Double and Triple Combinations of Broadly Neutralizing Antibodies Provide Efficient Neutralization of All HIV-1 Strains from the Global Panel
Abstract
:1. Introduction
2. Materials and Methods
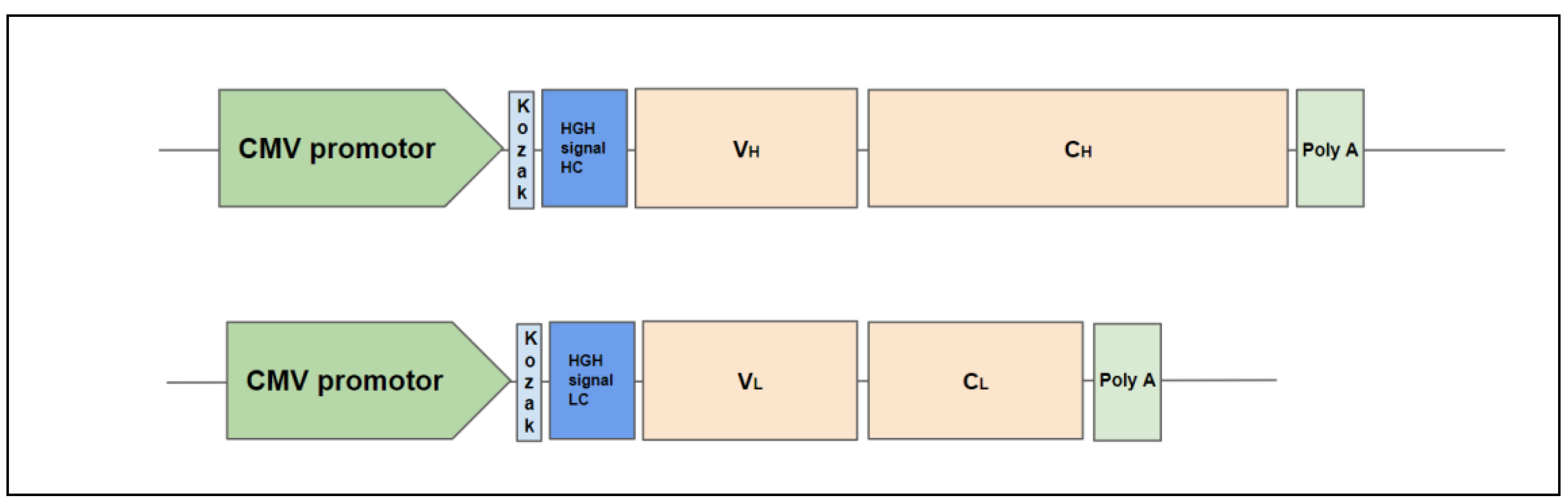
2.1. Design of Antibody Constructs
2.2. Cell Lines
2.3. Antibody Production
2.4. ELISA
2.5. Western Blot
2.6. Production of HIV-1 Pseudoviruses
2.7. Titration of HIV-1 Pseudoviruses
2.8. Neutralization Assay
2.9. Data Processing
3. Results
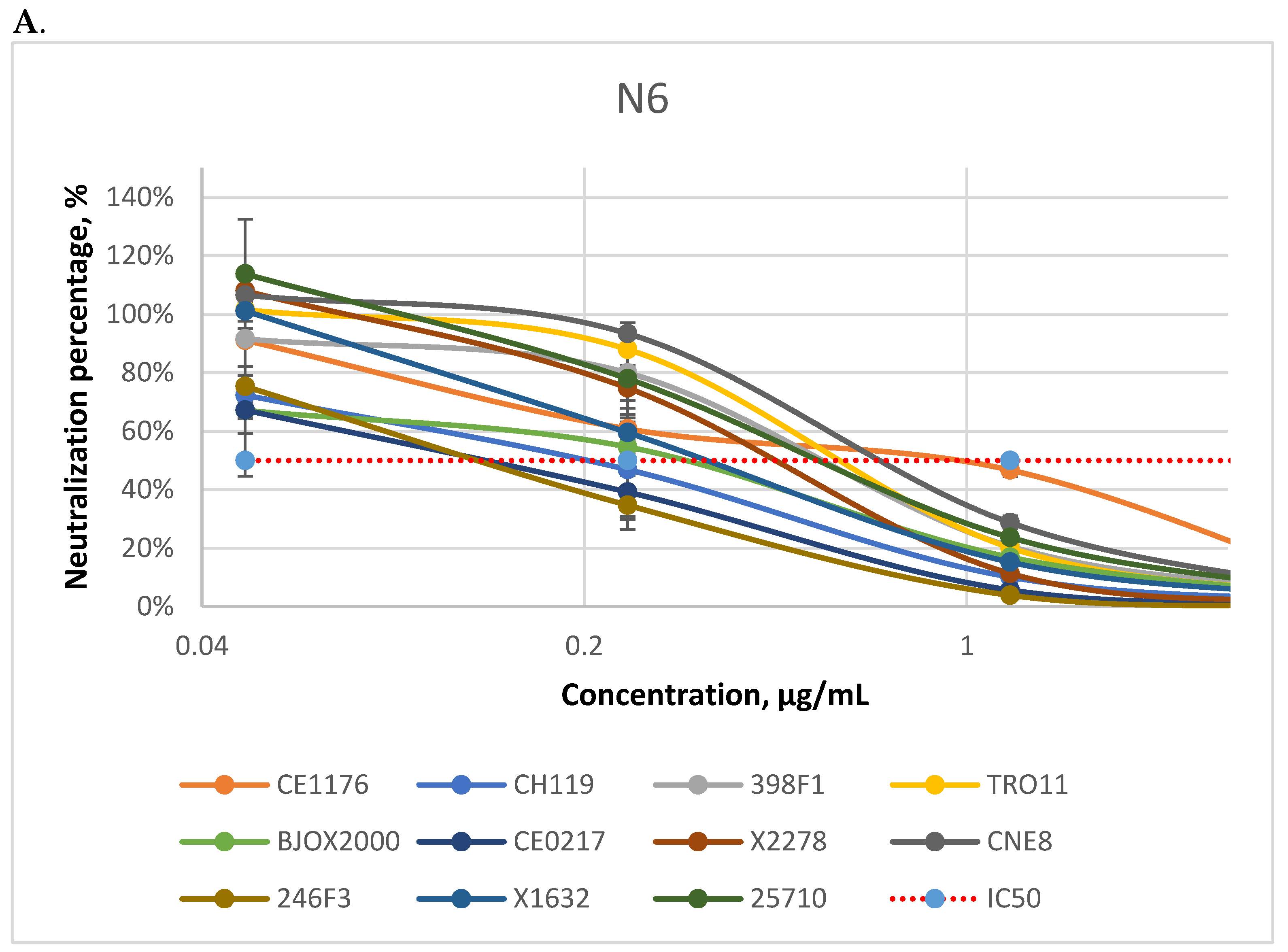
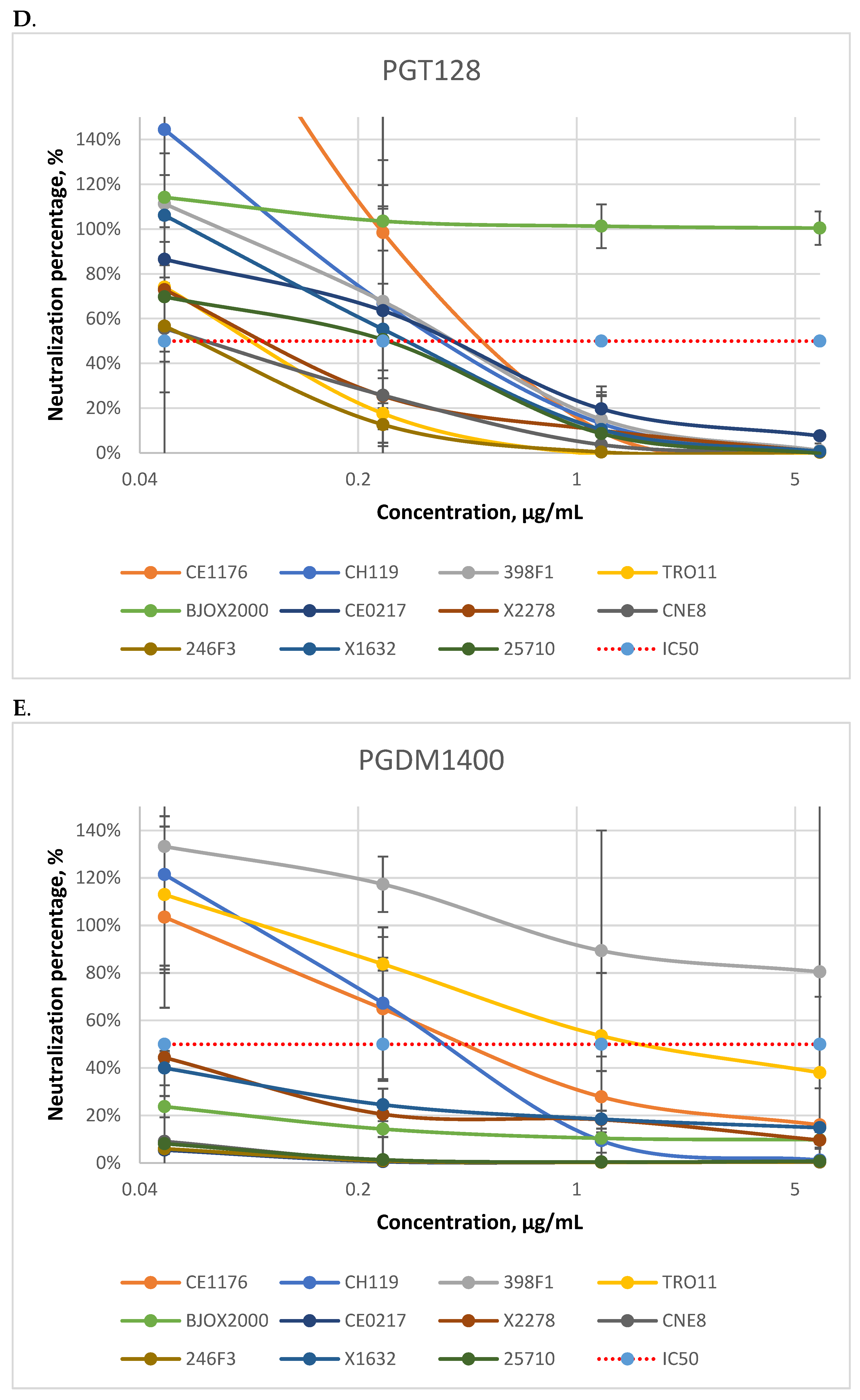
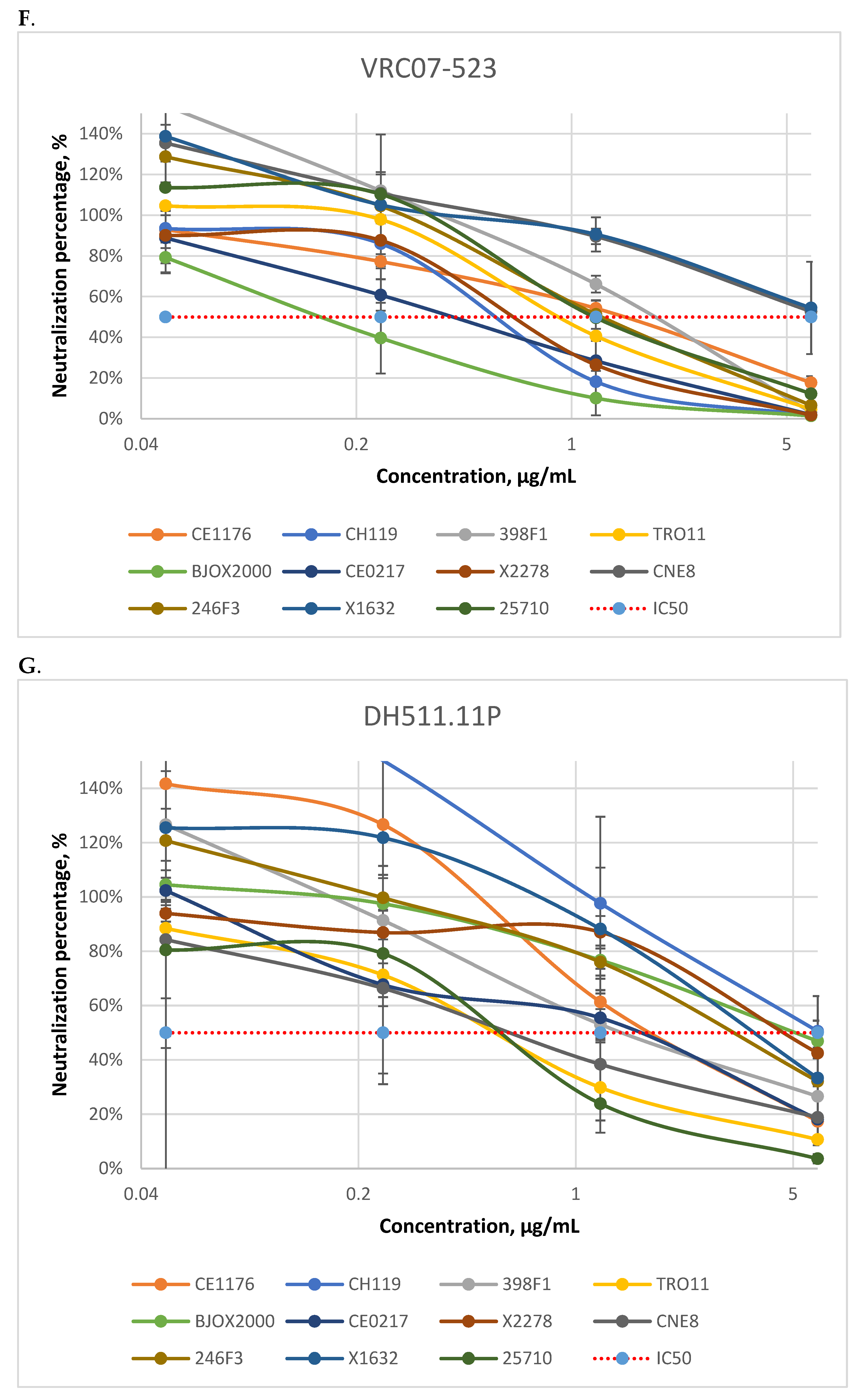
3.1. Neutralizing Activity of Antibodies
3.2. Antibody Combinations
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Appendix A


References
- UNAIDS. AIDS Statistics. Available online: https://www.unaids.org/ru/resources/fact-sheet (accessed on 25 April 2022).
- WHO. Available online: https://www.who.int/news-room/fact-sheets/detail/hiv-drugresistance (accessed on 25 April 2022).
- Scheid, J.F.; Mouquet, H.; Feldhahn, N.; Seaman, M.S.; Velinzon, K.; Pietzsch, J. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 2009, 458, 636–640. [Google Scholar] [CrossRef] [PubMed]
- Tiller, T.; Meffre, E.; Yurasov, S.; Tsuiji, M.; Nussenzweig, M.C.; Wardemann, H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 2008, 329, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Kepler, T.B.; Wiehe, K. Genetic and structural analyses of affinity maturation in the humoral response to HIV-1. Immunol. Rev. 2017, 275, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Caskey, M.; Klein, F.; Nussenzweig, M.C. Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic. Nat. Med. 2019, 25, 547–553. [Google Scholar] [CrossRef]
- Caskey, M.; Schoofs, T.; Gruell, H.; Settler, A.; Karagounis, T.; Kreider, E.F.; Murrell, B.; Pfeifer, N.; Nogueira, L.; Oliveira, T.Y.; et al. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 2017, 23, 185–191. [Google Scholar] [CrossRef]
- Derking, R.; Ozorowski, G.; Sliepen, K.; Yasmeen, A.; Cupo, A.; Torres, J.L.; Julien, J.-P.; Lee, J.H.; van Montfort, T.; de Taeye, S.W.; et al. Comprehensive Antigenic Map of a Cleaved Soluble HIV-1 Envelope Trimer. PLoS Pathog. 2015, 11, e1004767. [Google Scholar] [CrossRef]
- Glazkova, D.V.; Bogoslovskaya, E.V.; Shipulin, G.A.; Yudin, S.M. Broadly neutralizing antibodies for the treatment of HIV infection. HIV Infect. Immunosuppr. Disord. 2021, 13, 81–95. [Google Scholar] [CrossRef]
- Vazquez-Lombardi, R.; Nevoltris, D.; Luthra, A.; Schofield, P.; Zimmermann, C.; Christ, D. Transient expression of human antibodies in mammalian cells. Nat. Protoc. 2018, 13, 99–117. [Google Scholar] [CrossRef]
- De Camp, A.; Hraber, P.; Bailer, R.T.; Seaman, M.S.; Ochsenbauer, C.; Kappes, J.; Gottardo, R.; Edlefsen, P.; Self, S.; Tang, H.; et al. Global Panel of HIV-1 Env Reference Strains for Standardized Assessments of Vaccine-Elicited Neutralizing Antibodies. J. Virol. 2014, 88, 2489–2507. [Google Scholar] [CrossRef]
- Omelchenko, D.O.; Glazkova, D.V.; Bogoslovskaya, E.V.; Urusov, F.A.; Zhogina, Y.A.; Tsyganova, G.M.; Shipulin, G.A. Protection of Lymphocytes Against HIV using Lentivirus Vector Carrying a Combination of TRIM5α-HRH Genes and microRNA Against CCR5. Mol. Biol. 2018, 52, 251–261. [Google Scholar] [CrossRef]
- Sarzotti-Kelsoe, M.; Bailer, R.T.; Turk, E.; Lin, C.; Bilska, M.; Greene, K.M.; Gao, H.; Todd, C.A.; Ozaki, D.A.; Seaman, M.S.; et al. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J. Immunol. Methods 2014, 409, 131–146. [Google Scholar] [CrossRef] [PubMed]
- Montefiori, D.C. Measuring HIV Neutralization in a Luciferase Reporter Gene Assay. Methods Mol. Biol. 2009, 485, 395–405. [Google Scholar]
- Doria-Rose, N.A.; Bhiman, J.N.; Roark, R.S.; Schramm, C.A.; Gorman, J.; Chuang, G.Y.; Pancera, M.; Cale, E.M.; Ernandes, M.J.; Louder, M.K.; et al. New Member of the V1V2-Directed CAP256-VRC26 Lineage That Shows Increased Breadth and Exceptional Potency. J. Virol. 2016, 90, 76–91. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Kang, B.H.; Ishida, E.; Zhou, T.; Griesman, T.; Sheng, Z.; Wu, F.; Doria-Rose, N.A.; Zhang, B.; McKee, K.; et al. Identification of a CD4-Binding-Site Antibody to HIV that Evolved Near-Pan Neutralization Breadth. Immunity 2016, 45, 1108–1121. [Google Scholar] [CrossRef]
- Nishimura, Y.; Gautam, R.; Chun, T.W.; Sadjadpour, R.; Foulds, K.E.; Shingai, M.; Klein, F.; Gazumyan, A.; Golijanin, J.; Donaldson, M.; et al. Early antibody therapy can induce long-lasting immunity to SHIV. Nature 2017, 543, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Rudicell, R.S.; Kwon, Y.; do Ko, S.Y.; Pegu, A.; Louder, M.K.; Georgiev, I.S.; Wu, X.; Zhu, J.; Boyington, J.C.; Chen, X.; et al. Enhanced Potency of a Broadly Neutralizing HIV-1 Antibody In Vitro Improves Protection against Lentiviral Infection In Vivo. J. Virol. 2014, 88, 12669–12682. [Google Scholar] [CrossRef]
- Sok, D.; van Gils, M.J.; Pauthner, M.; Julien, J.P.; Saye-Francisco, K.L.; Hsueh, J.; Briney, B.; Lee, J.H.; Le, K.M.; Lee, P.S.; et al. Recombinant HIV envelope trimer selects for quaternary-dependent antibodies targeting the trimer apex. Proc. Natl. Acad. Sci. USA 2014, 111, 17624–17629. [Google Scholar] [CrossRef]
- Walker, L.M.; Huber, M.; Doores, K.J.; Falkowska, E.; Pejchal, R.; Julien, J.P.; Wang, S.-K.; Ramos, A.; Chan-Hui, P.-Y.; Moyle, M.; et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 2011, 477, 466–470. [Google Scholar] [CrossRef]
- Williams, L.D.; Ofek, G.; Schätzle, S.; McDaniel, J.R.; Lu, X.; Nicely, N.I.; Wu, L.; Lougheed, C.S.; Bradley, T.; Louder, M.K.; et al. Potent and broad HIV-neutralizing antibodies in memory B cells and plasma. Sci. Immunol. 2017, 2, eaal2200. [Google Scholar] [CrossRef]
- Scott, S.D. Rituximab: A New Therapeutic Monoclonal Antibody for Non-Hodgkin’s Lymphoma. Cancer Pract. 1998, 6, 195–197. [Google Scholar] [CrossRef]
- Webb, N.E.; Montefiori, D.C.; Lee, B. Dose–response curve slope helps predict therapeutic potency and breadth of HIV broadly neutralizing antibodies. Nat. Commun. 2015, 6, 8443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwong, P.D.; Mascola, J.R. HIV-1 Vaccines Based on Antibody Identification, B Cell Ontogeny, and Epitope Structure. Immunity 2018, 48, 855–871. [Google Scholar] [CrossRef]
- Klasse, P.J.; Ozorowski, G.; Sanders, R.W.; Moore, J.P. Env Exceptionalism: Why Are HIV-1 Env Glycoproteins Atypical Immunogens? Cell Host Microbe 2020, 27, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Binley, J.M.; Lybarger, E.A.; Crooks, E.T.; Seaman, M.S.; Gray, E.; Davis, K.L.; Decker, J.M.; Wycuff, D.; Harris, L.; Hawkins, N.; et al. Profiling the Specificity of Neutralizing Antibodies in a Large Panel of Plasmas from Patients Chronically Infected with Human Immunodeficiency Virus Type 1 Subtypes B and C. J. Virol. 2008, 82, 11651–11668. [Google Scholar] [CrossRef] [PubMed]
- Diskin, R.; Scheid, J.F.; Marcovecchio, P.M.; West, A.P.; Klein, F.; Gao, H.; Gnanapragasam, P.N.P.; Abadir, A.; Seaman, M.S.; Nussenzweig, M.C.; et al. Increasing the Potency and Breadth of an HIV Antibody by Using Structure-Based Rational Design. Science 2011, 334, 1289–1293. [Google Scholar] [CrossRef]
- Hsu, D.C.; Mellors, J.W.; Vasan, S. Can Broadly Neutralizing HIV-1 Antibodies Help Achieve an ART-Free Remission? Front. Immunol. 2021, 12, 710044. [Google Scholar] [CrossRef]
- Mendoza, P.; Gruell, H.; Nogueira, L.; Pai, J.A.; Butler, A.L.; Millard, K.; Lehmann, C.; Suarez, I.; Oliveira, T.Y.; Lorenzi, C.C.; et al. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 2018, 561, 479–484. [Google Scholar] [CrossRef]
- Hraber, P.; Rademeyer, C.; Williamson, C.; Seaman, M.S.; Gottardo, R.; Tang, H.; Greene, K.; Gao, H.; LaBranche, C.; Mascola, J.R.; et al. Panels of HIV-1 Subtype C Env Reference Strains for Standardized Neutralization Assessments. J. Virol. 2017, 91, e00991-17. [Google Scholar] [CrossRef]
- Liu, Q.; Lai, Y.T.; Zhang, P.; Louder, M.K.; Pegu, A.; Rawi, R.; Asokan, M.; Chen, X.; Shen, C.-H.; Chuang, G.-Y.; et al. Improvement of antibody functionality by structure-guided paratope engraftment. Nat. Commun. 2019, 10, 721. [Google Scholar] [CrossRef]
- Moshoette, T.; Ali, S.A.; Papathanasopoulos, M.A.; Killick, M.A. Engineering and characterising a novel, highly potent bispecific antibody iMab-CAP256 that targets HIV-1. Retrovirology 2019, 16, 31. [Google Scholar] [CrossRef]
- Wang, Z.; Barnes, C.O.; Gautam, R.; Cetrulo Lorenzi, J.C.; Mayer, C.T.; Oliveira, T.Y.; Ramos, V.; Cipolla, M.; Gordon, K.M.; Gristick, H.B.; et al. A broadly neutralizing macaque monoclonal antibody against the HIV-1 V3-Glycan patch. eLife 2020, 21, 9. [Google Scholar] [CrossRef] [PubMed]
- Klein, F.; Halper-Stromberg, A.; Horwitz, J.A.; Gruell, H.; Scheid, J.F.; Bournazos, S.; Mouquet, H.; Spatz, L.A.; Diskin, R.; Abadir, A.; et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 2012, 492, 118–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]

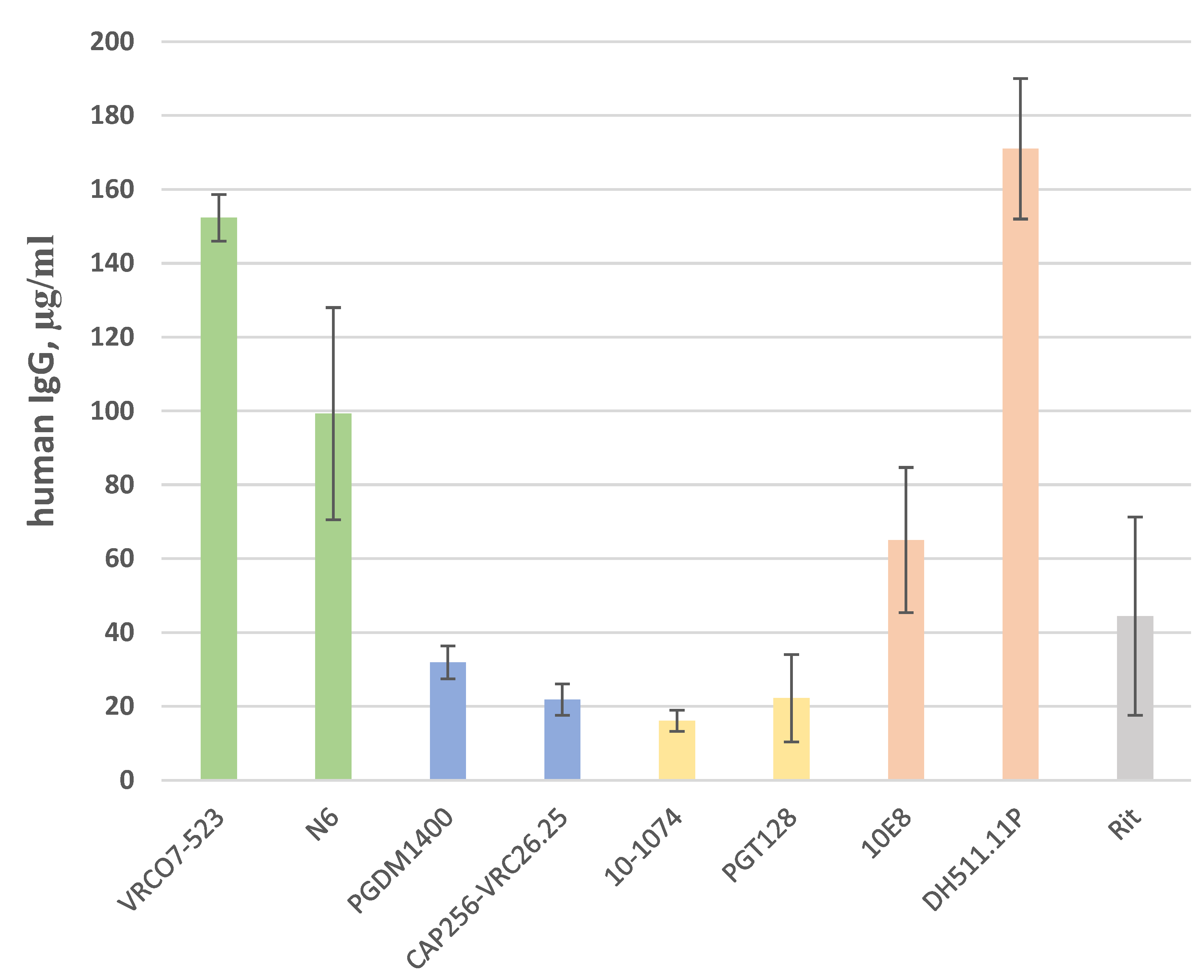

| bNAbs | 10E8 | DH511.11P | N6 | VRC07-523 | 10-1074 | PGT128 | CAP256-VRC26.25 | PGDM1400 | |
|---|---|---|---|---|---|---|---|---|---|
| Pseudovirus Strains | |||||||||
| CE1176(C) | 0.26 | 1.52 | 1.34 | 2.11 | 0.21 | 0.43 | >5 | 0.89 | |
| CH119 (CRF07) | 0.30 | >5 | 0.21 | 0.53 | 0.20 | 0.33 | 0.13 | 0.54 | |
| 398F1(A) | 0.49 | 2.05 | 0.87 | 1.72 | 0.17 | 0.32 | >5 | >5 | |
| TRO11 (B) | 0.82 | 0.67 | 0.70 | 0.60 | 0.11 | 0.08 | >5 | 2.11 | |
| BJOX2000 (CRF07) | 0.39 | 3.52 | 0.31 | 0.27 | >5 | >5 | <0.01 | <0.01 | |
| CE0217 (C) | >5 | 1.02 | 0.14 | 0.43 | <0.01 | 0.21 | <0.01 | <0.01 | |
| X2278 (B) | 0.36 | 2.39 | 0.56 | 0.56 | 0.11 | 0.07 | 0.01 | 0.01 | |
| CNE8 (CRF01) | 0.19 | 0.99 | 0.85 | >5 | >5 | 0.03 | 2.24 | <0.01 | |
| 246F3 (AC) | 0.46 | 2.04 | 0.16 | 1.24 | >5 | 0.05 | 0.18 | <0.01 | |
| X1632 (G) | 0.36 | >5 | 0.42 | >5 | 0.07 | 0.23 | <0.01 | 0.01 | |
| 25710 (C) | 0.03 | 0.46 | 0.72 | 1.47 | 0.30 | 0.11 | <0.01 | <0.01 | |
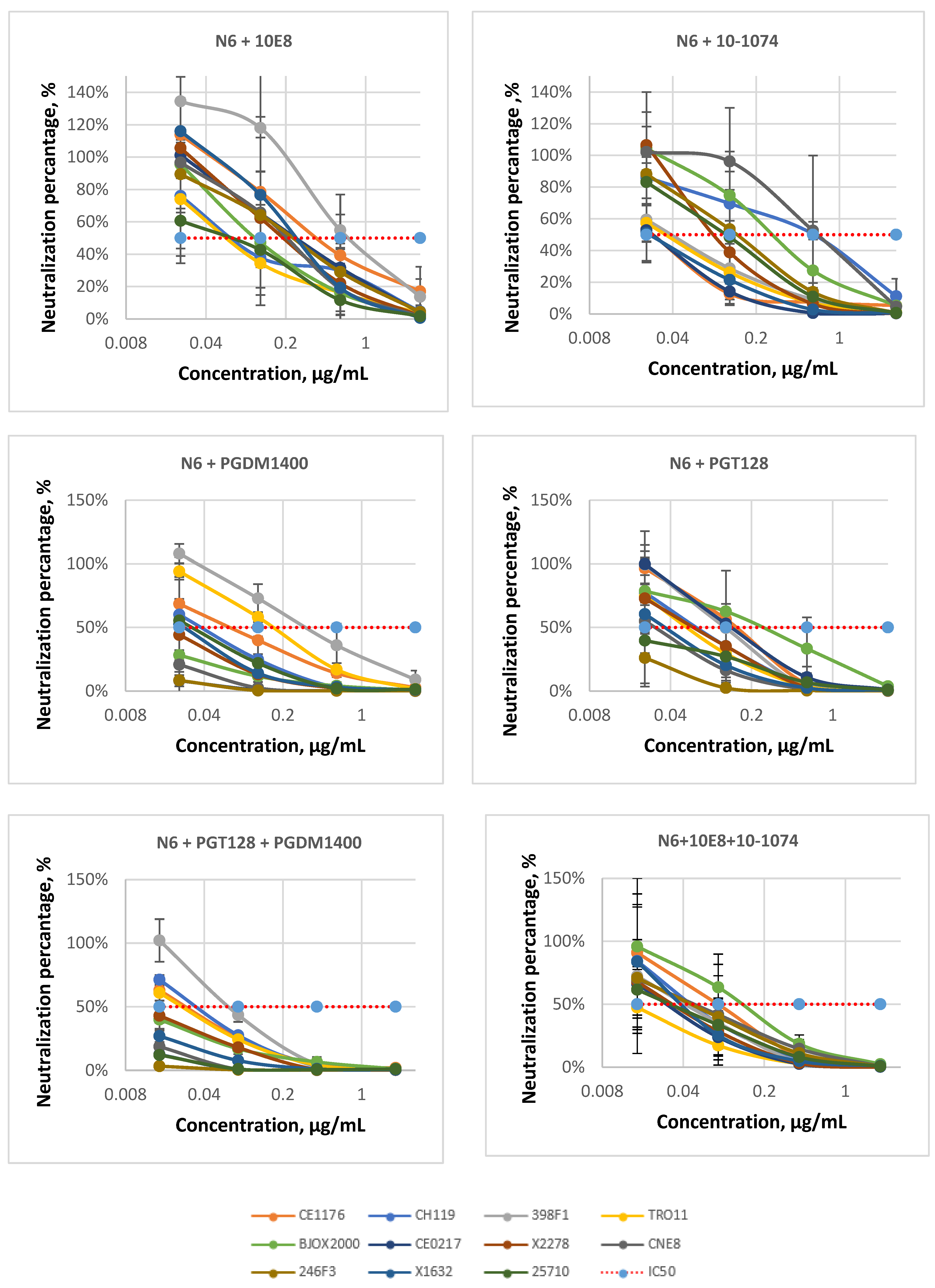
| bNAbs | N6 | 10-1074 | 10E8 | N6 + 10E8 | N6 + 10-1074 | N6 + 10-1074 + 10E8 | |
|---|---|---|---|---|---|---|---|
| Pseudovirus Strains | |||||||
| CE1176 (C) | 1.34 | 0.21 | 0.26 | 0.70 | 0.03 | 0.14 | |
| CH119 (CRF07) | 0.21 | 0.2 | 0.3 | 0.12 | 0.51 | 0.09 | |
| 398F1 (A) | 0.87 | 0.17 | 0.49 | 0.84 | 0.04 | 0.05 | |
| TRO11 (B) | 0.7 | 0.11 | 0.82 | 0.15 | 0.04 | 0.02 | |
| BJOX2000 (CRF07) | 0.31 | >5 | 0.39 | 0.71 | 0.30 | 0.15 | |
| CE0217 (C) | 0.14 | <0.01 | >5 | 0.33 | 0.02 | 0.03 | |
| X2278 (B) | 0.56 | 0.11 | 0.36 | 0.24 | 0.18 | 0.04 | |
| CNE8 (CRF01) | 0.85 | >5 | 0.19 | 0.27 | 0.48 | 0.07 | |
| 246F3 (AC) | 0.16 | >5 | 0.46 | 0.29 | 0.16 | 0.05 | |
| X1632 (G) | 0.42 | 0.07 | 0.36 | 0.33 | 0.03 | 0.08 | |
| 25710 (C) | 0.72 | 0.3 | 0.03 | 0.11 | 0.13 | 0.05 | |
| IC50 range | 0.14–1.34 | 0.01–5 | 0.03–5 | 0.11–0.84 | 0.02–0.51 | 0.02–0.15 | |
| bNAbs | N6 | 10-1074 | 10E8 | N6 + 10E8 | N6 + 10-1074 | N6 + 10-1074 + 10E8 | |
|---|---|---|---|---|---|---|---|
| Pseudovirus Strains | |||||||
| CE1176 (C) | 3.07 | 0.57 | 1.33 | 2.13 | 0.29 | 0.36 | |
| CH119 (CRF07) | 1.07 | 0.5 | 1.33 | 0.87 | 1.80 | 0.29 | |
| 398F1 (A) | 1.98 | 0.67 | 1.86 | 1.73 | 0.24 | 0.16 | |
| TRO11 (B) | 2.5 | 0.44 | 2.61 | 0.64 | 0.34 | 0.14 | |
| BJOX2000 (CRF07) | 0.8 | >5 | >5 | 2.63 | 1.23 | 0.61 | |
| CE0217 (C) | 0.9 | 0.09 | >5 | 1.18 | 0.21 | 0.22 | |
| X2278 (B) | 1.67 | 2.69 | 1.10 | 0.99 | 0.55 | 0.24 | |
| CNE8 (CRF01) | >5 | >5 | 1.24 | 0.83 | 1.75 | 0.39 | |
| 246F3 (AC) | 0.78 | >5 | 2.18 | 1.22 | 0.72 | 0.36 | |
| X1632 (G) | 1.62 | 0.31 | 1.58 | 0.87 | 0.26 | 0.23 | |
| 25710 (C) | 2.23 | 1.18 | 0.34 | 0.54 | 0.63 | 0.28 | |
| IC80 range | 0.78–5 | 0.09–5 | 0.34–5 | 0.54–2.63 | 0.21–1.80 | 0.14–0.61 | |
| bNAbs | N6 | PGT128 | PGDM1400 | N6 + PGT128 | N6 + PGDM1400 | N6 + PGDM1400 + PGT128 | |
|---|---|---|---|---|---|---|---|
| Pseudovirus Strains | |||||||
| CE1176 (C) | 1.34 | 0.43 | 0.89 | 0.18 | 0.05 | 0.03 | |
| CH119 (CRF07) | 0.21 | 0.33 | 0.54 | 0.09 | 0.03 | 0.04 | |
| 398F1 (A) | 0.87 | 0.32 | >5 | 0.16 | 0.30 | 0.11 | |
| TRO11 (B) | 0.7 | 0.08 | 2.11 | 0.14 | 0.21 | 0.03 | |
| BJOX2000 (CRF07) | 0.31 | >5 | <0.01 | 0.30 | 0.12 | <0.01 | |
| CE0217 (C) | 0.14 | 0.21 | <0.01 | 0.19 | <0.01 | <0.01 | |
| X2278 (B) | 0.56 | 0.07 | 0.01 | 0.08 | 0.01 | 0.01 | |
| CNE8 (CRF01) | 0.85 | 0.03 | <0.01 | 0.18 | 0.19 | <0.01 | |
| 246F3 (AC) | 0.16 | 0.05 | <0.01 | 0.18 | 0.16 | <0.01 | |
| X1632 (G) | 0.42 | 0.23 | 0.01 | 0.06 | 0.04 | <0.01 | |
| 25710 (C) | 0.72 | 0.11 | <0.01 | 0.06 | 0.06 | <0.01 | |
| IC50 range | 0.14–1.34 | 0.03–5 | 0.01–5 | 0.06–0.3 | 0.01–0.3 | 0.01–0.11 | |
| bNAbs | N6 | PGT128 | PGDM1400 | N6 + PGT128 | N6 + PGDM1400 | N6 + PGDM1400 + PGT128 | |
|---|---|---|---|---|---|---|---|
| Pseudovirus Strains | |||||||
| CE1176 (C) | 3.07 | 0.65 | 3.25 | 0.61 | 0.37 | 0.23 | |
| CH119 (CRF07) | 1.07 | 0.75 | 1.46 | 0.45 | 0.20 | 0.25 | |
| 398F1 (A) | 1.98 | 0.75 | >5 | 0.36 | 0.50 | 0.24 | |
| TRO11 (B) | 2.5 | 0.28 | 3.80 | 0.56 | 0.69 | 0.22 | |
| BJOX2000 (CRF07) | 0.8 | >5 | 0.30 | 1.17 | 0.30 | 0.12 | |
| CE0217 (C) | 0.9 | 1.00 | <0.01 | 0.70 | <0.01 | <0.01 | |
| X2278 (B) | 1.67 | 0.44 | 0.36 | 0.42 | 0.11 | 0.12 | |
| CNE8 (CRF01) | >5 | 0.32 | 0.01 | 0.53 | 0.57 | 0.01 | |
| 246F3 (AC) | 0.78 | 0.21 | <0.01 | 0.67 | 0.59 | <0.01 | |
| X1632 (G) | 1.62 | 0.69 | 1.14 | 0.27 | 0.14 | 0.04 | |
| 25710 (C) | 2.23 | 0.50 | <0.01 | 0.34 | 0.19 | <0.01 | |
| IC80 range | 0.78–5 | 0.21–5 | 0.01–5 | 0.27–1.17 | 0.01–0.69 | 0.01–0.25 | |
| Pseudoviruses | CE1176 | CH119 | 398F1 | TRO11 | BJOX2000 | CE0217 | X2278 | CNE8 | 246F3 | X1632 | 25710 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bNAbs | |||||||||||||
| 10E8 | [30] | ||||||||||||
| nd | nd | nd | [32] | ||||||||||
| nd | nd | nd | nd | nd | nd | [21] | |||||||
| Our data | |||||||||||||
| DH511.11P | nd | nd | nd | nd | nd | nd | [21] | ||||||
| Our data | |||||||||||||
| N6 | [31] | ||||||||||||
| Our data | |||||||||||||
| VRC07-523 | nd | nd | nd | nd | nd | [30] | |||||||
| [31] | |||||||||||||
| Our data | |||||||||||||
| 10-1074 | [30] | ||||||||||||
| nd | nd | nd | nd | nd | nd | nd | nd | nd | [33] | ||||
| Our data | |||||||||||||
| PGT128 | nd | nd | nd | nd | nd | nd | nd | nd | nd | [32] | |||
| [30] | |||||||||||||
| Our data | |||||||||||||
| CAP256-VRC26.25 | [30] | ||||||||||||
| Our data | |||||||||||||
| PGDM1400 | [30] | ||||||||||||
| Our data | |||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kochina, E.A.; Urusov, F.A.; Kruglov, A.A.; Glazkova, D.V.; Shipulin, G.A.; Bogoslovskaya, E.V. Double and Triple Combinations of Broadly Neutralizing Antibodies Provide Efficient Neutralization of All HIV-1 Strains from the Global Panel. Viruses 2022, 14, 1910. https://doi.org/10.3390/v14091910
Kochina EA, Urusov FA, Kruglov AA, Glazkova DV, Shipulin GA, Bogoslovskaya EV. Double and Triple Combinations of Broadly Neutralizing Antibodies Provide Efficient Neutralization of All HIV-1 Strains from the Global Panel. Viruses. 2022; 14(9):1910. https://doi.org/10.3390/v14091910
Chicago/Turabian StyleKochina, Evgeniya A., Felix A. Urusov, Artem A. Kruglov, Dina V. Glazkova, German A. Shipulin, and Elena V. Bogoslovskaya. 2022. "Double and Triple Combinations of Broadly Neutralizing Antibodies Provide Efficient Neutralization of All HIV-1 Strains from the Global Panel" Viruses 14, no. 9: 1910. https://doi.org/10.3390/v14091910
APA StyleKochina, E. A., Urusov, F. A., Kruglov, A. A., Glazkova, D. V., Shipulin, G. A., & Bogoslovskaya, E. V. (2022). Double and Triple Combinations of Broadly Neutralizing Antibodies Provide Efficient Neutralization of All HIV-1 Strains from the Global Panel. Viruses, 14(9), 1910. https://doi.org/10.3390/v14091910





