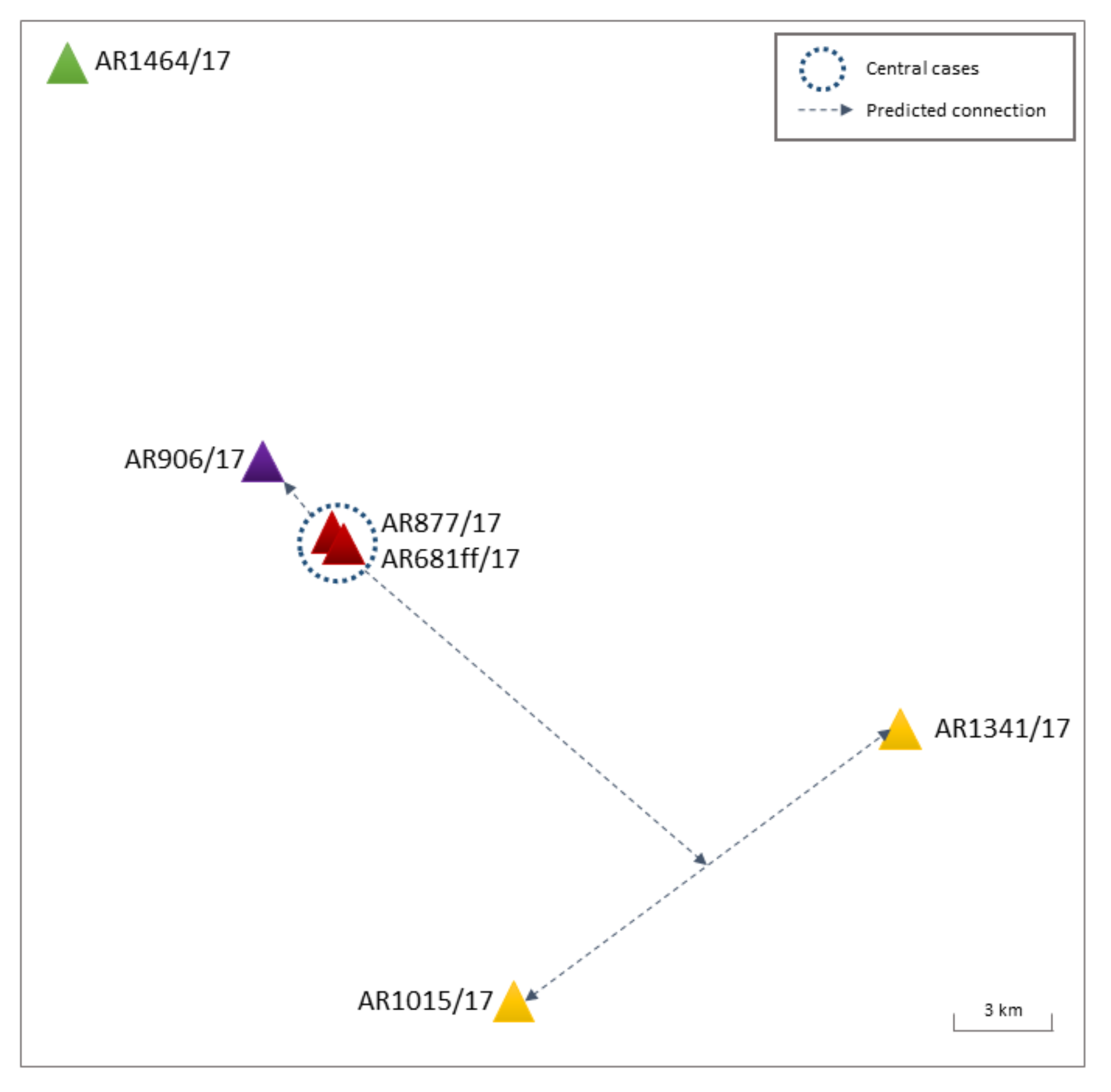
Farm-to-Farm Transmissions within Ger-11-16
Circulation Period of Ger-11-16
3.2.2. H5N8 Ger-12-16.1
Epidemiological Data, Genetic Distinction, and Phylogenetic Analysis
3.3. Cluster I and II: Cluster Development and Modification by Genetic Drift—H5N8 Ger-10-16.2
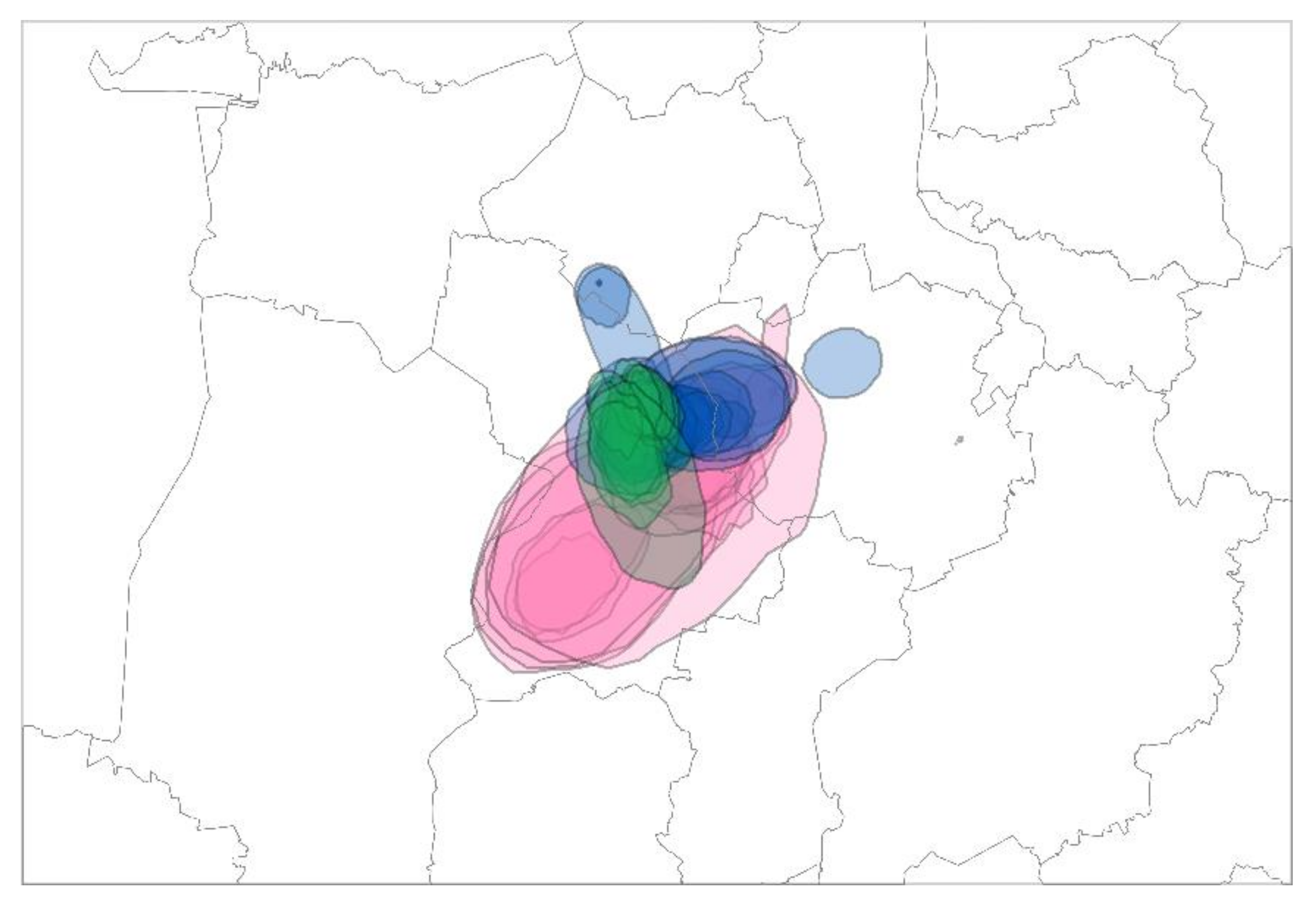
3.3.1. Cluster I—“Brandenburg (BB)”
3.3.2. Cluster II—“Cloppenburg/Oldenburg (CLOL)”
3.4. Cluster III: Genetic Modification by Reassortment in Poultry Holdings—H5N5 Ger-12-16-N5.1 and Ger-12-16-N5.2
3.4.1. Epidemiological Data
3.4.2. Genetic Modification by Reassortment
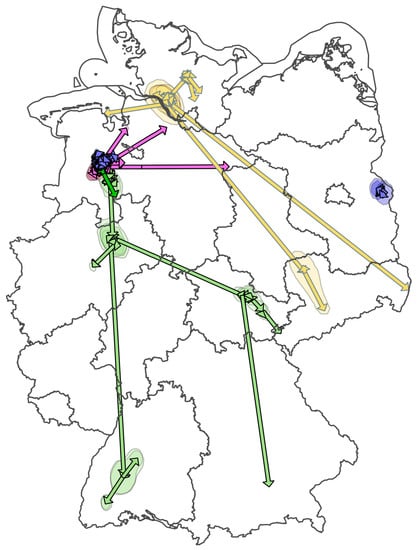
3.5. Cluster IV, V, and VI: Cluster Development and Distant Spread—H5N8 Ger-10-20-N8
Epidemiological Data and Time-Scaled Phylogeny Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pohlmann, A.; Starick, E.; Harder, T.; Grund, C.; Höper, D.; Globig, A.; Staubach, C.; Dietze, K.; Strebelow, G.; Ulrich, R.G.; et al. Outbreaks among Wild Birds and Domestic Poultry Caused by Reassorted Influenza A(H5N8) Clade 2.3.4.4 Viruses, Germany, 2016. Emerg. Infect. Dis. 2017, 23, 633–636. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Subbarao, K.; Cox, N.J.; Guo, Y. Genetic characterization of the pathogenic influenza A/Goose/Guangdong/1/96 (H5N1) virus: Similarity of its hemagglutinin gene to those of H5N1 viruses from the 1997 outbreaks in Hong Kong. Virology 1999, 261, 15–19. [Google Scholar] [CrossRef]
- Fusaro, A.; Monne, I.; Mulatti, P.; Zecchin, B.; Bonfanti, L.; Ormelli, S.; Milani, A.; Cecchettin, K.; Lemey, P.; Moreno, A.; et al. Genetic Diversity of Highly Pathogenic Avian Influenza A(H5N8/H5N5) Viruses in Italy, 2016–2017. Emerg. Infect. Dis. 2017, 23, 1543–1547. [Google Scholar] [CrossRef]
- Meier, S.; Hussy, D.; Hofmann, M.; Renzullo, S.; Vogler, B.; Sigrist, B.; Hoop, R.; Albini, S. Outbreak of Highly Pathogenic Avian Influenza H5N8 in November 2016 in Wild Birds in Switzerland. Schweiz. Arch. Tierheilkd. 2017, 159, 663–667. [Google Scholar] [CrossRef] [PubMed]
- Alarcon, P.; Brouwer, A.; Venkatesh, D.; Duncan, D.; Dovas, C.I.; Georgiades, G.; Monne, I.; Fusaro, A.; Dan, A.; Smietanka, K.; et al. Comparison of 2016-17 and Previous Epizootics of Highly Pathogenic Avian Influenza H5 Guangdong Lineage in Europe. Emerg. Infect. Dis. 2018, 24, 2270–2283. [Google Scholar] [CrossRef] [PubMed]
- Guinat, C.; Nicolas, G.; Vergne, T.; Bronner, A.; Durand, B.; Courcoul, A.; Gilbert, M.; Guerin, J.L.; Paul, M.C. Spatio-temporal patterns of highly pathogenic avian influenza virus subtype H5N8 spread, France, 2016 to 2017. Euro Surveill 2018, 23, 1700791. [Google Scholar] [CrossRef]
- Pohlmann, A.; Starick, E.; Grund, C.; Höper, D.; Strebelow, G.; Globig, A.; Staubach, C.; Conraths, F.J.; Mettenleiter, T.C.; Harder, T.; et al. Swarm incursions of reassortants of highly pathogenic avian influenza virus strains H5N8 and H5N5, clade 2.3.4.4b, Germany, winter 2016/17. Sci. Rep. 2018, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Pohlmann, A.; Hoffmann, D.; Grund, C.; Koethe, S.; Hussy, D.; Meier, S.M.; King, J.; Schinkoethe, J.; Ulrich, R.; Harder, T.; et al. Genetic Characterization and Zoonotic Potential of Highly Pathogenic Avian Influenza Virus A(H5N6/H5N5), Germany, 2017-2018. Emerg. Infect. Dis. 2019, 25, 1973–1976. [Google Scholar] [CrossRef]
- Lewis, N.S.; Banyard, A.C.; Whittard, E.; Karibayev, T.; Al Kafagi, T.; Chvala, I.; Byrne, A.; Meruyert Akberovna, S.; King, J.; Harder, T.; et al. Emergence and spread of novel H5N8, H5N5 and H5N1 clade 2.3.4.4 highly pathogenic avian influenza in 2020. Emerg. Microbes Infect. 2021, 10, 148–151. [Google Scholar] [CrossRef]
- King, J.; Harder, T.; Globig, A.; Stacker, L.; Gunther, A.; Grund, C.; Beer, M.; Pohlmann, A. Highly pathogenic avian influenza virus incursions of subtype H5N8, H5N5, H5N1, H5N4, and H5N3 in Germany during 2020-21. Virus Evol. 2022, 8, veac035. [Google Scholar] [CrossRef]
- Verhagen, J.H.; van der Jeugd, H.P.; Nolet, B.A.; Slaterus, R.; Kharitonov, S.P.; de Vries, P.P.; Vuong, O.; Majoor, F.; Kuiken, T.; Fouchier, R.A. Wild bird surveillance around outbreaks of highly pathogenic avian influenza A(H5N8) virus in the Netherlands, 2014, within the context of global flyways. Euro Surveill. 2015, 20, 21–32. [Google Scholar] [CrossRef] [PubMed]
- El-Shesheny, R.; Barman, S.; Feeroz, M.M.; Hasan, M.K.; Jones-Engel, L.; Franks, J.; Turner, J.; Seiler, P.; Walker, D.; Friedman, K.; et al. Genesis of Influenza A(H5N8) Viruses. Emerg. Infect. Dis. 2017, 23, 1368–1371. [Google Scholar] [CrossRef] [PubMed]
- Poen, M.J.; Venkatesh, D.; Bestebroer, T.M.; Vuong, O.; Scheuer, R.D.; Oude Munnink, B.B.; de Meulder, D.; Richard, M.; Kuiken, T.; Koopmans, M.P.G.; et al. Co-circulation of genetically distinct highly pathogenic avian influenza A clade 2.3.4.4 (H5N6) viruses in wild waterfowl and poultry in Europe and East Asia, 2017–2018. Virus. Evol. 2019, 5, vez004. [Google Scholar] [CrossRef]
- Global Consortium for H5N8 and Related Influenza Viruses. Role for migratory wild birds in the global spread of avian influenza H5N8. Science 2016, 354, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Bodewes, R.; Kuiken, T. Changing Role of Wild Birds in the Epidemiology of Avian Influenza A Viruses. Adv. Virus Res. 2018, 100, 279–307. [Google Scholar] [CrossRef] [PubMed]
- Ku, K.B.; Park, E.H.; Yum, J.; Kim, J.A.; Oh, S.K.; Seo, S.H. Highly pathogenic avian influenza A(H5N8) virus from waterfowl, South Korea, 2014. Emerg. Infect. Dis. 2014, 20, 1587–1588. [Google Scholar] [CrossRef]
- Zhou, L.C.; Liu, J.; Pei, E.L.; Xue, W.J.; Lyu, J.M.; Cai, Y.T.; Wu, D.; Wu, W.; Liu, Y.Y.; Jin, H.Y.; et al. Novel Avian Influenza A(H5N8) Viruses in Migratory Birds, China, 2013–2014. Emerg. Infect. Dis. 2016, 22, 1121–1123. [Google Scholar] [CrossRef]
- Lee, D.H.; Bertran, K.; Kwon, J.H.; Swayne, D.E. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J. Vet. Sci. 2017, 18, 269–280. [Google Scholar] [CrossRef]
- Harder, T.C.; Maurer-Stroh, S.; Pohlmann, A.; Starick, E.; Höreth-Böntgen, D.; Albrecht, K.; Pannwitz, G.; Teifke, J.; Gunalan, V.; Lee, R.T.; et al. Influenza A(H5N8) Virus Similar to Strain in Korea Causing Highly Pathogenic Avian Influenza in Germany. Emerg. Infect. Dis. 2015, 21, 860–863. [Google Scholar] [CrossRef]
- Lee, D.H.; Bahl, J.; Torchetti, M.K.; Killian, M.L.; Ip, H.S.; DeLiberto, T.J.; Swayne, D.E. Highly Pathogenic Avian Influenza Viruses and Generation of Novel Reassortants, United States, 2014-2015. Emerg. Infect. Dis. 2016, 22, 1283–1285. [Google Scholar] [CrossRef]
- Lee, D.H.; Sharshov, K.; Swayne, D.E.; Kurskaya, O.; Sobolev, I.; Kabilov, M.; Alekseev, A.; Irza, V.; Shestopalov, A. Novel Reassortant Clade 2.3.4.4 Avian Influenza A(H5N8) Virus in Wild Aquatic Birds, Russia, 2016. Emerg. Infect. Dis. 2017, 23, 359–360. [Google Scholar] [CrossRef]
- Li, M.; Liu, H.; Bi, Y.; Sun, J.; Wong, G.; Liu, D.; Li, L.; Liu, J.; Chen, Q.; Wang, H.; et al. Highly Pathogenic Avian Influenza A(H5N8) Virus in Wild Migratory Birds, Qinghai Lake, China. Emerg. Infect. Dis. 2017, 23, 637–641. [Google Scholar] [CrossRef]
- Chen, J.; Liang, B.; Hu, J.; Liu, H.; Sun, J.; Li, M.; Chen, Q.; He, Y.; Liu, D. Circulation, Evolution and Transmission of H5N8 virus, 2016–2018. J. Infect. 2019, 79, 363–372. [Google Scholar] [CrossRef]
- Beerens, N.; Heutink, R.; Bergervoet, S.A.; Harders, F.; Bossers, A.; Koch, G. Multiple Reassorted Viruses as Cause of Highly Pathogenic Avian Influenza A(H5N8) Virus Epidemic, the Netherlands, 2016. Emerg. Infect. Dis. 2017, 23, 1974–1981. [Google Scholar] [CrossRef] [PubMed]
- Marchenko, V.; Goncharova, N.; Susloparov, I.; Kolosova, N.; Gudymo, A.; Svyatchenko, S.; Danilenko, A.; Durymanov, A.; Gavrilova, E.; Maksyutov, R.; et al. Isolation and characterization of H5Nx highly pathogenic avian influenza viruses of clade 2.3.4.4 in Russia. Virology 2018, 525, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Napp, S.; Majo, N.; Sanchez-Gonzalez, R.; Vergara-Alert, J. Emergence and spread of highly pathogenic avian influenza A(H5N8) in Europe in 2016-2017. Transbound. Emerg. Dis. 2018, 65, 1217–1226. [Google Scholar] [CrossRef] [PubMed]
- Olsen, B.; Munster, V.J.; Wallensten, A.; Waldenstrom, J.; Osterhaus, A.D.; Fouchier, R.A. Global patterns of influenza a virus in wild birds. Science 2006, 312, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Cattoli, G.; Fusaro, A.; Monne, I.; Capua, I. H5N1 Virus Evolution in Europe-An Updated Overview. Viruses 2009, 1, 1351–1363. [Google Scholar] [CrossRef]
- Takekawa, J.Y.; Newman, S.H.; Xiao, X.; Prosser, D.J.; Spragens, K.A.; Palm, E.C.; Yan, B.; Li, T.; Lei, F.; Zhao, D.; et al. Migration of waterfowl in the East Asian flyway and spatial relationship to HPAI H5N1 outbreaks. Avian Dis. 2010, 54, 466–476. [Google Scholar] [CrossRef]
- Saito, T.; Tanikawa, T.; Uchida, Y.; Takemae, N.; Kanehira, K.; Tsunekuni, R. Intracontinental and intercontinental dissemination of Asian H5 highly pathogenic avian influenza virus (clade 2.3.4.4) in the winter of 2014-2015. Rev. Med. Virol. 2015, 25, 388–405. [Google Scholar] [CrossRef]
- Ip, H.S.; Dusek, R.J.; Bodenstein, B.; Torchetti, M.K.; DeBruyn, P.; Mansfield, K.G.; DeLiberto, T.; Sleeman, J.M. High Rates of Detection of Clade 2.3.4.4 Highly Pathogenic Avian Influenza H5 Viruses in Wild Birds in the Pacific Northwest During the Winter of 2014-15. Avian Dis. 2016, 60, 354–358. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, D.; Poen, M.J.; Bestebroer, T.M.; Scheuer, R.D.; Vuong, O.; Chkhaidze, M.; Machablishvili, A.; Mamuchadze, J.; Ninua, L.; Fedorova, N.B.; et al. Avian Influenza Viruses in Wild Birds: Virus Evolution in a Multihost Ecosystem. J. Virol. 2018, 92, e00433-18. [Google Scholar] [CrossRef]
- Poen, M.J.; Bestebroer, T.M.; Vuong, O.; Scheuer, R.D.; van der Jeugd, H.P.; Kleyheeg, E.; Eggink, D.; Lexmond, P.; van den Brand, J.M.A.; Begeman, L.; et al. Local amplification of highly pathogenic avian influenza H5N8 viruses in wild birds in the Netherlands, 2016 to 2017. Euro Surveill. 2018, 23, 17–00449. [Google Scholar] [CrossRef]
- Lycett, S.J.; Pohlmann, A.; Staubach, C.; Caliendo, V.; Woolhouse, M.; Beer, M.; Kuiken, T.; Global Consortium for H5N8 and Related Influenza Viruses. Genesis and spread of multiple reassortants during the 2016/2017 H5 avian influenza epidemic in Eurasia. Proc. Natl. Acad. Sci. USA 2020, 117, 20814–20825. [Google Scholar] [CrossRef]
- Mulatti, P.; Fusaro, A.; Scolamacchia, F.; Zecchin, B.; Azzolini, A.; Zamperin, G.; Terregino, C.; Cunial, G.; Monne, I.; Marangon, S. Integration of genetic and epidemiological data to infer H5N8 HPAI virus transmission dynamics during the 2016–2017 epidemic in Italy. Sci. Rep. 2018, 8, 18037. [Google Scholar] [CrossRef]
- Swieton, E.; Smietanka, K. Phylogenetic and molecular analysis of highly pathogenic avian influenza H5N8 and H5N5 viruses detected in Poland in 2016–2017. Transbound. Emerg. Dis. 2018, 65, 1664–1670. [Google Scholar] [CrossRef]
- Swieton, E.; Fusaro, A.; Shittu, I.; Niemczuk, K.; Zecchin, B.; Joannis, T.; Bonfante, F.; Smietanka, K.; Terregino, C. Sub-Saharan Africa and Eurasia Ancestry of Reassortant Highly Pathogenic Avian Influenza A(H5N8) Virus, Europe, December 2019. Emerg. Infect. Dis. 2020, 26, 1557–1561. [Google Scholar] [CrossRef]
- Naguib, M.M.; Graaf, A.; Fortin, A.; Luttermann, C.; Wernery, U.; Amarin, N.; Hussein, H.A.; Sultan, H.; Al Adhadh, B.; Hassan, M.K.; et al. Novel real-time PCR-based patho- and phylotyping of potentially zoonotic avian influenza A subtype H5 viruses at risk of incursion into Europe in 2017. Euro Surveill. 2017, 22, 30435. [Google Scholar] [CrossRef]
- Hoffmann, E.; Stech, J.; Guan, Y.; Webster, R.G.; Perez, D.R. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 2001, 146, 2275–2289. [Google Scholar] [CrossRef] [PubMed]
- King, J.; Harder, T.; Beer, M.; Pohlmann, A. Rapid multiplex MinION nanopore sequencing workflow for Influenza A viruses. BMC Infect Dis. 2020, 20, 648. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Bielejec, F.; Rambaut, A.; Suchard, M.A.; Lemey, P. SPREAD: Spatial phylogenetic reconstruction of evolutionary dynamics. Bioinformatics 2011, 27, 2910–2912. [Google Scholar] [CrossRef]
- King, J.; Schulze, C.; Engelhardt, A.; Hlinak, A.; Lennermann, S.L.; Rigbers, K.; Skuballa, J.; Staubach, C.; Mettenleiter, T.C.; Harder, T.; et al. Novel HPAIV H5N8 Reassortant (Clade 2.3.4.4b) Detected in Germany. Viruses 2020, 12, 281. [Google Scholar] [CrossRef]
- King, J.; Harder, T.; Conraths, F.J.; Beer, M.; Pohlmann, A. The genetics of highly pathogenic avian influenza viruses of subtype H5 in Germany, 2006–2020. Transbound. Emerg. Dis. 2021, 68, 1136–1150. [Google Scholar] [CrossRef] [PubMed]
- Bergervoet, S.A.; Ho, C.K.Y.; Heutink, R.; Bossers, A.; Beerens, N. Spread of Highly Pathogenic Avian Influenza (HPAI) H5N5 Viruses in Europe in 2016–2017 Appears Related to the Timing of Reassortment Events. Viruses 2019, 11, 501. [Google Scholar] [CrossRef]
- Denzin, N.; Bolling, M.; Pohlmann, A.; King, J.; Globig, A.; Conraths, F.J. Investigation into a Superspreading Event of the German 2020–2021 Avian Influenza Epidemic. Pathogens 2022, 11, 309. [Google Scholar] [CrossRef] [PubMed]
- Wilking, H.; Ziller, M.; Staubach, C.; Globig, A.; Harder, T.C.; Conraths, F.J. Chances and limitations of wild bird monitoring for the avian influenza virus H5N1—Detection of pathogens highly mobile in time and space. PLoS ONE 2009, 4, e6639. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Beerens, N.; Heutink, R.; Pritz-Verschuren, S.; Germeraad, E.A.; Bergervoet, S.A.; Harders, F.; Bossers, A.; Koch, G. Genetic relationship between poultry and wild bird viruses during the highly pathogenic avian influenza H5N6 epidemic in the Netherlands, 2017–2018. Transbound. Emerg. Dis. 2019, 66, 1370–1378. [Google Scholar] [CrossRef] [PubMed]

| Reassortant | Subtype | First Date | Last Date | Total | Poultry | Wild |
|---|---|---|---|---|---|---|
| Ger-11-16-N8 | H5N8 | 7 November 2016 | 22 August 2017 | 34 | 25 | 9 |
| Ger-12-16-N8.1 | H5N8 | 1 January 2017 | 24 February 2017 | 8 | 2 | 6 |
| Ger-12-16-N8.2 | H5N8 | 13 December 2016 | 9 May 2017 | 71 | 61 | 10 |
| Ger-12-16-N5.1 | H5N5 | 13 December 2016 | 23 January 2017 | 7 | 2 | 5 |
| Ger-12-16-N5.2 | H5N5 | 22 January 2017 | 9 March 2017 | 22 | 15 | 7 |
| Ger-10-20-N8 | H5N8 | 26 October 2020 | 20 July 2021 | 161 | 122 | 39 |
| Ger-02-21-N8 | H5N8 | 26 February 2021 | 10 March 2021 | 3 | 3 | 0 |
| Ger-03-21-N8 | H5N8 | 2 March 2021 | 2 March 2021 | 1 | 1 | 0 |
| Ger-10-20-N5 | H5N5 | 26 October 2020 | 9 November 2020 | 2 | 1 | 1 |
| Ger-12-20-N3 | H5N3 | 14 December 2020 | 26 January 2021 | 5 | 0 | 5 |
| Ger-02-21-N4 | H5N4 | 17 February 2021 | 17 February 2021 | 2 | 0 | 2 |
| Ger-02-21-N1 | H5N1 | 17 February 2021 | 17 July 2021 | 9 | 4 | 5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
King, J.; Staubach, C.; Lüder, C.; Koethe, S.; Günther, A.; Stacker, L.; Rubbenstroth, D.; Dietze, K.; Grund, C.; Conraths, F.J.; et al. Connect to Protect: Dynamics and Genetic Connections of Highly Pathogenic Avian Influenza Outbreaks in Poultry from 2016 to 2021 in Germany. Viruses 2022, 14, 1849. https://doi.org/10.3390/v14091849
King J, Staubach C, Lüder C, Koethe S, Günther A, Stacker L, Rubbenstroth D, Dietze K, Grund C, Conraths FJ, et al. Connect to Protect: Dynamics and Genetic Connections of Highly Pathogenic Avian Influenza Outbreaks in Poultry from 2016 to 2021 in Germany. Viruses. 2022; 14(9):1849. https://doi.org/10.3390/v14091849
Chicago/Turabian StyleKing, Jacqueline, Christoph Staubach, Christiane Lüder, Susanne Koethe, Anne Günther, Lina Stacker, Dennis Rubbenstroth, Klaas Dietze, Christian Grund, Franz J. Conraths, and et al. 2022. "Connect to Protect: Dynamics and Genetic Connections of Highly Pathogenic Avian Influenza Outbreaks in Poultry from 2016 to 2021 in Germany" Viruses 14, no. 9: 1849. https://doi.org/10.3390/v14091849
APA StyleKing, J., Staubach, C., Lüder, C., Koethe, S., Günther, A., Stacker, L., Rubbenstroth, D., Dietze, K., Grund, C., Conraths, F. J., Harder, T., Beer, M., & Pohlmann, A. (2022). Connect to Protect: Dynamics and Genetic Connections of Highly Pathogenic Avian Influenza Outbreaks in Poultry from 2016 to 2021 in Germany. Viruses, 14(9), 1849. https://doi.org/10.3390/v14091849